Background: Heat shock proteins bind TPR-containing proteins to facilitate client folding.
Results: The TPR domain of PP5 and the C-terminal IEEVD of Hsp70 are important for binding.
Conclusion: Hsp70 binds through the TPR domain and activates PP5 phosphatase activity.
Significance: Small molecules to inhibit Hsp70-PP5 interaction may be an alternative approach for cancer therapy.
Keywords: Hsp90, NMR, Peptides, Protein Phosphatase, Protein-protein Interactions, Cancer Signaling, Co-chaperones, Heat Shock Protein 70, Split Luciferase, Tethered Binding
Abstract
Protein phosphatase 5 (PP5) is auto-inhibited by intramolecular interactions with its tetratricopeptide repeat (TPR) domain. Hsp90 has been shown to bind PP5 to activate its phosphatase activity. However, the functional implications of binding Hsp70 to PP5 are not yet clear. In this study, we find that both Hsp90 and Hsp70 bind to PP5 using a luciferase fragment complementation assay. A fluorescence polarization assay shows that Hsp90 (MEEVD motif) binds to the TPR domain of PP5 almost 3-fold higher affinity than Hsp70 (IEEVD motif). However, Hsp70 binding to PP5 stimulates higher phosphatase activity of PP5 than the binding of Hsp90. We find that PP5 forms a stable 1:1 complex with Hsp70, but the interaction appears asymmetric with Hsp90, with one PP5 binding the dimer. Solution NMR studies reveal that Hsc70 and PP5 proteins are dynamically independent in complex, tethered by a disordered region that connects the Hsc70 core and the IEEVD-TPR contact area. This tethered binding is expected to allow PP5 to carry out multi-site dephosphorylation of Hsp70-bound clients with a range of sizes and shapes. Together, these results demonstrate that Hsp70 recruits PP5 and activates its phosphatase activity which suggests dual roles for PP5 that might link chaperone systems with signaling pathways in cancer and development.
Introduction
Protein phosphatase 5 (PP5)2 is a member of the PPP family of serine/threonine-specific phosphatases and has been linked to signaling pathways that control growth arrest, apoptosis, and DNA damage repair (1–3). Specifically, PP5 plays important roles in regulating the dynamic phosphorylation of p53, ASK-1, MAPK, and many other signaling components (2, 5, 6). PP5 also has been implicated in the regulation of glucocorticoid receptor, although the mechanism is controversial (1, 7). Additionally, PP5 levels are elevated in human breast cancer (8). Together, these studies have suggested that PP5 may be a novel target for anti-cancer therapies (9). However, the catalytic subunit of PPP phosphatases is highly conserved, and it has been difficult to develop selective, competitive inhibitors of these enzymes (10). In addition to its catalytic domain, PP5 is the only member of the PPP family that contains an N-terminal tetratricopeptide repeat (TPR) domain (11, 12). TPR domains are assembled from repeats of an amphipathic antiparallel helix that assemble into superhelical structures bearing a concave central groove (13–16). PP5 has been shown to interact with the molecular chaperones heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) (17–21). Specifically, the TPR domain of PP5 binds to cytoplasmic Hsp90 homologs, Hsp90α (stress-inducible) and Hsp90β (constitutively active), through a conserved MEEVD motif that is located at the end of the C termini of these chaperones (17–20). Although biochemical data illustrates that an MEEVD peptide has high affinity for the TPR domain of PP5 (∼50 nm), solution phase NMR studies revealed that this interaction is highly dynamic with only few enduring contacts (19, 20). Comparatively less is known about how PP5 interacts with Hsp70. Co-immunoprecipitation studies suggest that PP5 binds Hsp70 (21), but it is not yet clear how PP5 interacts with this chaperone or whether the TPR domain is involved. Based on the Hsp90-PP5 complex, it is likely that this interaction occurs through the IEEVD motif at the C termini of the cytoplasmic Hsp70 family members, including heat shock cognate 70 (Hsc70; HSPA8) and heat shock protein 72 (Hsp72; HSPA1A).
PP5 belongs to a family of TPR domain-containing co-chaperones that includes Hop (Hsp70/90 organizing protein), CHIP (carboxyl terminus of Hsp70 interacting protein) and a number of immunophilins such as FKBP52 (FK506 binding protein 52 kDa). Members of this co-chaperone family bind to Hsp70 and/or Hsp90 at these chaperones' C-terminal EEVD motifs. In turn, the TPR co-chaperones are important regulators of chaperone function (16) (22, 23). For example, complexes between CHIP and either Hsp70 or Hsp90 are linked to the ubiquitination and therefore the proteasomal degradation of chaperone-bound clients. Likewise, a complex between these chaperones and HOP is critical to the folding of some clients such as nuclear hormone receptors (24–26). Additionally, FKBP52 couples clients of Hsp70 and Hsp90 to the cytoskeleton (27). However, less is known about the Hsp70-PP5 and Hsp90-PP5 complexes and their potential roles in the protein homeostasis network. One important clue comes from observations that the TPR domain and the C-terminal catalytic subunit of PP5 have an auto-inhibitory function, suppressing phosphatase activity. Indeed, binding of Hsp90 to the TPR domain has been reported to weakly activate PP5 (14). However, it is not yet clear whether Hsp70 also binds the TPR domain or whether this interaction activates PP5.
Toward these questions, we characterized the interaction of Hsp70 and Hsp90 with PP5, using a panel of cell-based assays and biophysical methods. These studies confirmed that PP5 binds Hsp70 and Hsp90 through the canonical EEVD motifs. However, we found that C-terminal peptides derived from Hsp90α/β bind to PP5 10-fold tighter than C-terminal peptides derived from Hsc70/Hsp72. Despite the weaker affinity of Hsp70 for PP5, this chaperone was far more effective at stimulating the phosphatase activity of PP5. Additionally, solution phase NMR studies showed that Hsp70 and PP5 move independently of each other in the bound complex, suggesting that the disordered C terminus of Hsp70 allows the activated PP5 to “sample” a relatively large area around the chaperone. This ultra-structure might be important in allowing PP5 to act on a wide range of chaperone clients. Together, these results suggest that the Hsp70-PP5 complex is a potent phosphatase that might link chaperone systems with signaling pathways in cancer and development.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following sources: pLentilox vectors (University of Michigan Vector Core); pMSCG9 vector (Clay Brown, Center for Structural Biology, University of Michigan); restriction endonucleases (New England Biolabs); HEK293 (American Type Culture Collection); Dulbecco's modified Eagle's medium (Invitrogen, 11965-092); fetal bovine serum (10082-147); antibiotic-antimycotic (Invitrogen, 15240–062); six-well tissue culture plate (BD Falcon, 3046); polybrene linker (Santa Cruz Biotechnology, sc-134220) Renilla GLO luciferase kit (Promega, Madison, WI); and p-nitrophenyl phosphate phosphatase substrate kit (Thermo Scientific, 37620).
