Background: Fast cardiolipin-mediated proton translocation from pumps to ATP synthase has been hypothesized.
Results: Mutational loss of membrane cardiolipin did not significantly affect alkaliphile ATP synthesis but other cardiolipin roles were observed.
Conclusion: Cardiolipin contributes to respiratory complex stability, thus indirectly to oxidative phosphorylation, and to stationary phase survival.
Significance: Clarification of cardiolipin roles in bacterial physiology can have pharmacological impact.
Keywords: Bacillus, Bacteria, Cardiolipin, Membrane, Phospholipid, Oxidative Phosphorylation
Abstract
Cardiolipin (CL), a membrane phospholipid in bacteria and mitochondria, has been hypothesized to facilitate movement of protons on the outer surface of membranes in support of respiration-dependent ATP synthesis, oxidative phosphorylation (OXPHOS). If so, the high levels of membrane CL found in alkaliphilic bacteria, such as Bacillus pseudofirmus OF4, might facilitate its robust OXPHOS at pH 10.5, where the bulk protonmotive (PMF) force is low. To address the role of CL in Bacillus pseudofirmus OF4, we studied strains in which genes (cls) potentially encoding a CL synthase (CLs) were deleted: three single (ΔclsA, ΔclsB, and ΔclsC), one double (ΔclsA/B), and one triple (ΔclsA/B/C) mutant. Two-dimensional thin layer chromatography analyses of lipid extracts from 32P-labeled strains showed that the wild-type CL content was 15% of total phospholipids at pH 10.5 versus 3% at pH 7.5 during log phase. The % CL was higher (28–33%) at both pH values during stationary phase. The clsA gene plays a major role in CL biosynthesis as no detectable CL was found in ΔclsA-containing mutants, whereas the CL precursor phosphatidylglycerol was elevated. The ΔclsB mutant exhibited no significant reduction in CL, but clsB expression was up-regulated and appeared to support growth at pH 7.5. In the absence of detectable CL, the alkaliphile showed no significant deficits in non-fermentative growth, respiration-dependent ATP synthesis, or salt tolerance. Minor deficits in respiration and ATP synthase assembly were noted in individual mutants. In long term survival experiments, significant growth defects were found in ΔclsA strains and the ΔclsC strain at pH 10.5.
Introduction
Cardiolipin (CL)2 is an anionic glycerophospholipid with four acyl chains. The presence of CL is primarily associated with membranes of bacteria and mitochondria that carry out F1F0-ATP synthase-dependent synthesis of ATP that is energized by electrochemical ion gradients generated by respiratory chain activity, i.e. OXPHOS. CL is linked to Barth syndrome, a human metabolic disorder in which mutations in taffazin lead to reduced CL, increased lyso-CL, and associated deficits in mitochondrial function in OXPHOS (1–3). Recently, it has also been shown that CL loss in yeast leads to defects in cellular iron homeostasis (4), which may reflect the importance of the energy status for this function. Similarly, the greater sensitivity to high salinity reported in CL-deficient mutants of Bacillus subtilis and a deficit in managing prolonged salinity in Staphylococcus aureus (5, 6) could reflect the OXPHOS deficits because a high salinity challenge requires an energy-demanding response. The specific CL roles most frequently reported in connection with OXPHOS-related deficits are roles in stabilizing respiratory complexes and supercomplexes in both eukaryote mitochondria and bacteria (7–11). Although CL is integral to the structural organization of individual mitochondrial respiratory complexes (12), the CL-precursor phosphatidylglycerol (PG) appears to substitute for CL (11). However, higher order organization of the respiratory chain into supercomplexes is highly dependent on CL (10). CL forms microdomains in regions of negative membrane curvature (13–16), which have been shown to promote polar localization of osmo-sensitive transporter ProP from Escherichia coli (17, 18). Drosophila mutants with severe reductions in flight muscle mitochondrial CL exhibit deficits in the oligomerization and order of ATP synthase assemblies and formation of the regions of high membrane curvature characteristic of mitochondrial cristae (19). CL is also found near the entrance of a proton uptake pathway in respiratory Complex III (20–22). Digestion of bound CL in bovine Complex III inactivates the complex, which can be reversed by addition of CL (23). However, bound CL in a Rhodobacter cytochrome c oxidase complex has been shown to be replaceable with other lipids without an impact on structure or function (24), whereas CL was required for full activity of a mammalian Complex IV (25). In both E. coli and B. subtilis, CL has been shown to be dispensable (26, 27), so flexibility in accommodating CL loss may be greater in prokaryotes than eukaryotes.
One of the goals of this study was to test a hypothesis developed by Haines and Dencher (28–30) that CL could play a direct role in proton translocation on the membrane surface during OXPHOS by acting as a proton sink. Peter Mitchell's (31) ground-breaking chemiosmotic hypothesis posited that during generation of a protonmotive force (PMF) by proton-pumping respiratory chain complexes, pumped protons equilibrate with the bulk liquid phase outside the coupling membrane and are then re-captured for entry into the ATP synthase during OXPHOS. However, recent experimental and computational data have indicated that pumped protons can move rapidly on the membrane surface or near the membrane surface and reach the ATP synthase before their full equilibration, thus making the effective PMF larger than the PMF calculated using the bulk phase proton concentration to calculate the pH gradient component (32–35). There is not yet a complete consensus on how the protons move on the surface and the roles of various variables in “trapping” them near the surface, e.g. whether CL or other specific charged molecules at the surface are involved. A major model system in which robust OXPHOS occurs under conditions of low bulk PMF, is alkaliphilic Bacillus pseudofirmus OF4, a genetically tractable alkaliphile that carries out OXPHOS more effectively at an external pH of 10.5 than at 7.5, although the bulk PMF during growth in media at pH 10.5 is much lower than at pH 7.5 because the cytoplasmic pH is 8.3 under those conditions. As a consequence, this organism provides a setting in which to test whether CL is a likely participant in the near-surface proton translocation that is expected to be crucial for alkaliphile OXPHOS (36–38). Haines and Dencher (28–30) proposed that unusual features of the CL head group could enable a cluster of such head groups to pick up protons and then serve as a rapid and direct conduit of protons from proton pumping respiratory chain complexes to the ATP synthase. If this is a key mechanism, mutational loss of the three Cls enzymes identified in the B. pseudofirmus OF4 genome (39) would be expected to have a major negative impact on OXPHOS capacity at high pH. Study of the individual and collective roles of the Cls enzymes is also of general interest to explore in an alkaliphile that grows well, both fermentatively and non-fermentatively, in a pH range from 7.5 to >11.2. The three paralogous Cls enzymes may well have distinct roles at different pH values, and may reveal roles apart from OXPHOS for the high CL content that is found in B. pseudofirmus OF4 (40).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. The primers used in this study are available on request. The wild-type (WT) strain is an alkaliphilic B. pseudofirmus 811M strain (41), a derivative of B. pseudofirmus OF4, whose whole genome was recently sequenced (39). Three genes potentially encoding a Cls enzyme were identified and designated as clsA, clsB, and clsC, respectively. The cls deletion mutants, including three single deletion mutants, one double mutant (deletion of clsA and clsB), and one triple mutant (deletion of clsA, clsB, and clsC), were constructed in the native alkaliphile host as described (42). Briefly, to construct the ΔclsA strain, an upstream and a downstream flanking region about 1000 bp of the clsA gene were amplified with B. pseudofirmus 811M genomic DNA as the template, and cloned into pGEM3Zf(+) (Promega) and pG+host4 (Appligene, Pleasanton, CA) sequentially. The resulting pG+host4 construct was transformed into B. pseudofirmus 811M strain. The ΔclsA strain was constructed after a single crossover step and a double crossover recombination step. The deletion region was verified by DNA sequencing performed by Genewiz, Inc. (South Plainfield, NJ). The ΔclsB and ΔclsC strains were constructed in a similar way except that the amplified clsC upstream and downstream fragments were ligated with low copy plasmid pMW118 (Nippon Gene, Tokyo, Japan) instead of pGEM3Zf(+) to avoid mutation. The double ΔclsA/B strain was constructed by deleting the clsB gene in the ΔclsA strain and the triple ΔclsA/B/C strain was constructed by deleting the clsC gene in the ΔclsA/B strain.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Properties | Ref. |
|---|---|---|
| B. pseudofirmus 811M | ||
| Wild type, 811M | A methionine auxotroph of B. pseudofirmus OF4 | 41 |
| ΔclsA | Deletion of clsA gene | This study |
| ΔclsB | Deletion of clsB gene | This study |
| ΔclsC | Deletion of clsC gene | This study |
| ΔclsA/B | Deletion of clsA and clsB | This study |
| ΔclsA/B/C | Deletion of clsA, clsB, and clsC | This study |
| ΔFo | Deletion of atpB-F | 44 |
| E. coli BKT12 strain | E. coli triple deletion of clsA, clsB and clsC | 43 |
| Plasmids | ||
| pBAD-TOPO | Expression vector, Apr | Invitrogen |
| pBAD-TOPO/lacZ/V5-His | A control vector containing the gene for β-galactosidase | Invitrogen |
| pBAD-Bp-ClsA | pBAD-TOPO containing clsA from B. pseudofirmus 811M | This study |
| pBAD-Bp-ClsB | pBAD-TOPO containing clsB from B. pseudofirmus 811M | This study |
| pBAD-Bp-ClsC | pBAD-TOPO containing clsC from B. pseudofirmus 811M | This study |
| pBAD-Bp-ClsC-His | pBAD-TOPO containing B. pseudofirmus 811M clsC with His-tagged at 3′ end | This study |
The three individual cls genes from B. pseudofirmus 811M were also amplified, and then ligated with pBAD-TOPO vector (Invitrogen) for complementation studies. The resulting constructs were designated as pBAD-Bp-ClsA, pBAD-Bp-ClsB, and pBAD-Bp-ClsC, respectively. These three recombinant plasmids and a pBAD-TOPO/lacZ/V5-His control (Invitrogen) were transformed into a CL-deficient E. coli BKT12 strain (43) and expressed by overnight induction with 0.2% arabinose. A His-tagged pBAD-Bp-ClsC-His construct was prepared to check clsC gene expression after it failed to show complementation in E. coli BKT12.
