Abstract
Endocrine therapy resistance in estrogen receptor alpha positive (ERα+) breast cancers remains a major obstacle for maintaining efficacy of targeted therapies. We investigated the significance and the mechanisms involved in cMYC over-expression in a MCF7 derived panel of ERα+ breast cancer cells which can proliferate in the absence of estrogen with different sensitivities to anti-hormone therapies. We show that all the resistant cell lines tested over-express cMYC as compared to parental MCF7 cells and its inhibition lead to the differential blocking of estrogen-independent proliferation in resistant cells. Further investigation of the resistant cell line, MCF7:5C, suggested transcriptional de-regulation of cMYC gene was responsible for its over-expression. Chromatin immuno-precipitation assay revealed markedly higher recruitment of phosphorylated serine-2 carboxy-terminal domain (CTD) of RNA polymerase-II at the proximal promoter of cMYC gene, which is responsible for transcriptional elongation of the cMYC RNA. The level of CDK9, a factor responsible for the phosphorylation of serine-2 of RNA polymerase II CTD, was found to be elevated in all the resistant cell lines. Pharmacological inhibition of CDK9 not only reduced the transcripts and the protein levels of cMYC in MCF7:5C cells but also selectively inhibited the estrogen-independent growth of all the resistant cell lines. This study describes the up-stream molecular events involved in the transcriptional over-expression of cMYC gene in breast cancer cells proliferating estrogen-independently and identifies CDK9 as a potential novel drug target for therapeutic intervention in endocrine-resistant breast cancers.
Keywords: Aromatase inhibitor, cyclin dependent kinase-9, Breast Cancer, Endocrine therapy resistance, cMYC
Introduction
Resistance to endocrine therapies (tamoxifen and aromatase inhibitors) represents a major clinical concern for the survivorship of the estrogen receptor positive (ER+) breast cancer patients [1-3]. The majority of hormone receptor positive advanced breast cancer (ABC) patients report disease progression within 2-3 years of endocrine therapy treatment [4-6]. Recent clinical studies have found over-expression of the cMYC oncogene and the genes regulated by cMYC as one of the major predictor in the aromatase inhibitor resistant breast cancers [7-9] whereas its over-expression is sufficient to confer resistance to anti-estrogens [10]. Besides endocrine resistance, cMYC oncoprotein have been found to regulate the expression of “poor-outcome” signature genes responsible for metastasis [11]. Gain of cMYC is also associated with the progression of invasive ductal carcinoma (IDC) from the ductal carcinoma in situ (DCIS) [12] and amplification of cMYC in breast cancer is significantly associated with risk of relapse and death [13]. It is therefore appropriate to study the underlying molecular mechanisms which contribute to estrogen independence and acquired resistance to identify novel therapeutic targets for the endocrine therapy resistant breast cancers.
Although targeting cMYC represents an obvious therapeutic opportunity to block the growth of the resistant breast cancer cells, this has not been successful due to the lack of a drug-able domain in its ‘basic helix-loop-helix’ structure [14]. Additionally, unacceptable toxicity is associated with cMYC inhibition, as the protein is critically involved in proliferation and regeneration of normal adult tissues [15,16]. Other approaches such as synthetic lethality [17] and modulating chromatin-dependent signal transduction have been used to circumvent direct targeting of cMYC [18].
To determine the relevance and mechanism of cMYC over-expression in imparting estrogen-independence to the endocrine-resistant breast cancer cells we used a panel of MCF7 ERα+ breast cancer cells which are known to proliferate in the absence of estrogen and exhibit different sensitivities to the anti-hormone therapies. The different MCF7 cell line derivatives used were MCF7:5C [19], MCF7:2A [20], MCF7/LCC1 [21], MCF7/LCC2 [22] and MCF7/LCC9 [23,24]. All these cells mimic aromatase inhibitor resistance as they can grow in an estrogen-deprived condition. In addition, MCF7:5C and LCC2 cells are also resistant to anti-estrogens, 4-hydroxy - tamoxifen (4OHT) whereas LCC9 cells demonstrate resistance to 4OHT and fulvestrant. All these cell lines cells showed high expression of cMYC protein as compared to parent MCF7 cells and estrogen-independent growth of all the resistant cells was drastically inhibited by a cMYC inhibitor, 10058-F4 (F4). For focused studies we chose MCF7:5C cells as we have extensive experience with this cell line and the LCC1, LCC2 and LCC9 cells showed modest estrogen stimulation of growth [21,23,22] despite being estrogen-independent. On the other hand MCF7:5C cells undergo apoptosis after estrogen treatment [25,26]. This is a documented response clinically, following the development of anti-hormone resistance [27].