Plasmid Construction
C-terminal Renilla luciferase (CRL, residues 1–229) and full-length Hsp70 or Hsp90 (upstream of CRL) were PCR-amplified and subcloned into a pLentilox RSV-2 dsRed vector using the following restriction site design: BamHI-Hsp72-Xba1-CRL-Not1 and Xma1-HSP90α-BamHI-CRL-Xba1. In these fusion constructs, the stop codon of Hsp70/90 was deleted. Similarly, N-terminal Renilla luciferase (NRL, resides 230–311) and full-length PP5 (downstream of NRL) were amplified and subcloned into the pLentilox RSV vector using the following restriction site design: BamHI-NRL-Xba1-PP5-Not1. In this construct, the NRL stop codon was deleted. All fusion constructs (Hsp70/90-CRL and NRL-PP5) contained a GGGGSGGGGS (G4S)2 peptide linker between the protein of interest and the Renilla luciferase reporter (28). After all sequences were confirmed at the University of Michigan DNA sequencing core, lentiviral particles containing these constructs were purchased (University of Michigan Vector Core) to create a stable cell lines for additional studies.
Cell Transduction and SRL-PFAC Assay
HEK293 cells were plated using DMEM (Invitrogen) with 10% FBS and no antibiotics into a six-well tissue culture plate. Cells were allowed to adhere and grow for 1 day (∼70% confluent) before transduction. The next day, fresh media without antibiotics (1.35 ml), 0.15 ml of 10× lentiviral particles (either Hsp70-CRL, Hsp90-CRL, or NRL-PP5), and polybrene linker to a final concentration of 8 μg/ml was added to each well. After an incubation period of 8 h at 37 °C and 5% CO2, the medium was replaced with DMEM with 10% FBS and 1% antibiotic-antimycotic. For cells that contained both HSP70/90-CRL and NRL-PP5 viral particles, the above procedure first performed with Hsp90/Hsp70-CRL viral particles. Using these cells, the same procedure was repeated a second time; however, NRL-PP5 lentiviral particle was added. The HSP70/90-CRL and PP5-NRL viral particles contained dsRED and GFP, respectively. Cells containing Hsp90-CRL, Hsp70-CRL, NRL-PP5, Hsp90-CRL + NRL-PP5, and Hsp70-CRL + NRL-PP5 constructs were then seeded into a 24-well plate at a density of 5,000 cell/wells and allowed to grow overnight at 37 °C and 5% CO2. The following day, the medium was removed, and the cells were washed with phosphate-buffered saline. Luciferase activity determined using the Renilla-GLO luciferase assay system kit. Briefly, following washing, 100 μl of 1× passive lysis buffer was added to each well and plates were allowed to shake for 15 min at room temperature. Afterward, 1× luciferase substrate was added, and the luminescence was measured using Biotek Synergy 2 plate reader.
Protein Purification
Full-length PP5 and Hsp90 were expressed in Escherichia coli BL21(DE3) cells from pMCSG9 plasmids. A fresh colony was grown in terrific broth medium supplemented with 50 mg/liter ampicillin at 37 °C with shaking at 250 rpm until A600 reached ∼0.8 and protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside (final concentration of 1 mm). The temperature was reduced to 18 °C, and the culture was allowed to shake overnight. Cells were harvested by centrifugation (4000 × g, 10 min, 4 °C). Cell pellets was suspended in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole (pH 8.0)) and sonicated on ice, and clarified by centrifugation at 15,000 × g for 30 min. The His-tagged proteins were purified using a nickel-nitrilotriacetic acid (Qiagen) column. The eluted protein was subjected to dialysis (PP5 buffer, 40 mm Tris-HCl, pH 7.4, 10% glycerol, 1 mm DTT, and HSP90 buffer, 20 mm Tris-HCl, pH 7.4, 20 mm NaCl, 10% glycerol, 1 mm DTT). Proteins were treated with His-tagged tobacco etch virus protease TEV protease (1 μm) overnight at 4 °C to remove tags. This process was repeated a second time prior to extensive dialysis and removal of any residual His-tagged protein by nickel-nitrilotriacetic acid column. Human Hsp70s (pMCSG7 vector) were expressed in E. coli BL21(DE3) cells using terrific broth medium supplemented with 50 mg/liter ampicillin at 37 °C with shaking at 250 rpm until A600 of ∼0.8 was reached. Additionally, Hsc70 protein was isotopically labeled for all NMR experiments. For labeled Hsc70, the BL21 cells were grown M9 media with15NH4Cl (Sigma Aldrich). The temperature was reduced to 18 °C, and the culture was allowed to shake overnight. Cells were harvested by centrifugation (4000 × g, 10 min, 4 °C). All Hsp70s were purified as described (29), and His tags were removed via TEV protease. Final purification was performed on an ATP agarose column. All protein concentrations were measure using the Pierce BCA protein assay kit according to the manufacturer's protocol. To verify Hsc70 was not aggregated, the raw fluorescence values of the parallel and perpendicular intensity using fluorescence polarization shows no indication of aggregation. In addition, non-binding and binding tracer in fluorescence polarization was also used to confirm Hsc70 was not aggregated. NMR structure showed no sign of Hsc70 aggregation.
Peptide Synthesis
All peptides were synthesized manually or with an ABI 433 peptide synthesizer using Fmoc chemistry with 2-chlorotrityl resin as the solid support. Either DIC/HOAt or HOBt/HBTU was used as the coupling reagent. Following completion of the peptide, a cleavage mixture composed of TFA/triisopropylsilane/H2O (19 ml:0.5 ml:0.5 ml) removed the peptide from the resin as well as any side chain protecting groups. The resulting solution was evaporated, and the crude peptide was precipitated with diethyl ether. Peptides were purified via RP-HPLC (Waters, Sunfire Prep C18, 19 mm × 150 mm, 5 μm) and confirmed by electrospray ionization mass spectroscopy (30).
Fluorescence Polarization Assay
All fluorescence polarization experiments were conducted in 384-well, black, low volume, round-bottomed plates (Corning) using a BioTeck Synergy 2 plate reader (Winooski, VT). For binding experiments, to each well was added increasing amounts of protein and the 5-carboxyfluorescein-labeled Hsp70/90 C-terminal probe/tracer (20 nm). For competition studies, each well had PP5 protein at a concentration equivalent to the Kd, 5-carboxyfluorescein-labeled peptide was held constant at 20 nm, and varying concentrations of unlabeled peptide was added to compete off labeled peptide. All wells had a final volume of 20 μl in the assay buffer (40 mm Tris-HCl, pH 7.4, 10% glycerol, 1 mm DTT). The plate was allowed to incubate at room temperature for 5 min to reach equilibrium. The polarization values in millipolarization units were measured at an excitation wavelength at 485 nm and an emission wavelength at 528 nm. An equilibrium binding isotherm was constructed by plotting the fluorescence polarization reading as a function of the protein concentration at a fixed concentration of tracer (20 nm). All experimental data were analyzed using Prism software (version 5.0, Graphpad Software, San Diego, CA) and WinNonlin (version 5.3).