Growth Media
Two types of media were used: both were buffered at pH 7.5 or 10.5 for particular experiments. The carbon source was either malate (to 50 mm) to support non-fermentative growth or glucose (to 50 mm) to support fermentative growth. The semi-defined medium with the above respective carbon sources are referred to as MYE (malate-yeast extract) and GYE (glucose-yeast extract) medium (44). A defined medium in which glutamine and alanine provided the nitrogen source (QA) was also used at pH 7.5 or 10.5 and contained either 50 mm malate (M-QA) or glucose (G-QA) (45).
Growth Experiments
Growth curve experiments were conducted in the media and at the pH values specified in the legends for the particular experiments. The pre-cultures, unless specified, were grown in the same media in which they would ultimately be grown overnight and then diluted to an A600 of 0.2 with fresh medium; then 10 μl of the diluted culture was inoculated into 190 μl of medium in a 96-well plate (Greiner Bio-one, Germany). The plate was incubated with shaking at 30 °C in a PowerWave XS2 microplate spectrophotometer (BioTek Instruments, Inc.), and the A600 was recorded hourly. For long term survival growth tests, the WT strain and cls deletion strains were pre-grown overnight in MYE or M-QA media at pH 7.5 or 10.5. Ten μl of each pre-culture (A600 ∼ 2) was diluted into 2 ml of fresh corresponding media in a 15-ml tube. The tubes were incubated at either 30 or 37 °C with shaking at 250 rpm. Growth was monitored by recording the A600 in a Shimadzu UV-1601 spectrophotometer at the indicated time. Appropriate dilutions were made so that the A600 was within the range of 0.3 to 0.5. For growth on the MYE medium, a strain deleted in F0 genes of the ATP synthase operon (ΔF0), which does not grow non-fermentatively, was included as a control and the pre-culture was grown in GYE medium at pH 7.5. The modest growth exhibited by the ΔF0 mutant on malate was subtracted from the raw A600 data to calculate the non-fermentative growth.
Phospholipid Analysis
For determinations of phospholipid composition, cells were grown in the presence of 5 μCi/ml of 32PO4, which was added either during overnight growth or after dilution of an overnight culture to an A600 of 0.05 to initiate logarithmic growth. Uniformly labeled cells were harvested by centrifugation and phospholipids were extracted as described previously (43). Analyses of the phospholipids were carried out either by silica gel one-dimensional thin-layer chromatography (TLC), using chloroform/methanol/water/NH4OH (60:37.5:3:1, v/v/v/v) as solvent 1 (46), or by two-dimensional TLC using solvent 2 (chloroform/methanol/NH4OH (130:60:8, v/v/v)) in the first dimension (vertical direction) and solvent 3 (chloroform-methanol/acetic acid/water (170:25:25:6, v/v/v/v)) in a second dimension (horizontal direction).
For two-dimensional TLC, 20 × 20-cm TLC plates of Merck Kiesel Gel 60 (0.25 mm) were used. Radiolabeled lipids were visualized and quantified using a Personal Molecular ImagerTM FX (Bio-Rad). Stored images were processed and quantified using Quantity One software for scanning and analysis of the captured PhosphorImages (Bio-Rad). Phospholipid content is expressed as mol % of total phospholipid, after correcting for two phosphates per molecule of CL, and is based on the intensity of the captured signal generating a latent image of the radiolabeled spot on the Phosphorscreen. The results presented are representative of two or more independent determinations.
Isolation of Everted Membrane Vesicles, Assays of ATPase Activity, Quantification of β-Subunit Content by Western Blot and Assays of ATP- and NADH-driven Proton Pumping Activity
The WT and cls deletion strains were grown on MYE medium at pH 10.5 and the ΔF0 strain was grown on GYE medium at pH 10.5. The cells were harvested at 4–5 h when A600 was around 1.5. Everted membrane vesicles were prepared as described previously (47). Protein concentrations were determined by Lowry assays (48). Everted membrane vesicles were diluted to 20 mg of protein/ml with a solution containing 20% glycerol, 10 mm Tris-HCl, pH 8, 5 mm MgCl2, and stored at −80 °C for further experiments. Octyl glucoside-stimulated ATPase activities were assayed as described previously (47). Membrane vesicles and cytoplasmic fractions were resolved by 11% SDS-PAGE gel (49), and Western blot analyses were performed as described previously (47). The amount of β subunit was quantified by ImageJ 1.47 software and described as % of WT, with WT set 100%. NADH-driven proton pumping assays were carried out similarly to the ATP-driven proton pumping assays except that 0.5 mm NADH was used to initiate the reaction (42).
Assays of Respiratory Chain Components
All enzyme assays were conducted in a Shimadzu UV-1601 UV-Visible spectrophotometer at room temperature. The assay buffer was 50 mm Tris-HCl, pH 8, and the assay volume was 1 ml using 50 or 100 μg of everted membrane vesicle protein. NADH oxidase assays were conducted by monitoring the decrease of A340 over time in the presence of 0.2 mm NADH. The NADH-ferricyanide oxidoreductase activity was measured at 420 nm in a buffer containing 1 mm NADH, 1 mm K3Fe(CN)6, and 10 mm KCN as described (50). Succinate dehydrogenase activity was monitored by following the phenazine methosulfate-coupled reduction of 2,6-dichloroindophenol at 600 nm (51). The reaction mixture, consisting of 10 mm succinate, 50 μg of vesicles, and 10 mm KCN, was preincubated for 5 min at room temperature. Then, 0.07 mm 2,6-dichloroindophenol and 1.625 mm phenazine methosulfate were added to initiate the reaction. N,N,N′,N′-Tetramethyl-p-phenylenediamine (TMPD) oxidase was measured by monitoring the increase in A562 nm in the presence of 0.25 mm TMPD (52). The extinction coefficients (mm−1 cm−1) used for activity calculations were 6.2 at 340 nm, 1 at 420 nm, 21 at 600 nm, and 10.5 at 562 nm. One unit (U) was defined as 1 μmol of substrate reduced or oxidized per minute per mg of protein.
Heme Staining and Spectral Analyses of Cytochrome Content
20 or 30 μg of everted membrane vesicle protein were separated by native 15% PAGE (49). The gels were immersed in 100 ml of staining solution containing 0.5 mg/ml of 3,3′,5,5′-tetramethylbenzidine, 50% methanol, 1 m sodium acetate, pH 4.7, for 30 min at room temperature in the dark (53), with slow shaking, after which H2O2 was added to 0.5%. The stained bands appeared in 5 min and the gels were scanned. The bands were quantified by ImageJ 1.47 software and described as % of WT, with WT set at 100%. Cytochromes were determined in a Shimadzu UV-2501PC UV-Visible recording spectrophotometer. Everted membrane vesicles were diluted to 4 or 5 mg/ml with 50 mm Tris-HCl, pH 8, 0.1% dodecyl maltoside in the cuvette. Ascorbate/TMPD-reduced minus air-oxidized spectra were used to quantitate cytochromes c and aa3; dithionite was then added to the reduced samples to measure the cytochrome b spectra (as the dithionite minus ascorbate/TMPD difference spectrum). For the quantification of cytochromes, the following wavelength pairs and millimolar extinction coefficients were used: (a+a3), ΔA600–620, Δϵ = 20.5 mm−1; b, ΔA560–575, Δϵ = 17.5 mm−1; c, ΔA551–538, Δϵ = 17.3 mm−1 (54).
ATP Synthesis from ADP + Pi-loaded Right-side Out Vesicles
The WT, ΔclsA, and ΔclsA/B/C strains were grown to an A600 around 0.6 in 1 liter of MYE at pH 10.5. The right-side out membrane vesicles loaded with ADP plus Pi were prepared as described previously (47). For the ATP synthesis reaction, 500 μg of vesicle protein was added into 1 ml of pH 7.5 or 10.5 assay buffer as described previously (47), and the reaction was initiated by adding 0.1 mm phenazine methosulfate and 10 mm ascorbate. At 10 s, part of each reaction mixture (200 μl) was removed and transferred to a pre-cooled solution of 30% perchloric acid. After neutralization, the amount of ATP was determined by the luciferin-luciferase method described previously (55); fluorescence was recorded by a chemiluminometer (Sirius L Tube Luminometer, Berthold, Germany). The amount of ATP synthesized was calculated from a standard curve. For each experimental set, the background ATP was measured using non-energized vesicles.