This study dissects the upstream molecular mechanism involved in the transcriptional over-expression of cMYC oncogene in the endocrine-therapy resistant cells, which imparts estrogen-independence. In addition, we present CDK9 as a potential target for therapeutic intervention which can suppress the deregulated transcriptional over-expression of cMYC leading to complete inhibition of estrogen-independent proliferation of the endocrine-resistant breast cancer cells.
Materials and Methods
Cell Culture and Reagents
Cell culture media were purchased from Invitrogen Inc. (Grand Island, NY) and fetal calf serum (FCS) was obtained from HyClone Laboratories (Logan, UT). The ERα+ breast cancer cells MCF-7:WS8 (mentioned as MCF7) and estrogen-deprived MCF7:5C and MCF7:2A cells were derived from MCF7 cells obtained from the Dr. Dean Edwards, San Antonio, Texas as reported previously [19]. The MCF7/LCC1, LCC2 and LCC9 were obtained from the shared tissue culture facility of the Lombardi comprehensive cancer center. The cell lines were authenticated by DNA fingerprinting. All the cells except MCF7 cells were maintained in phenol red-free RPMI media (Invitrogen Inc, Grand Island, NY) supplemented with 10% charcoal dextran treated FCS, 6 ng/ml bovine insulin and penicillin and streptomycin. MCF7 cells were maintained in phenol red containing media with 10% FCS. Three to four days prior to harvesting the MCF7 cells were cultivated in phenol red-free media containing 10% charcoal dextran treated FCS. cMYC inhibitor, 10058-F4 was purchased from Sigma-Aldrich (St. Louis, MO) and CDK9 inhibitor, CAN 508 (cat # 238811), was purchased from EMD Chemicals Inc. (San Diego, CA). All the experiments were performed at least three times, in triplicate to confirm the results.
Cell growth assay
The cell growth assays were performed by measuring the total DNA per well in 24 well plates. Twenty to twenty five thousand cells were plated per well and treatment with indicated concentrations of compounds was started after 24 hours, in triplicates. Media with specific treatments were changed every 48 hours. The cells were harvested in hypotonic buffer solution followed by sonication after indicated time points. Total DNA was measured using a fluorescent dye (Hoechst 33258) in the DNA quantitation kit (Cat # 170-2480; Bio-Rad, Hercules, CA, USA) according to manufacturer’s instructions.
RNA isolation and real time PCR
TRIzol reagent (Invitrogen,Carlsbad, CA) and RNAeasy kit (Qiagen, Valencia, CA, USA) were used to isolate total RNA according to the manufacturer’s instructions. Real-time PCR was performed as previously described [28]. Briefly, cDNA was generated from RNA using High capacity cDNA reverse transcription kits (Applied Biosystems, Foster City, CA). Subsequently the cDNA was diluted and RT-PCR was performed using ABI Prism 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA). The change in expression of transcripts was determined as described previously using the ribosomal protein 36B4 mRNA as the internal control [28]. The primer sequences for the cMYC mRNA was 5′GCCAGCTCTCCACACATCAG 3′ (forward); 5′ TCTTGGCAGCAGGATAGTCCTT 3′ (reverse).
Cell cycles analysis
The cells were treated with vehicle (0.1% dimethyl sulfoxide (DMSO)), or cMYC inhibitor 10058-F4 at indicated concentrations and the cells were harvested and gradually fixed with 75% EtOH on ice. Cells were stained with propidium iodide (PI), and analyzed using a fluorescence-activated cell sorter (FACS) flow cytometer (Becton Dickinson, San Jose, CA), and the data analysis was performed by CellQuest software. All experiments were performed in triplicates and the graphs shown in the figures are representative of them.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as described previously [28] with minor changes. Cells were cross-linked with paraformaldehyde and nuclei were isolated from cells which were re-suspended in SDS-lysis buffer followed by sonication and centrifugation. The supernatant were diluted 1:10 with ChIP dilution buffer. For the immuno-clearing and pull down of the immuno-complexes, protein A magnetic beads (Upstate cell signaling solutions, Temecula CA, USA) were linked to rabbit IgG raised against mouse IgM. This modification was essential to ensure effective pull-down by the anti-bodies against serine-2 phospho (Covance, Cat # MMS 129R; H5) and serine-5 phospho (Covance, Cat # MMS 134R; H14) RNA polymerase II. The beads bound to immuno-complexes were thereafter washed and precipitates were extracted twice using freshly made 1% SDS and 0.1M NaHCO3 followed by de-crosslinking. The DNA fragments were purified using Qiaquick PCR purification kit (Qiagen, Valencia, CA, USA). RT-PCR was performed using 2μL isolated DNA, using primers specific for cMYC proximal promoter. The primers used (forward: GAGCAGCAGAGAAAGGGAGA; reverse: CAGCCGAGCACTCTAGCTCT) recognizes a region ~150bp upstream of transcription start site (TSS) of cMYC gene. The data is presented as percent input of starting chromatin input after subtracting the percent input pull down of the negative control (normal mouse IgM).