Protein NMR Experiments
NMR data were collected using an Agilent/Varian NMR System with a room temperature triple resonance probe, interfaced to an Oxford instruments 18.7 tesla magnet (1H 800 MHz). Backbone assignments for the C terminus of Hsc70 were reported previously (38). Experiments for studying the interaction of PP5 with Hsc70 were carried out using full-length PP5 and full-length 15N-labeled Hsc70 in the following buffer: 50 mm HEPES, 75 mm NaCl, 1 mm ADP, 5 mm MgCl2, 0.02% NaN3, 0.01% Triton, pH 7.4, 30 °C. The TROSY spectrum with 1:0 Hsc70/PP5 was recorded in 10 h with a sample of 254 μm Hsc70. The spectrum with 1:1 Hsc70/PP5 was recorded in 22 h with a sample of 169 μm Hsc70 and 149 μm PP5. The two spectra have the same intrinsic signal to noise ratio ((254/169)2 = 2.25).
Size Exclusion Chromatography and Multi-angle Light Scattering (SEC-MALS)
PP5-Hsp70 and PP5-Hsp90 complexes were formed by incubating proteins at equal molar concentrations (10 μm) in binding buffer (100 mm KCl, 20 mm HEPES (pH 7.5), 7 mm β-mercaptoethanol) at room temperature for 30 min. Identification and molecular weight determination of complexes was achieved through SEC (Wyatt WTC-050S5 and WTC-030S5 columns) with an Akta micro FPLC (GE Healthcare) and in-line DAWN HELEOS MALS and Optilab rEX differential refractive index detectors (Wyatt Technology Corp.). SEC was performed in 100 mm KCl, 20 mm HEPES (pH 7.5). Data were analyzed by the ASTRA software package (version 6, Wyatt Technology Corp.). The two spectra have the same intrinsic signal to noise ratio ((254/169)2 = 2.25).
p-Nitrophenyl Phosphate Assay
Purified PP5, Hsp70, and Hsp90 were immobilized in 4HBX 96-well plates (Thermo Scientific) and diluted with ELISA buffer (BioLegend). Equal molar concentration of protein was added to each well, and the plate was incubated overnight at 4 °C. Protein concentrations ranged from 50 to 0.5 μm. The following day, p-nitrophenyl phosphate (Fisher Scientific) was used according to the manufacturer's instructions. Once p-nitrophenyl phosphate substrates were added to the plates, they were incubated at 37 °C for 1 h. After color change, the OD405 was measured on the Biomatrix Plate Reader Synergy 2. The enzymatic activity was calculated using the following equation,
 |
where ϵ is the molar extinction coefficient, which equals 1.78 × 104 m−1 × cm−1.
RESULTS
SRL-PFAC Confirms That Hsp70 and Hsp90 Bind to PP5 in Cells
Previous co-immunoprecipitation studies have suggested that PP5 interacts with both Hsp70 and Hsp90 in cells (21). To confirm this result, we utilized the SRL-PFAC system, which has proven to be a powerful method for studying protein-protein interactions in cells (31). In this assay, the full-length Renilla luciferase gene is divided into inactive halves, the NRL (residues 1–229) and the CRL (residues 230–311). The NRL and CRL will reconstitute functional luciferase if they are brought into close proximity. To explore whether Hsp90 and Hsp70 bind PP5, we created constructs that would express NRL-PP5, Hsp70-CRL, or Hsp90-CRL fusion proteins (Fig. 1A). We anticipated that a luminescence signal would be detected only if Hsp90 or Hsp70 interacts with PP5 (Fig. 1B). When HEK293 cells were transduced with viral vectors expressing either NRL-PP5, Hsp70-CRL, or Hsp90-CRL alone, low luciferase activity was measured (Fig. 1C). However, co-transduction with either the NRL-PP5+Hsp70-CRL pair or the NRL-PP5+Hsp90-CRL pair led to enhanced luciferase activity (Fig. 1C), consistent with the interaction of PP5 with both chaperones in cells. We also examined at a control pair (NRL-PP5+HOP-CRL) if nonspecific complementation would occur. As expected, no complementation of two luciferase fragments (NRL and CRL with low luciferase activity) was observed (data not shown).
FIGURE 1.
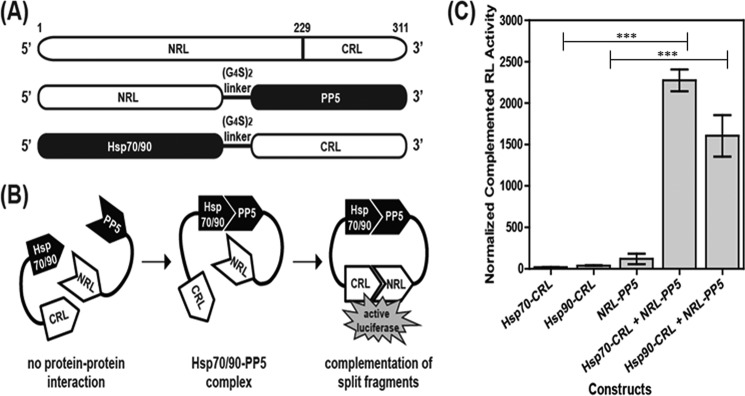
SRL-PFACs confirm that both Hsp70-PP5 and Hsp90-PP5 interact. A, schematic diagram of plasmid constructs. The two interacting proteins PP5 and Hsp70/90 are fused to NRL (amino acids 1–229) and CRL (amino acids 230–311) portion of the RL, respectively, through a (G4S)2 peptide linker. B, schematic diagram of the SRL-PFAC system for monitoring complex formation between Hsp70/90 and PP5. Interactions between Hsp70/90 and PP5 bring CRL and NRL in close proximity, ultimately resulting in the complementation of RL enzyme activity and photon production in the presence of the substrate coelenterazine. C, SRL-PFAC system is sensitive for monitoring complex formation of Hsp70/90 and PP5 and shows highly complemented RL activity and low background. HEK293 cells were transduced with either Hsp70-CRL, Hsp90-CRL, NRL-PP5, Hsp70-CRL+NRL-PP5, or Hsp90-CRL+NRL-PP5. Data are presented as mean ± S.D. (n = 3). ***, p < 0.001 compared with controls.