RNA Isolation and Quantitative Real-time PCR (qPCR)
The WT and cls deletion strains were grown on MYE medium at pH 7.5 or 10.5. The cells were harvested when the A600 reached 0.4. RNA was isolated and 200 ng of total RNA was used for reverse transcription with an iScript cDNA synthesis kit (Bio-Rad, number 170–8891) as described previously (42). Relative and absolute qPCR experiments were carried out in 384-well plates at the qPCR shared resource facility (Icahn School of Medicine at Mount Sinai, New York). Primers were designed by using Primer3 software (56). PCR were set up as described previously (42). Samples were run in triplicate with no template and no reverse transcriptase as controls. Data were analyzed using SDS 2.2.1 software (Applied Biosystems). For the relative qPCR, the fold-changes in gene expression were calculated according to the ΔΔ threshold cycle method (57) after normalization using gyrB and recA as reference genes. These two reference genes gave similar results. For the absolute qPCR determinations, standard curves were constructed for each test gene (clsA, clsB, and clsC) according to Refs. 42 and 58, and the transcript copies were calculated from the linear relationship between the threshold cycle (Ct) and the value of the natural log of gene concentration (copies/μl). The gyrB transcript was used as the internal standard.
RESULTS
Three Putative Alkaliphile Cls Proteins and Their Complementation Profiles in E. coli
The membranes of alkaliphilic B. pseudofirmus OF4 had earlier been shown to contain large amounts of CL at stationary phase at both pH 7.5 and 10.5 (40). Here, we first examined the % CL content among the phospholipids in the WT strain grown to exponential growth phase at pH 7.5 and 10.5. The % CL in the phospholipid panel at pH 10.5 was about 5 times higher than at pH 7.5 (Table 2). The lower % CL observed at the lower pH, pH 7.5, was generally closer to the % CL reported for the exponential phase cells of neutralophiles, such as B. subtilis (5, 14) and E. coli (43). Three cls genes had been identified in the genome of B. pseudofirmus OF4 (39), with locus tags of BpOF4_01900, BpOF4_07070, and BpOF4_03205. The corresponding genes are designated clsA, clsB, and clsC, respectively; the clsA gene had already been identified by Guo and Tropp (59) to encode a Cls. Bp-ClsA and Bp-ClsB have two conserved HKD motifs (HXK(X)4D(X)6G(X)2N motif), a feature of the phospholipase D family (60), whereas Bp-ClsC shows some variations in the two HKD motifs. The predicted transmembrane helices for Bp-ClsA, ClsB, and ClsC are three, two, and one, respectively, as determined using TMHMM 2.0 software. Bp-ClsA shows significant sequence similarity to YwnE from B. subtilis (45% identity; 215/482 amino acids), and to Cls1 and Cls2 from S. aureus, with identities of 40 and 43%, respectively, Bp-ClsB has a high sequence similarity to YwjE of B. subtilis (41% identity, 155/380 amino acids) as well as YwnE from B. subtilis (42% identity, 155/373 amino acids). Bp-ClsC has a sequence similarity to YwiE of B. subtilis (23% identity, 84/364 amino acids). Among the three B. pseudofirmus OF4 Cls proteins, Bp-ClsA displays 39% identity to Bp-ClsB, and Bp-ClsC shows 28% identity to both Bp-ClsA and Bp-ClsC.
TABLE 2.
The major phospholipid composition of WT and cls deletion strains grown to mid-logarithmic phase at either pH 7.5 or 10.5 on MYE medium
| Total phospholipid |
||||
|---|---|---|---|---|
| PG | PE | CL | ||
| % | ||||
| WT | pH 7.5 | 81 | 16 | 3 |
| pH 10.5 | 77 | 8 | 15 | |
| ΔclsA | pH 7.5 | 84 | 16 | 0 |
| pH 10.5 | 91 | 9 | 0 | |
| ΔclsB | pH 7.5 | 79 | 17 | 4 |
| pH 10.5 | 76 | 8 | 16 | |
| ΔclsC | pH 7.5 | 81 | 15 | 4 |
| pH 10.5 | 75 | 10 | 15 | |
| ΔclsA/B | pH 7.5 | 83 | 17 | 0 |
| pH 10.5 | 92 | 8 | 0 | |
| ΔclsA/B/C | pH 7.5 | 83 | 17 | 0 |
| pH 10.5 | 92 | 8 | 0 | |
To test whether cls genes of B. pseudofirmus OF4 are functional in vivo, three candidate genes were cloned into E. coli expression vector pBAD-TOPO and transformed into a CL-deficient E. coli BKT12 strain (43). Lipid analyses of E. coli BKT12 transformants showed that both Bp-ClsA and Bp-ClsB produced CL (21% of total phospholipid), whereas no CL was observed for E. coli BKT12 transformant containing Bp-ClsC or the empty control (Fig. 1). These results suggest that Bp-clsA and Bp-clsB functionally complement the CL-deficient E. coli strain and thus encode Cls enzymes. The Cls activity indicated for Bp-ClsA in E. coli was consistent with earlier findings (59). As shown for the ywiE gene from B. subtilis (14), B. pseudofirmus OF4 clsC could not complement the CL production defect in the E. coli mutant. We further investigated Bp-clsC expression in E. coli BKT12 by introducing a His6 tag at the C-terminal of Bp-clsC, and no detectable expression of Bp-clsC was found with 0, 0.02, or 0.2% arabinose induction, indicating that failure of its complementation of CL production might be due to lack of expression.
FIGURE 1.
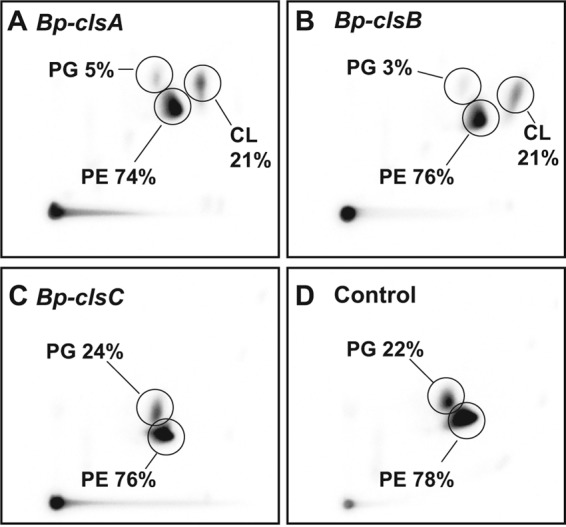
Complementation study on CL production of E. coli BKT12 strain expressing cls genes from B. pseudofirmus 811M. B. pseudofirmus 811M cls genes were cloned into pBAD-TOPO vector and overexpressed in CL-deficient E. coli BKT12 with 0.2% arabinose induction overnight in the presence of [32P]PO4. Lipids were extracted and analyzed as described under “Experimental Procedures.” The test plasmids are pBAD-Bp-ClsA (A), pBAD-Bp-ClsB (B), pBAD-Bp-ClsC (C), and pBAD-TOPO/lacZ/V5-His (D) as a control. The values shown are representative of three determinations.
Roles of the Three Synthase in CL Content of the Native Host
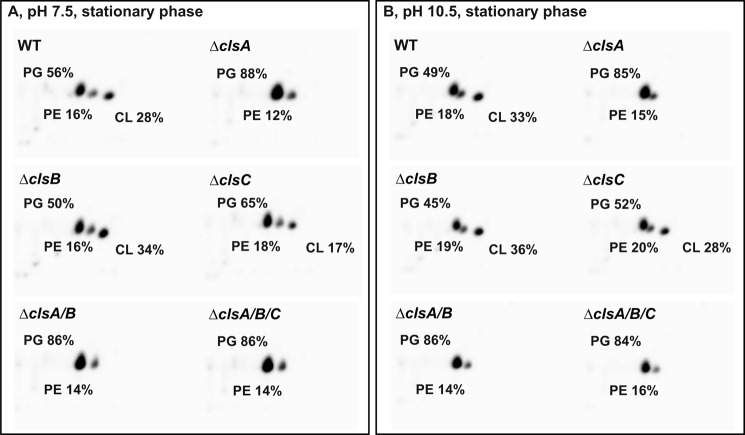
The lipid compositions of WT B. pseudofirmus OF4 and cls deletion strains grown to stationary phase on MYE medium were analyzed by two-dimensional TLC on silica gel plates (Fig. 2). The WT had CL contents of 28 and 33% of total phospholipid at pH 7.5 and 10.5, respectively. The CL contents at stationary phase were much higher than the values measured from log phase, suggesting that CL accumulated at the stationary phase, which is consistent with our previous observations (40) and reports of others (5, 14, 43). Therefore the initial assessment of CL deficiencies in the mutant panel was conducted on stationary cells. The ΔclsA mutant showed no detectable CL at either pH 7.5 or 10.5, and exhibited a significant increase in PG content to 88 or 85% of total phospholipid at pH 7.5 or 10.5, respectively. By comparison, the WT had PG contents of 56 and 49% of total phospholipid at pH 7.5 and 10.5, respectively. The absence of detectable CL in the ΔclsA strain was further confirmed in overexposed images (data not shown). The ΔclsB showed similar lipid profile and percentage of the total phospholipid to that obtained with the WT strain at both pH 7.5 and 10.5, indicating no contribution of clsB to CL production under these conditions. The ΔclsC mutant exhibited a significant decrease of CL content, relative to WT, from 28 to 17% at pH 7.5, but not at pH 10.5, suggesting that clsC plays a role in CL synthesis at pH 7.5 during stationary phase. The double ΔclsA/B and triple ΔclsA/B/C strains showed similar phospholipid profiles to the ΔclsA strain, with no detectable CL at both pH values tested. We also performed a similar phospholipid analysis for WT and cls mutants grown to exponential phase at both pH 7.5 and 10.5 (Table 2). The WT had much lower CL contents than noted above for stationary phase cells, with exponential phase values of only 3 and 15% of total phospholipid at pH 7.5 and 10.5, respectively, indicating an up-regulation of CL synthesis at high pH in exponential phase as well as stationary phase. The ΔclsA, ΔclsA/B, and the ΔclsA/B/C mutants all showed no detectable CL at either of the test pH values. By contrast, the ΔclsB and ΔclsC mutant strains exhibited phospholipid profiles similar to that of WT, suggesting that deletion of clsB or clsC did not significantly affect CL synthesis during exponential phase growth.