Western blotting
Whole cell protein lysates were isolated using RIPA buffer containing protease inhibitors (Roche Diagonistics, Mannheim, Germany) and phosphatase inhibitors I and II (EMD Chemicals Inc. San Diego, CA). 15-20μg of total protein was separated on the gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat dry milk in tris-buffered saline and probed with primary and secondary antibodies. Specific bands were visualized using west-pico chemi-luminescence (Thermo-Fisher, Rockford, IL, USA). The antibodies used: cMYC (# 5605), CDK9 (#2316), phospho-CDK9 (#2549), from Cell signaling Technologies (Danvers, MA); CTDP1 (# A301-172 A) Bethyl laboratories (Montgomery, TX); beta-actin (#A5441; Sigma, St. Louis, MO). The bands were scanned and quantified using imageJ software (National Institutes of Health, Bethesda, MD).
Relapse free survival (RFS) analysis
Kaplan-meier plots for RFS analysis were generated using the on-line tool “kmplot.com” which has the annotated data set from various breast cancer studies and allows studying single gene association with RFS outcome of the patients using user defined parameters. To evaluate the effect of cMYC overexpression on RFS of endocrine-therapy versus chemotherapy treated breast cancer patients we compared the top 25% patients expressing highest cMYC levels with the rest of the patient population. Two different plots were generated, one where the patients were treated with endocrine therapy (excludes chemotherapy) and the other with patients treated with chemotherapy (excludes endocrine-therapy). All other parameters were unchanged.
Statistics
Statistical significance of our data was assessed using the Student’s “t”-test wherever relevant. A p-value of < 0.05 was considered as statistically significant.
Results
Levels of cMYC and estrogen-independent growth of ERα+ endocrine resistant breast cancer cells
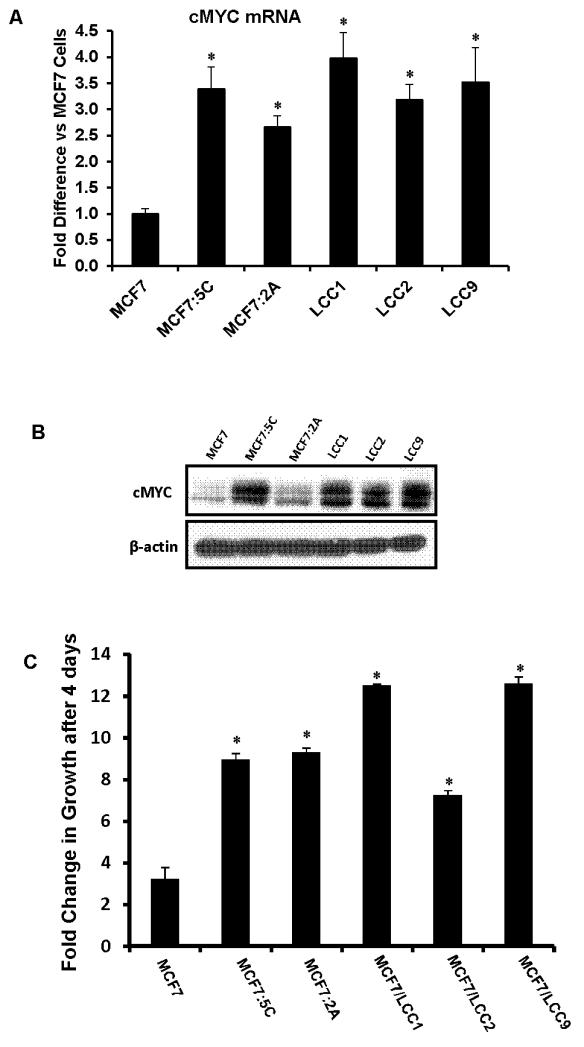
We found that all the endocrine-therapy resistant breast cancer cells used in this study, namely, MCF7:5C, MCF7:2A, MCF7/LCC1, MCF7/LCC2 and MCF7/LCC9 cells overexpress cMYCmRNA (Figure 1A) and protein (Figure 1B) as compared to parental MCF7 cells. All the resistant cells showed ~3-4 fold higher growth as compared to the parental MCF7 cells (Figure 1C) over a 4 day period. Cell cycle analysis of MCF7:5C cells revealed more than 2 fold higher “S” phase cells than in MCF7 cells (Supplementary figure S1B) and 5 fold higher proliferation over a six day period (Supplementary figure S1A).