PP5 Binds to the C Terminus of Hsp90 with Higher Affinity Than to Hsp70
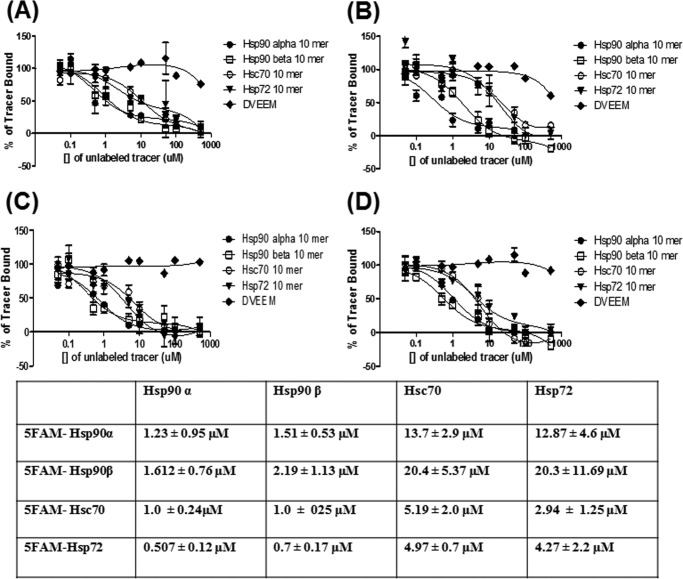
Binding of TPR co-chaperones such as HOP or CHIP to Hsp70 and Hsp90 is typically mediated by contacts between the TPR domain and the C-terminal EEVD motif that is shared by both chaperones (32–34). To explore the affinity of the Hsp70/90 C terminus (Table 1) for PP5, we developed a fluorescent MEEVD tracer and measured its binding to PP5 by fluorescence polarization. In this platform, the MEEVD tracer had a Kd value of 0.14 ± 0.005 μm (Fig. 2B), consistent with literature values (20). This interaction appeared to be specific, because the reverse tracer (DVEEM) had weak affinity for PP5 (Kd > 10 μm) (Fig. 2A). Next, we assessed the ability of an Hsp70-derived IEEVD tracer to bind PP5. This tracer had a ∼3-fold weaker affinity for PP5 (Kd = 0.426 ± 0.06 μm) (Fig. 2B). We were then curious to see whether the homologs of the chaperones might have different affinities for PP5. We found that 10-mer tracers derived from the C termini of Hsc70 and Hsp72 had comparable affinities for PP5, with Kd values of 1.06 ± 0.34 and 1.55 ± 0.43 μm, respectively (Fig. 2C). Additionally, the Hsp90α and Hsp90β tracers bound to PP5 with similar affinities, with Kd values of 0.079 ± 0.02 and 0.077 ± 0.02 μm, respectively (Fig. 2C). The longer 10-mer tracers also had affinities that were similar to those of their corresponding 5-mers, suggesting that most of the affinity of the interaction is engendered by the EEVD motif.
TABLE 1.
Peptides used in fluorescence polarization assay
Hsp70/90 C-terminal peptides used in fluorescence polarization assays are shown here. For binding experiments, each peptide tracer/probe contained a 5-carboxyfluorescein-labeled amino terminus. Labels were connected to peptides through an aminohexanoic acid linker. Additionally, for competitive binding experiments, each unlabeled peptide competitor contained a free amino-terminal end. Mutant residues in the C-terminal tracer are in boldface and underlined type.
| C-terminal peptide sequences | Corresponding human chaperones |
|---|---|
| MEEVD | Hsp90α/β |
| IEEVD | Hsc70 and Hsp72 |
| DDTSRMEEVD | Hsp90α |
| EDASRMEEVD | Hsp90β |
| SSGPTIEEVD | Hsc70 |
| GSGPTIEEVD | Hsp72 |
| DVEEM | Reverse peptide control |
| DDTSRIEEVD | Hsp90α mutant |
| GSGPTMEEVD | Hsp72 mutant |
FIGURE 2.
Fluorescence polarization confirms that Hsp90-PP5 has higher affinity than Hsp70-PP5. A, raw mP values were plotted with control peptide. No binding was observed with increasing concentrations of PP5 and control peptide at 20 nm. B, direct binding was measured with increasing concentration of PP5 and 5-mer peptides (20 nm). C, direct binding was measured using increasing concentrations of purified full-length PP5 with 5FAM 10-mer-labeled peptide (20 nm). D, to see how the residues Met and Ile contribute affinity to PP5, HSP70–90 peptides were synthesized so that HSP90 10-mer peptide contained an Ile instead of a Met and HSP70 10-mer peptide contained an Met instead of an Ile. Fluorescence polarization (FP) was performed with increasing concentrations of PP5, and both peptides were held constant at 20 nm. Data are presented as mean ± S.D. (n = 3).
The results of the fluorescence polarization experiments suggested that the methionine of the MEEVD motif in Hsp90α/β may increase affinity for PP5. To test this hypothesis in more detail, we constructed tracers in which this position was mutated. Specifically, the methionine of the Hsp90α tracer was mutated to an isoleucine and the corresponding isoleucine of the Hsp72 tracer was mutated to methionine. As expected, the Hsp72 mutant tracer had higher affinity than the Hsp90α mutant tracer (0.133 ± 0.03 and 0.42 ± 0.02 μm) (Fig. 2D). These results clearly showed that the methionine contributed to the greater affinity of Hsp90-derived peptides for PP5.
To further confirm these binding studies, we performed competition studies to compete unlabeled peptide with labeled peptide. The IC50 values for the isoforms of HSP90 again showed higher affinity than HSP70 peptides in all four tracer competition assays (Fig. 3, A–D). For these experiments, PP5 protein was added at a molar concentration that was equal to the binding affinity (Kd). Therefore, for the competition assay, Hsc70 and Hsp72 contained more PP5 protein compared with Hsp90 α and β due to the lower affinity, which can explain the similar IC50 among all four tracer assays. However, these results confirmed the binding experiments were correctly portraying the correct affinity.
FIGURE 3.
Competition studies confirm binding data. A, competition of 5FAM-HSP90α (20 nm) with increasing concentrations of unlabeled tracer (HSP90α, HSP90β, HSC70, and HSP72 peptides). B, competition of 5FAM-HSP90β (20 nm) with increasing concentrations of unlabeled tracer (HSP90α, HSP90β, HSC70, and HSP72 peptides). C, competition of 5FAM-HSC70 (20 nm) with increasing concentrations of unlabeled tracer (HSP90α, HSP90β, HSC70, and HSP72 peptides). D, competition of 5FAM-HSP72 (20 nm) with increasing concentrations of unlabeled tracer (HSP90α, HSP90β, HSC70, and HSP72 peptides). Data are presented as mean ± S.D. (n = 3).