FIGURE 2.
Comparative phospholipid profiles of WT and cls deletion strains grown to stationary phase on MYE medium at either pH 7.5 (A) or 10.5 (B). Cells were grown overnight to stationary phase and lipids were extracted and separated by two-dimensional TLC as described in the legend to Fig. 1. The values shown are representative of two or more determinations.
Expression Levels of the Three cls Genes
The expression levels of three cls genes were determined for cells grown at pH 7.5 and 10.5 by absolute quantification PCR. As shown in Fig. 3A, the transcript levels of clsA were constitutively high at both pH values relative to the other cls genes, consistent with a dominant role in CL synthesis. The level of clsB transcripts in cells grown at pH 7.5 was about twice the level in cells grown at pH 10.5, and the transcript levels of clsC were the same at both pH values and the lowest among the three cls genes. We further investigated whether deletion of one or more cls genes affected expression of the remaining cls genes at pH 7.5 and 10.5. At low pH, there were no very significant effects of the cls deletion mutants on expression of those remaining (Fig. 3B), and at high pH, the expression level of clsC was modestly up-regulated in the ΔclsA and ΔclsA/B mutant strains (Fig. 3C).
FIGURE 3.
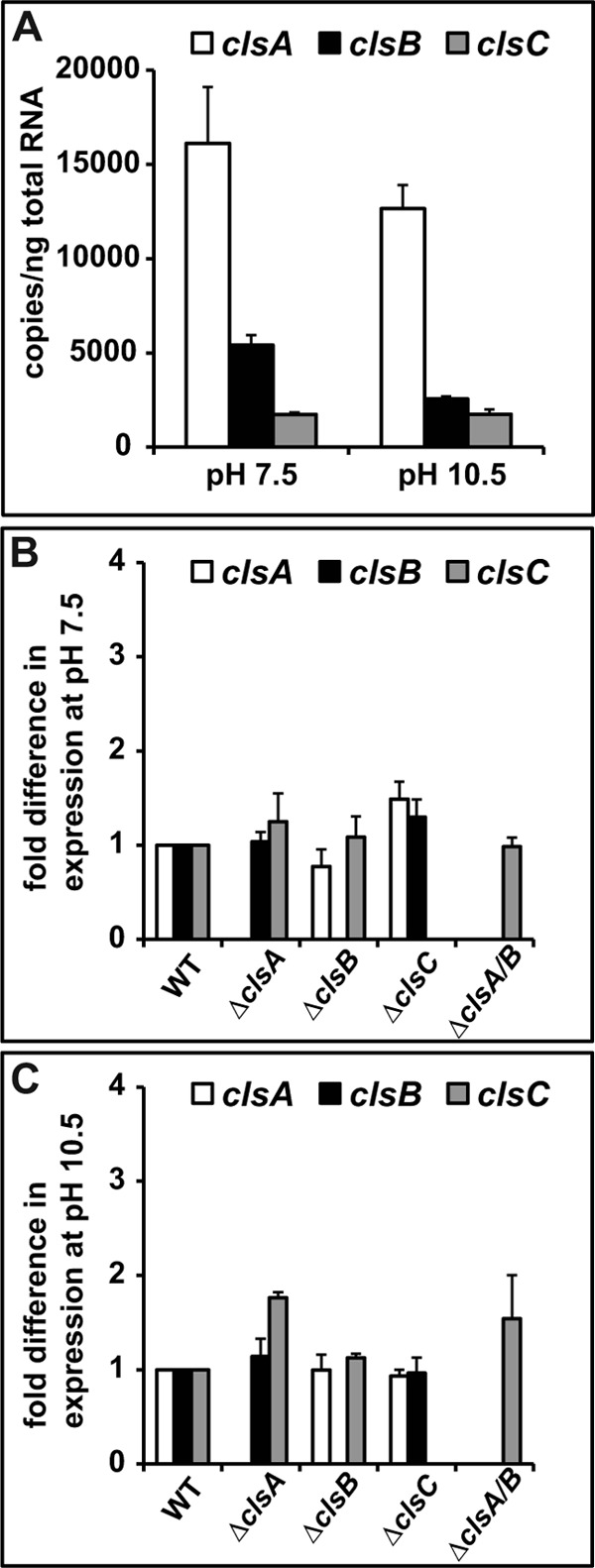
Analysis of cls gene expression in WT and cls deletion strains by qPCR. The experiments were conducted as described under “Experimental Procedures.” The gyrB gene was used as an internal reference gene. The values are averages of determinations from at least two independent experiments, and the error bars show the S.D. A, absolute quantification of WT cls gene transcripts at different pH values. The gyrB gene had values of (3.71 ± 0.04) × 104 and (3.52 ± 0.13) × 104 copies/ng of total RNA at pH 7.5 and 10.5, respectively. B, effects of deletion of a single cls gene or a double clsA and clsB on expression of the remaining cls genes in strains grown at pH 7.5 or 10.5 (C), the fold-difference is relative to the WT strain, with WT set to 1.
Effects of the Absence of CL on Non-fermentative Growth and OXPHOS
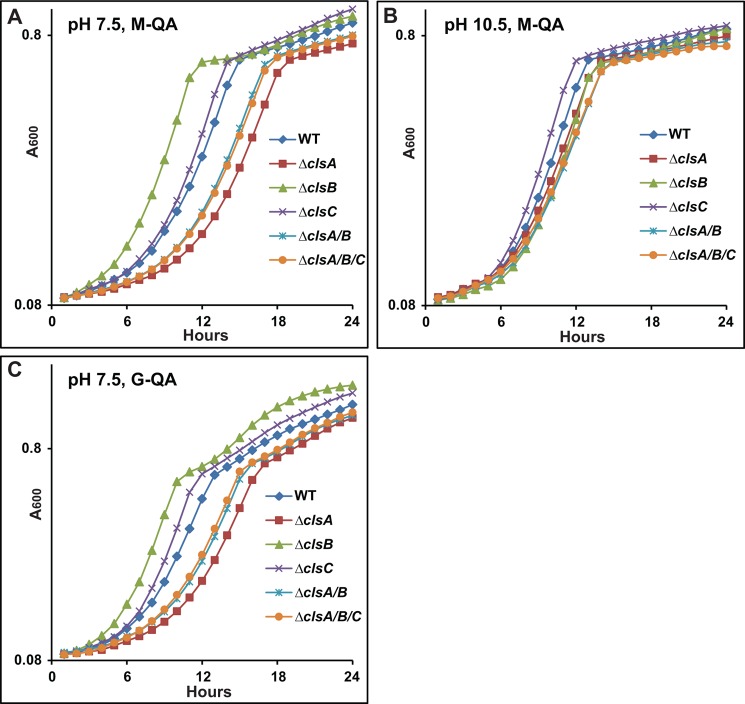
We then investigated whether the absence of detectable CL affects non-fermentative growth of B. pseudofirmus OF4, especially at high pH. First, we examined the growth of cls deletion mutants on semi-defined MYE medium, at both pH 7.5 and 10.5. The ATP synthase mutant ΔF0 strain was grown as a control to assess for fermentative growth supported by the presence of 0.1% yeast extract on MYE medium. At pH 7.5, the clsA and clsC mutants showed similar growth patterns to the WT, whereas the ΔclsB mutant showed a slightly lower level of growth at stationary phase (data not shown). At high pH, none of the mutants differed significantly from the WT strain in growth pattern (data not shown). We further measured the growth of cls mutants on QA medium at pH 7.5 and 10.5. QA is a defined medium that only supports non-fermentative growth when malate is the added carbon source (M-QA) and supports fermentative growth when glucose is added as the carbon source (G-QA). In M-QA medium at pH 7.5, the ΔclsB mutant exhibited a significantly shorter lag and more rapid exponential growth than the WT strain, the ΔclsC mutant grew just slightly faster than the WT, and all three mutants that had a clsA deletion showed a longer lag and slower exponential growth than the WT, especially the single ΔclsA mutant (Fig. 4A). At pH 10.5, in M-QA medium, there were only small differences among the growth patterns of the strains, with the ΔclsC strain again showing a slightly faster growth rate than WT, whereas all the other mutants grew slightly more slowly than WT (Fig. 4B). The impressive growth phenotypes of cls mutants, which were evident at pH 7.5 in M-QA but not on MYE medium, suggested that the phenotype might relate to particular properties of the QA medium rather than the non-fermentative carbon source. To test this, 50 mm glucose was substituted for malate as the carbon source in pH 7.5 G-QA. As shown in Fig. 4C, the pattern of phenotypes was indeed similar to that observed at pH 7.5 in M-QA medium except for an extra phase of modest additional growth, with a diauxic appearance, by the ΔclsB after the initial exponential phase in the G-QA. At pH 7.5, the spread of growth patterns suggests that stress from the high medium amine content leads to an increase in the value of Bp-ClsA relative to the other two synthases. However, the absence of detectable CL did not prevent non-fermentative growth of B. pseudofirmus OF4 in either MYE or M-QA media at pH 10.5, nor at pH 7.5.