Figure 1. Levels of cMYC and estrogen independent growth of endocrine therapy resistant breast cancer cells.
(A) cMYC mRNA levels were measured in different MCF7 derivative endocrine therapy resistant cells using RT-PCR. Data is represented as fold difference in cMYC mRNA versus MCF7 cells. (B) Western blot of cMYC protein in MCF7 and. Beta actin was used as a loading control. (C) Estrogen independent growth of MCF7 and other endocrine therapy resistant breast cancer cells over a 4 day period. Un-treated cells were grown and total DNA was measured on day 4 after seeding. The data is represented as fold change in growth versus day ‘0’. (* p<.05 versus MCF7 cells)
To determine if the high levels of cMYC mRNA was due to the elevated transcriptional activity or stability of the transcripts we performed a pulse chase assay and found that the cMYC mRNA had a similar rate of degradation in MCF7 and MCF7:5C cells (Supplementary Figure 3).
Inhibition or depletion of cMYC blocks estrogen-independent proliferation of ERα+ endocrine resistant cells cells
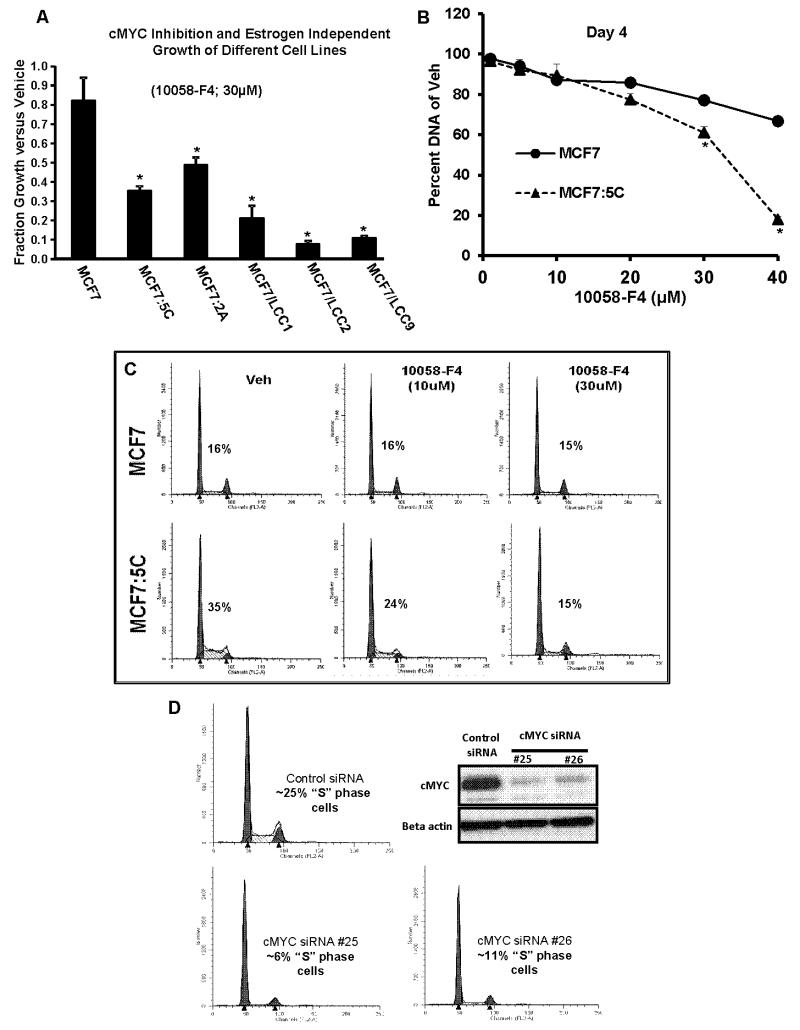
We determined the functional role of cMYC over-expression in estrogen-independent growth of the endocrine-therapy resistant breast cancer cells by blocking the cMYC action using a pharmacological inhibitor 10058-F4 which has been shown to specifically inhibit actions of cMYC by blocking its interaction with MAX [29] and stabilizing the MYC monomer [30]. cMYC inhibition with 30μM of 10058-F4 selectively inhibited 50% to 80% of the estrogen-independent growth of all the resistant cells (Figure 2A) whereas only 18% growth inhibition was observed in MCF7 cells. Further experiments with MCF7:5C cells showed that 10058-F4 was selectively able to inhibit its growth in a dose-dependent manner as compared to MCF7 cells over a four day period (Figure 2B). Cell cycle analysis confirmed that the decrease in proliferation resulted from a 57% reduction in the ‘S’ phase cells of the MCF7:5C cells (Figure 2C). In comparison, there was only 6% decrease in the ‘S’ phase cells of the parental MCF7 cells. We also used the targeted approach to confirm the role of cMYC in MCF7:5C cells, by depleting cMYC levels using short interfering RNA (siRNA). Two different siRNA against cMYC depleted the levels of its protein in MCF7:5C cells which led to 50-75% reduction in the number of ‘S’ phase cells (Figure 2D) with a concurrent inhibition of cell growth over a period of four days (Supplementary Figure S2A). Reduced phosphorylation of retinoblastoma protein (Supplementary Figure S2B) was also evident in the cells depleted of cMYC protein.