PP5 Binds Hsp70 and Hsp90 in Different Arrangements
To better understand how the chaperones bind PP5, we incubated full-length Hsp70 or Hsp90 with PP5 and analyzed the complexes by SEC-MALS to obtain an accurate measurement of the molecular weight and stoichiometry of the complexes (35). Additionally, both Hsp70 and Hsp90 are ATPases, and they undergo dramatic conformational changes in response to nucleotides (36, 37). When equimolar PP5 and Hsp70 are incubated and analyzed by SEC-MALS, a single peak predominates and is shifted in elution volume compared with individual runs of Hsp70 and PP5 (Fig. 4A), indicating the formation of an Hsp70-PP5 complex. The average molecular weight of this peak was determined to be 145 kDa, and based on a comparison with a 126-kDa molecular weight calculated from the sequence, this corresponds to a 1:1 complex of Hsp70:PP5. When the Hsp90 dimer and PP5 are incubated together, a single, shifted peak is observed as well, with a modest increase in molecular weight, 232 kDa compared with 190 kDa for Hsp90 alone (Fig. 4B). This corresponds to an average of less than one PP5 bound per Hsp90 dimer indicating the formation of an asymmetric complex of Hp90:PP5 with a 2:1 stoichiometry despite the presence of two MEEVD binding sites in the Hsp90 dimer. The Hsp90-PP5 interaction is likely weaker in affinity, resulting in a partial dissociation during the elution, explaining the lower average molecular weight. The presence of saturating amounts of ATP and ADP were tested but resulted in no changes the molecular weight of the Hsp90-PP5 complex. Differences in the average molecular weight measured by SEC-MALS compared with the protein sequence were likely due to minor presence of aggregated PP5 and Hsp90 tetramer species (data not shown). Overall, these results demonstrate that PP5 binds full-length Hsp70 and Hsp90 in vitro in different arrangements. Hsp70-PP5 is in a stable 1:1 complex, whereas Hsp90-PP5 is in a 2:1 arrangement and may be slightly weaker in affinity compared with Hsp70.
FIGURE 4.
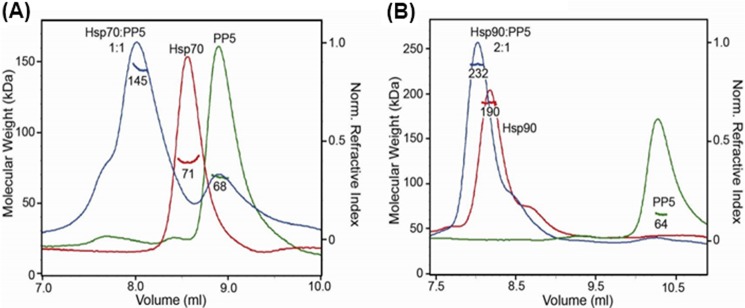
Hsp70-PP5 and Hsp90 interact with different stoichemetries. PP5 binds intact Hsp70 and Hsp90. A, SEC-MALS analysis of Hsp70:PP5 (blue) shows elution volume shift and molecular mass of 145 kDa, indicating stable 1:1 complex compared with Hsp70 (red) and PP5 (green) alone. B, Hsp90:PP5 (blue) is shifted in elution volume and determined to be a 2:1 complex at 232 kDa compared Hsp90 dimer alone (red). Molecular masses were determined from the Raleigh ratio, measured by static light scattering, and the protein concentration (right y axis), measured by a refractive index detector, are indicated. Molecular masses based on sequence are as follows (in kDa): 70 (Hsp70), 56 (PP5), and 170 (Hsp90 dimer). SEC columns WTC-030S5 and WTC-050S5 (Wyatt Technology) were used to optimally separate free Hsp70 and Hsp90 from PP5 complexes, respectively.
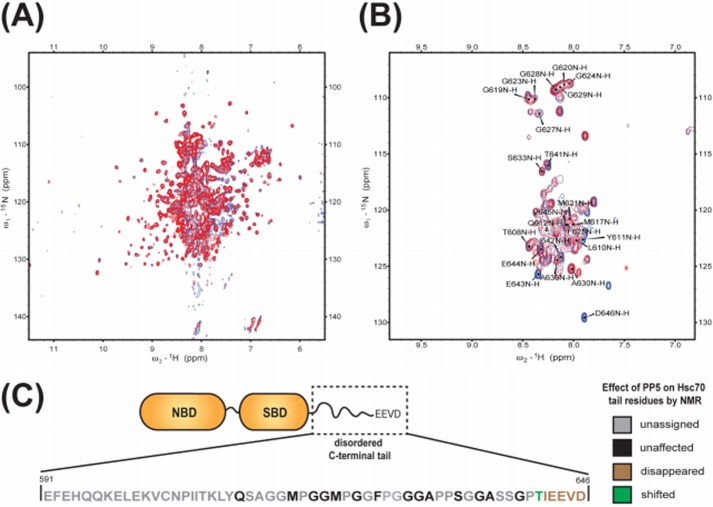
Hsc70 and PP5 Move Independently within the Complex
To explore the Hsp70-PP5 complex in greater detail, we utilized solution state NMR. A 1H-15N TROSY HSQC spectrum was collected for 15N-labeled Hsc70 (1–646) in the ADP-bound state (Fig. 5A in blue). As identified in our previous study where we investigated the interaction of Hsc70 with the highly homologous TPR protein CHIP (38), the middle of the TROSY spectrum (∼7.5 to 8.5 ppm) is dominated by intense sharp resonances with a signal-to-noise ratio (SNR) of ∼250:1 (Fig. 5A). These resonance were assigned using triple resonance experiments to residues 612–646 in C-terminal tail of Hsc70 as well as to a N-terminal extension containing a tag (38). The strong intensity of these resonances, as well as the lack of spectral dispersion, indicated that the C-terminal region (612–646) of Hsc70, including the IEEVD motif, is a dynamically disordered random coil. Based on comparison with published spectra of the isolated nucleotide-binding domain (NBD) and the substrate-binding domain (SBD), the much weaker signals with SNR ∼ 8:1 in the well dispersed part of the NMR spectrum in Fig. 5 originate from the 45-kDa Hsc70 NBD and 25-kDa SBD (38). In the ADP state, these domains are tethered by a ∼10-residue linker and move relatively independently (39–41). The large difference in peak intensity between the core region and the C terminus is due to the large difference in TROSY transfer efficiency in molecules with an effective molecular mass of 25–45 kDa and a flexible tail with a much smaller effective molecular mass (which one estimates to be ∼10 kDa on the basis of this intensity difference, see Table 2 and its legend). Increasing the contour level of the spectra focuses on the dynamically disordered region, which included the IEEVD motif (Fig. 5B).
FIGURE 5.
C-terminal residues of Hsc70 bind PP5. a, full 800 MHz TROSY spectra of 15N-labeled Hsc70 alone (blue) and in a 1:1 mixture with PP5 (red). b, high contour view of spectra with resonance assignments. c, schematic diagram of the disordered C-terminal tail of Hsc70. Unassigned residues are shown in gray. Residues from Hsc70 that are unaffected by binding the binding of PP5 are depicted in black; those that disappear or shift are shown in brown and green, respectively.
TABLE 2.