FIGURE 4.
Growth tests of WT and cls deletion strains on M-QA medium at pH 7.5 (A), pH 10.5 (B), or G-QA medium at pH 7.5 (C). The growth experiments were conducted as described under “Experimental Procedures.” The values are the average of at least two independent growth curves with triplicate repeats, and the S.D. were within 13%.
Although CL was not essential for non-fermentative growth at high pH, it was of interest to assess its impact on specific ATP synthase and respiratory chain functions that are important elements of OXPHOS at pH 10.5. We first determined the amount of β subunit of F1F0-ATP synthase in the membrane and cytoplasm fractions, because the incorporation of the catalytic F1 domain, which contains the β subunit, depends upon prior assembly of the membrane-associated F0 domain (42). As shown in Table 3, no significant differences on β subunit levels were found in the membrane. However, the ΔclsB mutant showed a significant, ∼2.5-fold, increase in the cytoplasmic β subunit content relative to the WT, consistent with an assembly or stability deficit (Table 3). There was also a significantly lower cytoplasmic β subunit content in the ΔclsC mutant relative to the WT standard, suggestive of a more efficient assembly process in that mutant than in the WT.
TABLE 3.
Characterization of cls deletion strains, relative to WT strain, in β subunit distribution, ATPase activity, and proton pumping activity
cls deletion strains and WT strains were grown on MYE medium at pH 10.5, whereas the ΔF0 strain, as a control, was grown on GYE medium at pH 10.5. The ΔF0 strain showed a 12 ± 3 and 1021 ± 203% of β-subunit content, relative to WT, in the membrane and cytoplasm fractions, respectively; for the WT, the β-subunit distribution was 88 ± 3% in the membrane and 12 ± 3% in the cytoplasm. The WT strain had a value of 1.1 ± 0.18 units (mg of membrane protein)−1 min−1 for octyl glucoside-stimulated ATPase activity. For the ACMA fluorescence quenching assay, the WT strain had a proton pumping activity of 44.9 ± 3.9 and 56 ± 4% in ATP-driven and NADH-driven assays, respectively. The data from ATPase and the ATP-driven proton pumping assay were corrected by subtracting the values of ΔF0 strain as background. The values are the averages of at least 3 independent preparations ±S.D.
| β-Subunit content |
Octyl glucoside-stimulated ATPase activity | ATP-driven proton pumping activity | NADH-driven proton pumping activity | ||
|---|---|---|---|---|---|
| Membrane | Cytoplasm | ||||
| % of WT | % of WT | ||||
| WT | 100 | 100 | 100 | 100 | 100 |
| ΔclsA | 93 ± 7 | 101 ± 11 | 103 ± 9 | 102 ± 10 | 97 ± 4 |
| ΔclsB | 102 ± 7 | 256 ± 47 | 116 ± 6 | 98 ± 12 | 95 ± 10 |
| ΔclsC | 98 ± 14 | 50 ± 18 | 94 ± 16 | 93 ± 9 | 99 ± 2 |
| ΔclsA/B | 97 ± 5 | 97 ± 16 | 99 ± 10 | 104 ± 1 | 92 ± 3 |
| ΔclsA/B/C | 86 ± 5 | 116 ± 27 | 86 ± 7 | 91 ± 15 | 93 ± 10 |
Assays of ATPase activity (the hydrolytic activity) also revealed no significant defects on the cls deletion mutants. All the mutants exhibited ATP-driven and NADH-driven proton pumping activities comparable with that of the WT. Because no detectable CL levels were found in ΔclsA-deleted strains, ΔclsA and ΔclsA/B/C strains were chosen for further experiments. We conducted several additional assays of membrane vesicles isolated from ΔclsA and ΔclsA/B/C strains in comparison with the WT control (Fig. 5). The capacities for ATP synthesis by the two mutants were determined in ADP + Pi-loaded right-side out membrane vesicles, yielding evidence that both mutants retained at least 90% of the WT ATP synthase activity at both pH 7.5 and 10.5 (Fig. 5A). This result indicated that any deficits in the respiratory chain did not have devastating effects on OXPHOS. To assess deficits that might be modestly reducing optimal growth, several respiratory complexes and segments were assayed in the same two mutants in comparison with the WT control. The two mutants showed comparable NADH oxidase activity, which assessed function of the whole respiratory chain with NADH as electron donor; there was a modest deficit observed with the ΔclsA mutant that was not observed in the triple mutant also containing that deletion (Fig. 5B). Comparable succinate dehydrogenase activity was observed in both mutants relative to WT (Fig. 5B). Similarly, with the TMPD oxidase assay used for measuring the activity of cytochrome caa3 oxidase, the proton-pumping terminal oxidase, the ΔclsA and ΔclsA/B/C strains had levels similar to those of WT. By contrast, both mutants had significant reductions on NADH ferricyanide activity; this assay assesses NADH dehydrogenase activity, which in B. pseudofirmus OF4, represents two NdhII type enzymes that are peripheral proteins that associate with the cytoplasmic side of the coupling membrane (61). The ΔclsA and ΔclsA/B/C had 64 and 59% of WT activity, respectively. The WT strain has four major heme staining polypeptides as described (62). Both ΔclsA and ΔclsA/B/C showed the expected four bands with no significant reductions in the amounts (Fig. 5C). This was consistent with spectral assays of cytochrome contents of ΔclsA and ΔclsA/B/C, in which only a slightly reduced cytochrome c relative to WT was observed in the ΔclsA/B/C mutant (Table 4).
FIGURE 5.