Summary of NMR of Hsc70-PP5
| Domain | Molecular mass | τc at 30 °C Ca | 1HN HSQC LWb | 1HN TROSY LWb | 15N TROSY LWb | TROSY transfer efficiencyc | Rel. peak heightd |
|---|---|---|---|---|---|---|---|
| kDa | ns | Hz | Hz | Hz | % | a.u. | |
| Tail | 10 | 7 | 20 | 16 | 0.5 | 39 | 1.0 × 10 |
| SBD | 25 | 14 | 40 | 32 | 0.9 | 15 | 1.9 × 10−1 |
| NBD | 45 | 23 | 65 | 51 | 1.5 | 4.7 | 3.8 × 10−2 |
| Hsc70 | 70 | 40 | 112 | 90 | 2.5 | 0.5 | 2.3 × 10−3 |
| PP5+tail | 70 | 40 | 112 | 90 | 2.5 | 0.5 | 2.3 × 10−3 |
| PP5+SBD | 85 | 45 | 126 | 100 | 2.8 | 0.3 | 1.1 × 10−3 |
| PP5+NBD | 105 | 55 | 154 | 122 | 3.5 | 0.07 | 2.4 × 10−4 |
| PP5+Hsc70 | 130 | 70 | 196 | 156 | 4.4 | 0.01 | 2.6 × 10−5 |
a The rotational correlation time τc was estimated from the molecular mass following Ref. 38.
b The average amide proton and nitrogen line widths (LW) at 800 MHz were calculated from coordinates of the crystal structure of ubiquitin as a model, using different rotational correlation times. The calculations took into account all dipole-dipole interactions with all magnetic nuclei in the molecule, and 1H CSA or 15N CSA relaxation. 1H-15N dipolar/1H CSA or 1H-15N/15N CSA cross-correlated R2 relaxation was taken into account for the columns marked “TROSY.” We assumed uniform 15N labeling, no 13C, or 2H labeling.
c In TROSY, there are three transfer periods with 1HN coherence, which all are tuned to 1/2JNH (5 ms). The transfer efficiency is reduced by 1H R2 relaxation. In total, I = I0 (exp(−3.1416 × LW × 0.005))3. The relevant line widths during these transfers are listed in the column marked “1HN HSQC LW.” Rel., relative.
d The peak height for 10 kDa was taken as a standard. The peak heights for other molecular weights were computed by taking the ratio of the relevant transfer efficiencies divided by the ratio of the relevant 1H TROSY linewidths (the latter affects peak height during data acquisition). The effect of increasing 15N line width on the TROSY peak intensity is minimal because of the short 15N acquisition time used and was not included in the calculation; a.u., absorbance units.
The 1H-15N TROSY HSQC spectrum of 15N-labeled Hsc70 with PP5 in a ∼1:1 ratio, shown in red in Fig. 5 has the same intrinsic SNR as the spectrum of uncomplexed Hsc70 (see “Experimental Procedures”). With a KD of Hsc70-PP5 binding of ∼1 μm (see above), 85% of the 169 μm Hsc70 should be complexed by 149 μm PP5. Significantly, most of the Hsc70 resonances in the presence of PP5 did not change from those of Hsc70 alone (comparing red and blue overlay, respectively; Fig. 5A). This result is unexpected. If the 60-kDa PP5 were to form a rigid 85-kDa complex with the 25-kDa SBD, the SBD TROSY resonance intensities in the complex should drop 180-fold and become invisible; if it were to form a 105-kDa complex with the 45-kDa NBD, the NBD TROSY resonance intensities should drop 160-fold and become invisible; if it were to form a 130-kDa triple complex with SBD and NBD, the latter resonance intensities would drop 7000 and 1500 times, respectively (see Table 2 and its legend for these calculations). In other words, if Hsc70 and PP5 were to form a rigid complex with either the SBD or NBD, the complexes TROSY-HSQC NMR spectrum would become undetectable at the conditions used. In these scenarios, the NMR spectrum of the 15% uncomplexed Hsc70 would also drop below the SNR limit. Hence, as we observe a largely unperturbed NMR spectrum for the Hsc70 core in the complex, we must conclude that the NMR data shows that the core regions of Hsc70 do not form stable complexes with PP5.
The NMR data do show that PP5 interacts with the C terminus of Hsc70. Several resonances in the high-level contour plot (Fig. 5B) disappeared and/or shifted in the 1:1 complex with PP5 (red spectrum). In particular, the intense resonances of the IEEVD motif residues disappeared completely from the NMR spectrum (Fig. 5, B and C), without new resonances appearing (also not at lower contour levels). This result is consistent with the dynamic C terminus of an effective molecular mass of 10 kDa being immobilized by the 58-kDa PP5 protein forming a ∼70 kDa complex. Table 2 shows that such a change in effective molecular weight results in a 430-fold reduction in TROSY peak intensity, which renders even peaks with an initial SNR of ∼250:1 invisible. We do not observe resonances for the ∼15% free Hsc70, which should have an intrinsic ∼40:1 SNR. It is most likely that the protein concentration estimation is not accurate and that there is no free Hsc70 in the sample.
Very significantly, the NMR spectra also show that the remaining peaks of the C-terminal region (residues 610–640) do not disappear. This strongly suggests that this area, located between the SBD of Hsc70 and the IEEVD motif, remains a dynamic random coil in the Hsc70-PP5 complex. Thus, the two proteins, although tightly bound through the IEEVD-TPR interaction, appear to move as dynamic, independent units tethered via Hsc70 residues 610–640.
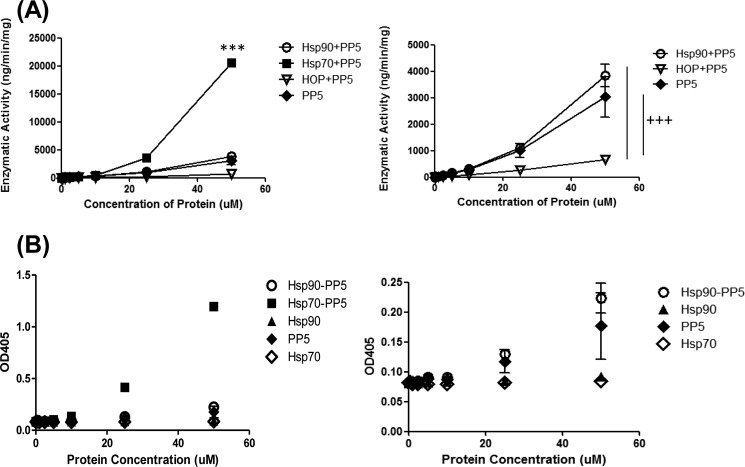
Hsp70 Stimulates the Phosphatase Activity of PP5
To further understand the functional consequence of the Hsp70-PP5 interaction, we utilized an in vitro phosphatase assay. Previous work illustrates that PP5 maintains a basal level of phosphatase activity, which can be weakly stimulated by Hsp90 (14). Thus, we assessed the ability of PP5 to hydrolyze the model substrate, p-nitrophenyl phosphate in the presence of Hsp70 or Hsp90. Importantly, neither Hsp70 nor Hsp90 interfered with the assay (Fig. 6B), allowing us to determine their effects on PP5. Moreover, PP5 basal activity was very weak (Fig. 6A), consistent with previous reports (4). Interestingly, Hsp70 stimulated the phosphatase activity of PP5, and this stimulatory activity was much greater than that of Hsp90 (Fig. 6A). In addition, we also tested whether 10mer C-terminal peptides (Hsp90α, Hsp90β, Hsc70, and Hsp72) would also stimulate PP5 activity. The data showed very little stimulation of PP5 activity when peptides were used (data not shown). These results suggest that the full length of Hsp70 may play a significant role in activating the enzymatic activity of PP5.
FIGURE 6.
Hsp70 preferentially stimulates PP5 phosphatase activity. p-Nitrophenylphosphate substrate were added at a concentration of 4.5 mm to each well. A represents enzymatic activity. B represents the raw OD values; this confirms that the phosphatase is not due to ATPase from Hsp70 or Hsp90 alone. Data are presented as mean ± S.D. (n = 3). ***, p < 0.001 compared with Hsp90-PP5; +++, p < 0.001 compared with Hop-PP5.
DISCUSSION
The chaperone activities of Hsp90 and Hsp70 are guided by interactions with co-chaperones, including members of the TPR domain family. In turn, Hsp90 and Hsp70 are thought to direct the activity of these co-chaperones toward specific clients. Our studies confirmed that PP5 is a bona fide member of the TPR co-chaperone family and that it binds to both Hsp90 and Hsp70 through its TPR domain. Furthermore, we found that Hsp90α and Hsp90β bind 10-fold tighter than Hsp70 family members. It is not yet clear whether this difference in affinity has physiological importance. However, our study suggests that methionine plays important role in the affinity. Binding studies showed that most of the affinity of the Hsp70-PP5 and Hsp90-PP5 interactions is engendered by polar contacts with the EEVD motif, and NMR studies confirmed that there are no significant stable interactions between other regions of the two proteins.
The independent motion (tethered binding) of Hsp70 and PP5 in the solution complex suggests that the folded regions of these proteins are able to sample a relatively wide area. In the fully extended form, the disordered C terminus of Hsp70 could be expected to extend nearly 40 Å. We speculate that this flexibility and length might be important in allowing PP5 to find phosphorylated residues in bound Hsp70 clients. Hsp70 clients are thought to include a number of kinases and transcription factors involved in apoptotic signaling. These clients have a wide range of sizes and shapes, so the flexibility of the tethered Hsp70-PP5 complex might be important in bringing activated PP5 in the vicinity of phosphorylated residues on these diverse targets.
Exclusive, tethered binding to the Hsc70 EEVD terminus was recently observed for CHIP, an E3 ubiquitin ligase containing a TPR domain highly homologous to that of PP5 (38). CHIP mediates broad-spectrum ubiquitination of Hsc70 client proteins destined for the proteasome. For CHIP, tethered binding was thought to be important to allow the ligase to ubiquitinate diverse Hsp70 clients. Similar mechanisms might also be important in the Hsp90-PP5 complex because Hsp90 also has a disordered region between its MEEVD motif and the folded portion of its C terminus.
We found that Hsp70 was a potent stimulator of the phosphatase activity of PP5. It is likely that binding of the IEEVD motif to the TPR disrupts the auto-inhibitory activity of the TPR domain, as has been observed in the Hsp90-PP5 system (14). This agrees with our observation that PP5 forms a stable 1:1 interaction with Hsp70. Based on our purification methods we expect Hsp70 favors the ADP-bound state in this complex. Because the ADP-bound form of Hsp70 has a tighter affinity for clients, this conformation might provide a way for stimulated PP5 to be held in proximity with Hsp70-bound clients for a length of time that is sufficient to allow dephosphorylation. In turn, this mechanism would limit the phosphatase activity of PP5 once it leaves the chaperone complexes, providing a reversible switch that responds to chaperone activity. Surprisingly, we found that in the context of the full-length protein, Hsp90 interaction appears asymmetric and likely weaker compared with Hsp70. This is in contrast to the 10-fold tighter affinity we identified for the Hsp90 MEEVD motif compared with the Hsp70 IEEVD. Thus, Hsp90 might use a different mechanism potentially involving client interactions or Hsp90 conformational changes that enable accessibility to the MEEVD to enhance PP5 interactions with phosphorylated clients.
Finally, these results point to the Hsp70-PP5 and Hsp90-PP5 protein-protein interactions as potential drug targets. PP5 has been proposed as an anti-tumor target (9), but the active sites of PPP family phosphatases are highly conserved and it has proven difficult to identify selective inhibitors. Based on our results, inhibitors of the protein-protein interactions between PP5 and the molecular chaperones might be an attractive alternative. Specifically, inhibitors of these protein-protein interactions might be expected to dysregulate kinase-phosphatase balance through multiple mechanisms, disconnecting PP5 from a major activation pathway and disrupting its chaperone-mediated ability to locate clients.
Acknowledgment
We thank Denzil Bernard for efforts with MD simulations.
This work was supported, in whole or in part, by National Institutes of Health R01 Grants CA120023 and NS059690.
- PP5
- protein phosphatase 5
- TPR
- tetratricopeptide repeat
- CRL
- C-terminal Renilla luciferase
- NRL
- N-terminal Renilla luciferase
- SEC-MALS
- size exclusion chromatography and multi-angle light scattering
- SNR
- signal-to-noise ratio
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- SRL-PFAC
- split Renilla luciferase protein fragment-assisted complementation.