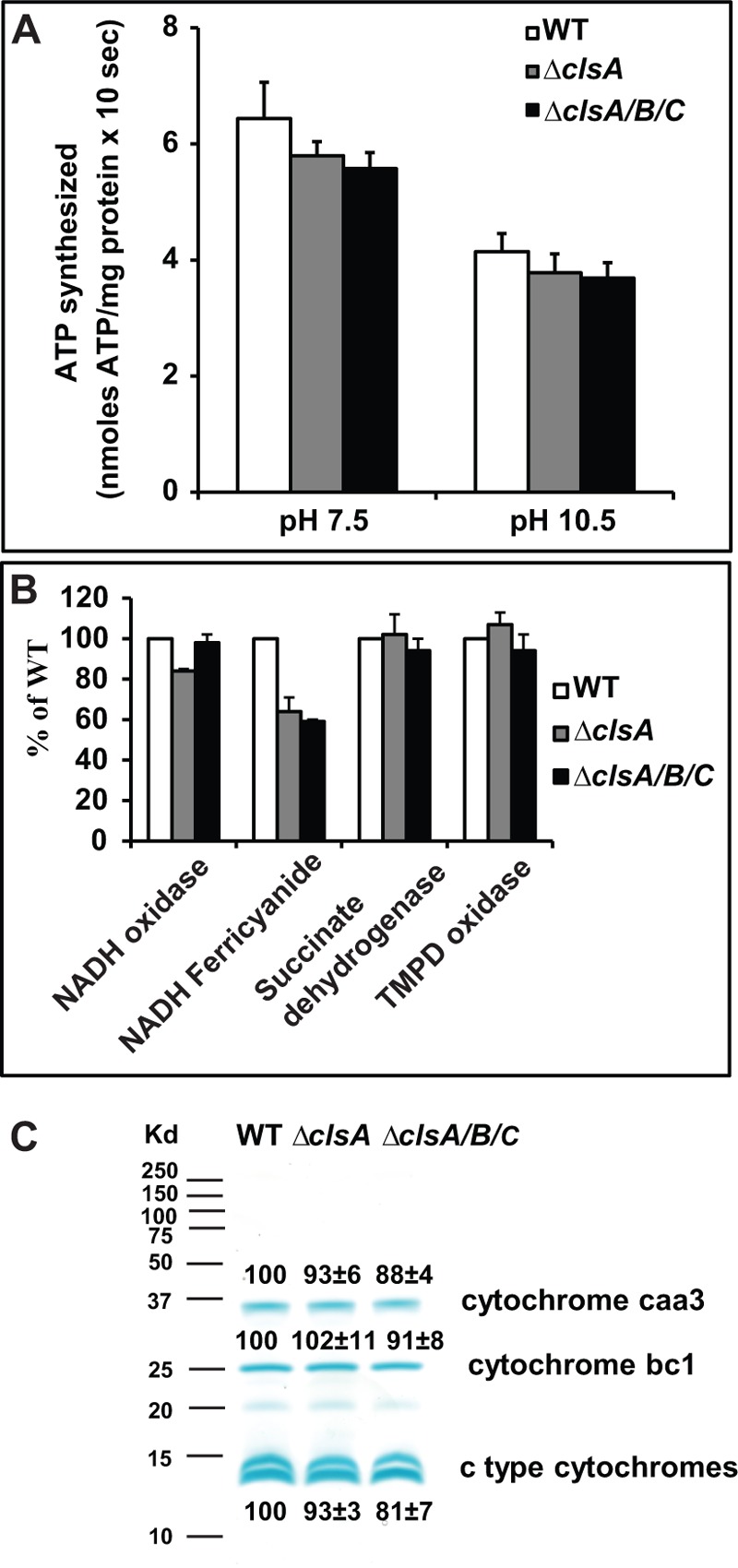
Functional characterization of WT, ΔclsA, and ΔclsA/B/C strains. A, ATP synthesis by ADP + Pi-loaded right-side out membrane vesicles. The strains were grown on MYE at pH 10.5. Assays were conducted as described under “Experimental Procedures.” The values are the averages of triplicate assays from three independent vesicle preparations and the error bars show the S.D. B, enzymatic activities of components of the respiratory chain. Everted membrane vesicles were prepared from cells grown on MYE at pH 10.5 and assays were conducted as described under “Experimental Procedures.” The values for mutants are described as % of WT, with WT set at 100%. The values are the averages of duplicate assays from two independent vesicle preparations, and the error bars show the S.D. The WT strain had values of 0.112 ± 0.003 and 0.283 ± 0.012 units for NADH oxidase and TMPD oxidase activity, respectively. The specific activities of the WT were 0.823 ± 0.014 and 0.561 ± 0.013 units/mg for NADH ferricyanide reductase and succinate dehydrogenase, respectively. C, heme-staining analysis of everted membrane vesicles. 30 μg of membrane vesicles were resolved on 15% polyacrylamide gels and stained with 3,3′,5,5′-tetramethylbenzidine + H2O2. The bands were quantified using ImageJ 1.47 software and the WT was designated 100%. Two c type cytochromes (bottom two bands) were quantified together. The values are the averages ± S.D. from two independent vesicle preparations.
TABLE 4.
Cytochrome content of WT, ΔclsA, and ΔclsA/B/C strains
Cells were grown to logarithmic phase on MYE medium at pH 10.5, and assayed as described under “Experimental Procedures.” The values are averages ± S.D. from at least two independent determinations.
| Cytochrome content |
|||
|---|---|---|---|
| Cytochrome aa3 | Cytochrome b nmol/mg of protein | Cytochrome c | |
| WT | 0.1 ± 0.016 | 0.29 ± 0.01 | 0.57 ± 0.06 |
| ΔclsA | 0.092 ± 0.023 | 0.29 ± 0.03 | 0.50 ± 0.08 |
| ΔclsA/B/C | 0.097 ± 0.016 | 0.26 ± 0.01 | 0.47 ± 0.08 |
Role of CL in Tolerance of High Salt and in Long Term Survival
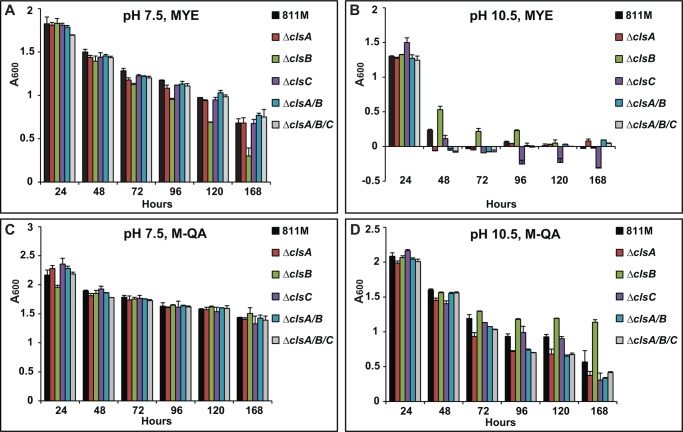
As noted earlier, CL deficiency is associated with reduced tolerance of salinity in B. subtilis and prolonged salinity stress in S. aureus (5, 6). It was therefore of interest to examine whether this pattern extended to alkaliphilic B. pseudofirmus OF4 as well. We conducted growth tests of cls deletion strains on high salt medium. As shown in Fig. 6, the WT had a longer lag time for initiation of growth in media with elevated NaCl concentrations than observed in its absence; the cls deletion strains did not differ greatly from WT at either 2 m NaCl on MYE at pH 7.5 (Fig. 6A), or at 1.5 m NaCl at pH 10.5 (Fig. 6B). The pattern of the alkaliphile response to elevated salinity indicated greater salt tolerance than observed with CL-less B. subtilis, which exhibited a lag and final growth deficit relative to WT in 1.5 m NaCl (5). The alkaliphile pattern was more similar to that of the salt-tolerant S. aureus strain, for which growth of cls mutants was not significantly different from growth of the WT (6). The S. aureus double cls1/cls2 mutant did, however, exhibit a significant drop in CFUs after 105 h incubation at around 2.5 m NaCl (6).
FIGURE 6.
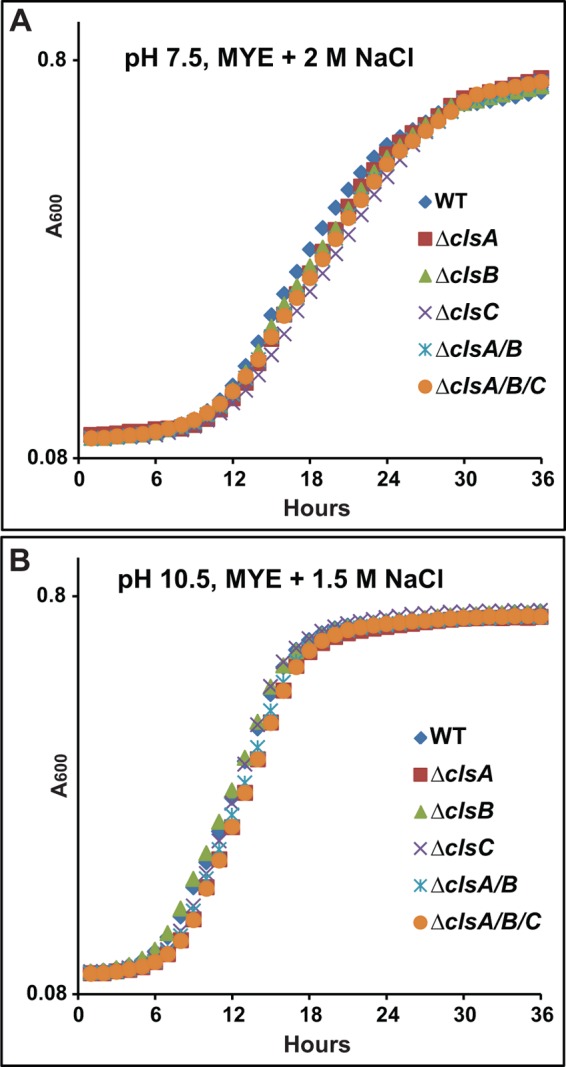
Growth of WT and cls deletion strains on high salt medium. The strains were pre-grown on MYE medium at either pH 7.5 or 10.5, and the growth experiments were conducted as described under “Experimental Procedures.” Strains were grown on 7.5 MYE + 2 m NaCl (A) or 10.5 MYE + 1.5 m NaCl (B). The values are the average of two independent growth curves with duplicate repeats, and the S.D. were within 9%.
CL has been shown to play a role in survival of cells in a prolonged incubation for E. coli (43, 63) as well as for S. aureus under elevated salt conditions (6). Here, we first examined the long term survival of alkaliphilic B. pseudofirmus OF4 cls mutants on MYE and QA media at 30 °C (Fig. 7). At 7.5 MYE, survival of the WT, as assessed by A600, declined with the increase in incubation time from 24 h, by which time the stationary phase is reached, to 168 h, and all the mutant strains except the ΔclsB mutant exhibited similar growth to the WT. The ΔclsB mutant exhibited an A600 that was only 71 and 44% of WT, at 120 and 168 h, respectively (Fig. 7A). The reduced A600 indicates that clsB plays a role in long term survival at pH 7.5. At pH 10.5, the WT strain exhibited a sharp decline in A600 between 24 and 48 h; interestingly, deletion of clsB appeared to offer some protection against that decline whereas, especially after 72 h, deletion of clsC alone seemed to exacerbate the decline relative to WT (Fig. 7B). Long term survival was also examined in the same panel of strains growing on the defined QA medium at 30 °C. At pH 7.5, all the mutant strains exhibited comparable growth to the WT, except for a small deficit in initial growth of the ΔclsB mutant (Fig. 7C). At pH 10.5, all the strains with clsA deletions exhibited a major decline in A600 by 96 h, with less than 80% of the WT A600 after 96 h incubation (Fig. 7D). The ΔclsC mutant more slowly reached the full decline shown by the clsA mutants by the end point of 168 h, whereas, by contrast, the ΔclsB mutant exhibited a significantly better survival than any of the other strains after 96 h (Fig. 7D). The generally greater retention of long term survival capacity in pH 10.5 M-QA media than in pH 10.5 MYE media was striking (compare Fig. 7, D with B). Finally, the same long term experiment was conducted at 37 °C (data not shown), under conditions in which the stationary phase is somewhat faster than at 30 °C. In pH 7.5 MYE, the strains with a clsA deletion exhibited an A600 decline to values less than 52% of WT growth at 72 h, a much more rapid decline than observed at 30 °C; strains with clsA deletions still showed even more profound declines in survival and there was still an indication that deletion of clsB favored survival up to 72 h at which point a deleterious effect of clsC deletion became evident. In M-QA medium, the survival deficits of the panel of strains were greater than at 30 °C; the ΔclsB decline as a function of temperature was significantly less than that of the other mutants.
FIGURE 7.
Long term survival growth of cls deletion strains at 30 °C. Strains were grown on MYE at pH 7.5 (A) or 10.5 (B), or M-QA at pH 7.5 (C) or 10.5 (D). The experiments were conducted as described under “Experimental Procedures.” The values are the averages of two independent growth experiments with duplicate repeats, and the error bars show the S.D.
DISCUSSION
A major finding of this study is that CL is not essential for OXPHOS and the non-fermentative growth it supports in alkaliphilic B. pseudofirmus OF4, during growth at either pH 7.5 or 10.5. This makes it unlikely that CL is a crucial participant in the proton movements on or near the membrane surface from proton-pumping respiratory chain complexes to the F1F0-ATP synthase complex. As in other studies in which CL has been reduced (14, 43), a compensatory rise in anionic PG was observed in the alkaliphile, but PG does not have the head group structure, which was hypothesized to underpin a specific mechanism that could account for the capacity of the alkaliphile for ATP synthesis at high pH (28).
Other mechanisms and bases for retention and translocation of protons before equilibration with the bulk have been proposed (33, 34). Further studies in this model system will explore the possibility that several such mechanisms make contributions in this extremophile. In keeping with early proposals of Williams (64, 65), several adaptations in the OXPHOS machinery itself have already been shown to be necessary for the OXPHOS capacity displayed by B. pseudofirmus OF4 (38, 66, 67).
The experiments conducted here reveal that Bp-ClsA is the major contributor to the CL content of B. pseudofirmus OF4. Consistent with that contribution was the observation that both the single ΔclsA and triple deletion mutants had a significant reduction in NADH dehydrogenase activity. This reduced activity did not ultimately cause a major defect in respiration as a whole, but may be caused by the reduced ability of the enzyme to bind well enough to the membrane surface in the absence of CL. Although ClsA is a dominant source of CL in the alkaliphile, Bp-ClsC makes an evident contribution, augmenting the role of Bp-ClsA during long term survival at pH 10.5 (Fig. 7). In addition, there are particular contexts in which either positive or negative effects on expression of Bp-ClsB are evident, i.e. a positive effect during growth at pH 7.5 (data not shown) and a negative effect during long term growth at pH 10.5 (Fig. 7). This greater role of Bp-ClsB at pH 7.5 than at pH 10.5 is consistent with an emerging pattern in B. pseudofirmus OF4 in which paralogous genes that may have arisen from gene duplications develop pH-specific functions. This was recently described for two yqjG/spoIIIJ homologues in the alkaliphile (42). B. pseudofirmus OF4 is “hard-wired” for alkaliphily, i.e. its capacities for growth at external pH values of 11.2 and above are accompanied by a cost in efficacy of some physiological systems, e.g. loss of motility, at pH 7.5 (68). However, in the realm of its membrane phospholipids, the current studies suggest that the major Bp-ClsA is in general well complemented by the mixture of contributions from the two other synthases in terms of meeting current demands to function well in the near neutral range of pH.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM28454 (to T. A. K.) and R37 GM20478 (to W. D.).
- CL
- cardiolipin
- Cls
- cardiolipin synthase
- OXPHOS
- oxidative phosphorylation
- PG
- phosphatidylglycerol
- PMF
- protonmotive force
- TMPD
- N,N,N′,N′-tetramethyl-p-phenylenediamine
- MYE
- malate-yeast extract
- GYE
- glucose-yeast extract
- qPCR
- quantitative PCR.
REFERENCES
- 1. Barth P. G., Scholte H. R., Berden J. A., Van der Klei-Van Moorsel J. M., Luyt-Houwen I. E., Van 't Veer-Korthof E. T., Van der Harten J. J., Sobotka-Plojhar M. A. (1983) An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 62, 327–355 [DOI] [PubMed] [Google Scholar]
- 2. McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T. (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 361, 462–469 [DOI] [PubMed] [Google Scholar]
- 3. Vartak R., Porras C. A., Bai Y. (2013) Respiratory supercomplexes. Structure, function and assembly. Protein Cell 4, 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patil V. A., Fox J. L., Gohil V. M., Winge D. R., Greenberg M. L. (2013) Loss of cardiolipin leads to perturbation of mitochondrial and cellular iron homeostasis. J. Biol. Chem. 288, 1696–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lopez C. S., Alice A. F., Heras H., Rivas E. A., Sanchez-Rivas C. (2006) Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152, 605–616 [DOI] [PubMed] [Google Scholar]
- 6. Tsai M., Ohniwa R. L., Kato Y., Takeshita S. L., Ohta T., Saito S., Hayashi H., Morikawa K. (2011) Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 11, 10.1186/1471-2180-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang M., Mileykovskaya E., Dowhan W. (2002) Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 [DOI] [PubMed] [Google Scholar]
- 8. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 9. Bogdanov M., Mileykovskaya E., Dowhan W. (2008) Lipids in the assembly of membrane proteins and organization of protein supercomplexes. Implications for lipid-linked disorders. Subcell. Biochem. 49, 197–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bazán S., Mileykovskaya E., Mallampalli V. K., Heacock P., Sparagna G. C., Dowhan W. (2013) Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J. Biol. Chem. 288, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang M., Mileykovskaya E., Dowhan W. (2005) Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Palsdottir H., Hunte C. (2004) Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]
- 13. Mileykovskaya E., Dowhan W. (2000) Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182, 1172–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawai F., Shoda M., Harashima R., Sadaie Y., Hara H., Matsumoto K. (2004) Cardiolipin domains in Bacillus subtilis marburg membranes. J. Bacteriol. 186, 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mukhopadhyay R., Huang K. C., Wingreen N. S. (2008) Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys. J. 95, 1034–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Renner L. D., Weibel D. B. (2011) Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 6264–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Romantsov T., Helbig S., Culham D. E., Gill C., Stalker L., Wood J. M. (2007) Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64, 1455–1465 [DOI] [PubMed] [Google Scholar]
- 18. Romantsov T., Guan Z., Wood J. M. (2009) Cardiolipin and the osmotic stress responses of bacteria. Biochim. Biophys. Acta 1788, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Acehan D., Malhotra A., Xu Y., Ren M., Stokes D. L., Schlame M. (2011) Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 100, 2184–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lange C., Nett J. H., Trumpower B. L., Hunte C. (2001) Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20, 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunte C., Palsdottir H., Trumpower B. L. (2003) Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett. 545, 39–46 [DOI] [PubMed] [Google Scholar]