REFERENCES
- 1. Zuo Z., Urban G., Scammell J. G., Dean N. M., McLean T. K., Aragon I., Honkanen R. E. (1999) Ser/Thr protein phosphatase type 5 (PP5) is a negative regulator of glucocorticoid receptor-mediated growth arrest. Biochemistry 38, 8849–8857 [DOI] [PubMed] [Google Scholar]
- 2. Morita K., Saitoh M., Tobiume K., Matsuura H., Enomoto S., Nishitoh H., Ichijo H. (2001) Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. EMBO J. 20, 6028–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ali A., Zhang J., Bao S., Liu I., Otterness D., Dean N. M., Abraham R.T., Wang X. F. (2004) Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 18, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen M. X., Cohen P. T. (1997) Activation of protein phosphatase 5 by limited proteolysis or the binding of polyunsaturated fatty acids to the TPR domain. FEBS Lett. 400, 136–140 [DOI] [PubMed] [Google Scholar]
- 5. Zhou G., Golden T., Aragon I. V., Honkanen R. E. (2004) Ser/Thr protein phosphatase 5 inactivates hypoxia-induced activation of an apoptosis signal-regulating kinase 1/MKK-4/JNK signaling cascade. J. Biol. Chem. 279, 46595–46605 [DOI] [PubMed] [Google Scholar]
- 6. Zuo Z., Dean N. M., Honkanen R. E. (1998) Serine/threonine protein phosphatase type 5 acts upstream of p53 to regulate the induction of p21(WAF1/Cip1) and mediate growth arrest. J. Biol. Chem. 273, 12250–12258 [DOI] [PubMed] [Google Scholar]
- 7. Chen M. S., Silverstein A. M., Pratt W. B., Chinkers M. (1996) The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J. Biol. Chem. 271, 32315–32320 [DOI] [PubMed] [Google Scholar]
- 8. Golden T., Aragon I. V., Rutland B., Tucker J. A., Shevde L. A., Samant R. S., Zhou G., Amable L., Skarra D., Honkanen R. E. (2008) Elevated levels of Ser/Thr protein phosphatase 5 (PP5) in human breast cancer. Biochim. Biophys. Acta 1782, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Golden T., Swingle M., Honkanen R. E. (2008) The role of serine/threonine protein phosphatase type 5 (PP5) in the regulation of stress-induced signaling networks and cancer. Cancer Metastasis Rev. 27, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McConnell J. L., Wadzinski B. E. (2009) Targeting protein serine/threonine phosphatases for drug development. Mol. Pharmacol. 75, 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bollen M., Stalmans W. (1992) The structure, role, and regulation of type-1 protein phosphatases. Crit. Rev. Biochem. Mol. Biol. 27, 227–281 [DOI] [PubMed] [Google Scholar]
- 12. Cohen P. T. (1997) Novel protein serine/threonine phosphatases: Variety is the spice of life. Trends Biochem. Sci. 22, 245–251 [DOI] [PubMed] [Google Scholar]
- 13. Zhang M. H., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Chaperoned ubiquitylation - crystal structures of the CHIPU box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 14. Yang J., Roe S. M., Cliff M. J., Williams M. A., Ladbury J. E., Cohen P. T., Barford D. (2005) Molecular basis for TPR domain-mediated regulation of protein phosphatase. EMBO J. 24, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang L., Liu Y. T., Hao R., Chen L., Chang Z., Wang Z. X., Wang H. R., Wu J. W. (2011) Molecular mechanism of the negative regulation of Smad1/5 protein by carboxyl terminus of Hsc70-interacting protein (CHIP). J. Biol. Chem. 286, 15883–15894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 17. Chen L., Qi H., Korenberg J., Garrow T. A., Choi Y. J., Shane B. (1996) Purification and properties of human cytosolic folylpoly-γ-glutamate synthetase and organization, localization, and differential splicing of its gene. J. Biol. Chem. 271, 13077–13087 [DOI] [PubMed] [Google Scholar]
- 18. Silverstein A. M., Galigniana M. D., Chen M. S., Owens-Grillo J. K., Chinkers M., Pratt W. B. (1997) Protein phosphatase 5 is a major component of glucocorticoid receptor.hsp90 complexes with properties of an FK506-binding immunophilin. J. Biol. Chem. 272, 16224–16230 [DOI] [PubMed] [Google Scholar]
- 19. Cliff M. J., Harris R., Barford D., Ladbury J. E., Williams M. A. (2006) Conformational diversity in the TPR domain-mediated interaction of protein phosphatase 5 with Hsp90. Structure 14, 415–426 [DOI] [PubMed] [Google Scholar]
- 20. Cliff M. J., Williams M. A., Brooke-Smith J., Barford D., Ladbury J. E. (2005) Molecular recognition via coupled folding and binding in a TPR domain. J. Mol. Biol. 346, 717–732 [DOI] [PubMed] [Google Scholar]
- 21. Zeke T., Morrice N., Vázquez-Martin C., Cohen P. T. (2005) Human protein phosphatase 5 dissociates from heat-shock proteins and is proteolytically activated in response to arachidonic acid and the microtubule-depolymerizing drug nocodazole. Biochem. J. 385, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Höhfeld J., Cyr D. M., Patterson C. (2001) From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 25. Qian S. B., McDonough H., Boellmann F., Cyr D. M., Patterson C. (2006) CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen S., Smith D. F. (1998) Hop as an adaptor in the heat shock protein 70 (Hsp70) and Hsp90 chaperone machinery. J. Biol. Chem. 273, 35194–35200 [DOI] [PubMed] [Google Scholar]
- 27. Heinlein C. A., Chang C. (2001) Role of chaperones in nuclear translocation and transactivation of steroid receptors. Endocrine 14, 143–149 [DOI] [PubMed] [Google Scholar]
- 28. Paulmurugan R., Massoud T. F., Huang J., Gambhir S. S. (2004) Molecular imaging of drug-modulated protein-protein interactions in living subjects. Cancer Res. 64, 2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang L., Thompson A. D., Ung P., Carlson H. A., Gestwicki J. E. (2010) Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK). J. Biol. Chem. 285, 21282–21291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amblard M., Fehrentz J. A., Martinez J., Subra G. (2006) Methods and protocols of modern solid phase peptide synthesis. Mol. Biotechnol. 33, 239–254 [DOI] [PubMed] [Google Scholar]
- 31. Jiang Y., Bernard D., Yu Y., Xie Y., Zhang T., Li Y., Burnett J. P., Fu X., Wang S., Sun D. (2010) Split Renilla luciferase protein fragment-assisted complementation (SRL-PFAC) to characterize Hsp90-Cdc37 complex and identify critical residues in protein/protein interactions. J. Biol. Chem. 285, 21023–21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu S. J., Liu F. H., Hu S. M., Wang C. (2001) Different combinations of the heat-shock cognate protein 70 (hsc70) C-terminal functional groups are utilized to interact with distinct tetratricopeptide repeat-containing proteins. Biochem. J. 359, 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu F. H., Wu S. J., Hu S. M., Hsiao C. D., Wang C. (1999) Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J. Biol. Chem. 274, 34425–34432 [DOI] [PubMed] [Google Scholar]
- 34. Russell L. C., Whitt S. R., Chen M. S., Chinkers M. (1999) Identification of conserved residues required for the binding of a tetratricopeptide repeat domain to heat shock protein 90. J. Biol. Chem. 274, 20060–20063 [DOI] [PubMed] [Google Scholar]
- 35. Southworth D. R., Agard D. A. (2011) Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell. 42, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Revington M., Zhang Y., Yip G. N., Kurochkin A. V., Zuiderweg E. R. (2005) NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J. Mol. Biol. 349, 163–183 [DOI] [PubMed] [Google Scholar]
- 37. Krukenberg K. A., Street T. O., Lavery L. A., Agard D. A. (2011) Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 44, 229–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith M. C., Scaglione K. M., Assimon V. A., Patury S., Thompson A. D., Dickey C. A., Southworth D. R., Paulson H. L., Gestwicki J. E., Zuiderweg E. R. (2013) The E3 ubiquitin ligase CHIP and the molecular chaperone Hsc70 form a dynamic, tethered complex. Biochemistry 52, 5354–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell. 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bertelsen E. B., Chang L., Gestwicki J. E., Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuiderweg E. R., Bertelsen E. B., Rousaki A., Mayer M. P., Gestwicki J. E., Ahmad A. (2013) Allostery in the Hsp70 chaperone proteins. Top Curr. Chem. 328, 99–153 [DOI] [PMC free article] [PubMed] [Google Scholar]