- 22. Wenz T., Hielscher R., Hellwig P., Schägger H., Richers S., Hunte C. (2009) Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim. Biophys. Acta 1787, 609–616 [DOI] [PubMed] [Google Scholar]
- 23. Gomez B., Jr., Robinson N. C. (1999) Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry 38, 9031–9038 [DOI] [PubMed] [Google Scholar]
- 24. Zhang X., Hiser C., Tamot B., Benning C., Reid G. E., Ferguson-Miller S. M. (2011) Combined genetic and metabolic manipulation of lipids in Rhodobacter sphaeroides reveals non-phospholipid substitutions in fully active cytochrome c oxidase. Biochemistry 50, 3891–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sedlák E., Robinson N. C. (1999) Phospholipase A2 digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38, 14966–14972 [DOI] [PubMed] [Google Scholar]
- 26. Kikuchi S., Shibuya I., Matsumoto K. (2000) Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J. Bacteriol. 182, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salzberg L. I., Helmann J. D. (2008) Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 190, 7797–7807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haines T. H. (1983) Anionic lipid headgroups as a proton-conducting pathway along the surface of membranes. A hypothesis. Proc. Natl. Acad. Sci. U.S.A. 80, 160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haines T. H., Dencher N. A. (2002) Cardiolipin. A proton trap for oxidative phosphorylation. FEBS Lett. 528, 35–39 [DOI] [PubMed] [Google Scholar]
- 30. Haines T. H. (2009) A new look at cardiolipin. Biochim. Biophys. Acta 1788, 1997–2002 [DOI] [PubMed] [Google Scholar]
- 31. Mitchell P. (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 [DOI] [PubMed] [Google Scholar]
- 32. Mulkidjanian A. Y., Heberle J., Cherepanov D. A. (2006) Protons @ interfaces. Implications for biological energy conversion. Biochim. Biophys. Acta 1757, 913–930 [DOI] [PubMed] [Google Scholar]
- 33. Springer A., Hagen V., Cherepanov D. A., Antonenko Y. N., Pohl P. (2011) Protons migrate along interfacial water without significant contributions from jumps between ionizable groups on the membrane surface. Proc. Natl. Acad. Sci. U.S.A. 108, 14461–14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brändén M., Sandén T., Brzezinski P., Widengren J. (2006) Localized proton microcircuits at the biological membrane-water interface. Proc. Natl. Acad. Sci. U.S.A. 103, 19766–19770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sandén T., Salomonsson L., Brzezinski P., Widengren J. (2010) Surface-coupled proton exchange of a membrane-bound proton acceptor. Proc. Natl. Acad. Sci. U.S.A. 107, 4129–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krulwich T. A. (1995) Alkaliphiles. “Basic” molecular problems of pH tolerance and bioenergetics. Mol. Microbiol. 15, 403–410 [DOI] [PubMed] [Google Scholar]
- 37. Hicks D. B., Liu J., Fujisawa M., Krulwich T. A. (2010) F1F0-ATP synthases of alkaliphilic bacteria. Lessons from their adaptations. Biochim. Biophys. Acta 1797, 1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krulwich T. A., Ito M. (2013) Prokaryotic alkaliphiles. In The Prokaryotes (Rosenberg E., Delong E. F., Lory S., Stackebrandt E., Thompson F., eds) 4th Ed, pp 441–469, Springer, New York [Google Scholar]
- 39. Janto B., Ahmed A., Ito M., Liu J., Hicks D. B., Pagni S., Fackelmayer O. J., Smith T. A., Earl J., Elbourne L. D., Hassan K., Paulsen I. T., Kolstø A. B., Tourasse N. J., Ehrlich G. D., Boissy R., Ivey D. M., Li G., Xue Y., Ma Y., Hu F. Z., Krulwich T. A. (2011) Genome of alkaliphilic Bacillus pseudofirmus OF4 reveals adaptations that support the ability to grow in an external pH range from 7.5 to 11.4. Environ. Microbiol. 13, 3289–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clejan S., Krulwich T. A., Mondrus K. R., Seto-Young D. (1986) Membrane lipid composition of obligately and facultatively alkalophilic strains of Bacillus spp. J. Bacteriol. 168, 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clejan S., Guffanti A. A., Cohen M. A., Krulwich T. A. (1989) Mutation of Bacillus firmus OF4 to duramycin resistance results in substantial replacement of membrane lipid phosphatidylethanolamine by its plasmalogen form. J. Bacteriol. 171, 1744–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J., Hicks D. B., Krulwich T. A. (2013) Roles of AtpI and two YidC-type proteins from alkaliphilic Bacillus pseudofirmus OF4 in ATP synthase assembly and nonfermentative growth. J. Bacteriol. 195, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan B. K., Bogdanov M., Zhao J., Dowhan W., Raetz C. R., Guan Z. (2012) Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 16504–16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z., Hicks D. B., Guffanti A. A., Baldwin K., Krulwich T. A. (2004) Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of Alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and non-fermentative growth at pH 10.5. J. Biol. Chem. 279, 26546–26554 [DOI] [PubMed] [Google Scholar]
- 45. Wei Y., Southworth T. W., Kloster H., Ito M., Guffanti A. A., Moir A., Krulwich T. A. (2003) Mutational loss of a K+ and NH4+ transporter affects the growth and endospore formation of alkaliphilic Bacillus pseudofirmus OF4. J. Bacteriol. 185, 5133–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fine J. B., Sprecher H. (1982) Unidimensional thin-layer chromatography of phospholipids on boric acid-impregnated plates. J. Lipid Res. 23, 660–663 [PubMed] [Google Scholar]
- 47. Liu J., Fujisawa M., Hicks D. B., Krulwich T. A. (2009) Characterization of the functionally critical AXAXAXA and PXXEXXP motifs of the ATP synthase c-subunit from an alkaliphilic Bacillus. J. Biol. Chem. 284, 8714–8725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 49. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 50. Swartz T. H., Ito M., Ohira T., Natsui S., Hicks D. B., Krulwich T. A. (2007) Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J. Bacteriol. 189, 3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hatefi Y. (1978) Resolution of complex II and isolation of succinate dehydrogenase (EC 1.3.99.1). Methods Enzymol. 53, 27–35 [DOI] [PubMed] [Google Scholar]
- 52. Sakamoto J., Matsumoto A., Oobuchi K., Sone N. (1996) Cytochrome bd-type quinol oxidase in a mutant of Bacillus stearothermophilus deficient in caa3-type cytochrome c oxidase. FEMS Microbiol. Lett. 143, 151–158 [DOI] [PubMed] [Google Scholar]
- 53. Guikema J. A., Sherman L. A. (1981) Electrophoretic profiles of cyanobacterial membrane polypeptides showing heme-dependent peroxidase activity. Biochim. Biophys. Acta Bioenergetics 637, 189–201 [Google Scholar]
- 54. Quirk P. G., Guffanti A. A., Plass R. J., Clejan S., Krulwich T. A. (1991) Protonophore-resistance and cytochrome expression in mutant strains of the facultative alkaliphile Bacillus firmus OF4. Biochim. Biophys. Acta 1058, 131–140 [DOI] [PubMed] [Google Scholar]
- 55. Stanley P. E., Williams S. G. (1969) Use of the liquid scintillation spectrometer for determining adenosine triphosphate by the luciferase enzyme. Anal. Biochem. 29, 381–392 [DOI] [PubMed] [Google Scholar]
- 56. Rozen S., Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 [DOI] [PubMed] [Google Scholar]
- 57. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 58. Chini V., Foka A., Dimitracopoulos G., Spiliopoulou I. (2007) Absolute and relative real-time PCR in the quantification of tst gene expression among methicillin-resistant Staphylococcus aureus. Evaluation by two mathematical models. Lett. Appl. Microbiol. 45, 479–484 [DOI] [PubMed] [Google Scholar]
- 59. Guo D., Tropp B. E. (1998) Cloning of the Bacillus firmus OF4 cls gene and characterization of its gene product. Biochim. Biophys. Acta 1389, 34–42 [DOI] [PubMed] [Google Scholar]
- 60. Guo D., Tropp B. E. (2000) A second Escherichia coli protein with CL synthase activity. Biochim. Biophys. Acta 1483, 263–274 [DOI] [PubMed] [Google Scholar]
- 61. Liu J., Krulwich T. A., Hicks D. B. (2008) Purification of two putative type II NADH dehydrogenases with different substrate specificities from alkaliphilic Bacillus pseudofirmus OF4. Biochim. Biophys. Acta 1777, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hicks D. B., Krulwich T. A. (1995) The respiratory chain of alkaliphilic bacteria. Biochim. Biophys. Acta 1229, 303–314 [DOI] [PubMed] [Google Scholar]
- 63. Hiraoka S., Matsuzaki H., Shibuya I. (1993) Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 336, 221–224 [DOI] [PubMed] [Google Scholar]
- 64. Williams R. J. (1978) The multifarious couplings of energy transduction. Biochim. Biophys. Acta 505, 1–44 [DOI] [PubMed] [Google Scholar]
- 65. Williams R. J. (1988) Proton circuits in biological energy interconversions. Annu. Rev. Biophys. Biophys. Chem. 17, 71–97 [DOI] [PubMed] [Google Scholar]
- 66. Preiss L., Yildiz O., Hicks D. B., Krulwich T. A., Meier T. (2010) A new type of proton coordination in an F1F0-ATP synthase rotor ring. PLoS Biol. 8, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Preiss L., Klyszejko A. L., Hicks D. B., Liu J., Fackelmayer O. J., Yildiz Ö., Krulwich T. A., Meier T. (2013) The c-ring stoichiometry of ATP synthase is adapted to cell physiological requirements of alkaliphilic Bacillus pseudofirmus OF4. Proc. Natl. Acad. Sci. U.S.A. 110, 7874–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krulwich T. A., Liu J., Morino M., Fujisawa M., Ito M., Hicks D. B. (2011) Adaptive mechanisms of extreme alkaliphiles. In Extremophiles Handbook (Horikoshi K., Antranikian G., Bull A., Robb F., Stetter K., eds) pp. 120–139, Springer, Heidelberg [Google Scholar]