Figure 2. Inhibition or depletion of cMYC blocks estrogen independent growth of endocrine therapy resistant breast cancer cells.
(A) Total DNA was measured from the MCF7 and the resistant breast cancer cells after four days of treatment with 30μM, cMYC inhibitor (10058-F4). (* p<.05 versus MCF7 cells) (B) Total DNA was measured from the MCF7 and MCF7:5C cells after four days of treatment with cMYC inhibitor (10058-F4) with indicated concentration. (* p<.05 versus MCF7 cells) (C) “S” phase cells were assessed using cell cycle analysis of MCF7 and MCF7:5C cells treated with indicated concentration of cMYC inhibitor for 24 hrs. The numbers on each graph represents the percentage of “S” phase cells. (D) Assessment of “S” phase cells using cell cycle analysis 48 hours after siRNA mediated depletion of cMYC using two different siRNA (#25 and #26). The inset shows the western blot of cMYC protein levels after depletion of cMYC.
cMYC gene expression correlates with RFS in endocrine therapy but not chemotherapy treated patients
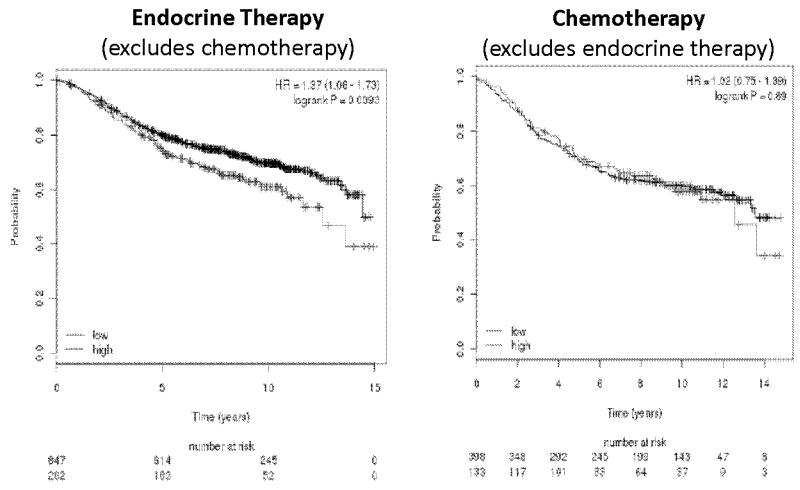
The Kaplan-meier plots were generated for cMYC gene association with RFS of early breast cancer patients who received endocrine-therapy or chemotherapy only as an adjuvant treatment. We used the on-line tool (www.kmplot.com) which has a combined data set from various annotated breast cancer studies and can be used to study the association of a single gene with patients outcome using various user defined parameters [31]. The top 25% percent highest cMYC expressing patients (top quartile) were compared with the rest of the 75%. Kaplan-Meier plots (Figure 3) reveal that high levels of cMYC expression is associated with poor RFS (P value; 0.0093) in 1129 patients treated with endocrine therapy only (Tamoxifen or AIs) whereas this association was not observed in the 531 patients (P value; 0.89) treated with chemotherapy only.
Figure 3. cMYC gene expression correlates with relapse free survival (RFS) in endocrine therapy but not chemotherapy treated patients.
The Kaplan-Meier plots show the association of cMYC gene expression and RFS in endocrine therapy or chemotherapy treated ERα+ breast cancer patients. The top 25% percent highest expressing cMYC patients (top quartile; in red) were compared with the rest of the 75% patient population (in black).
Recruitment of phospho-serine-2 and phospho-serine-5 RNA polymerase II at the cMYC promoter in MCF7:5C and MCF7 Cells
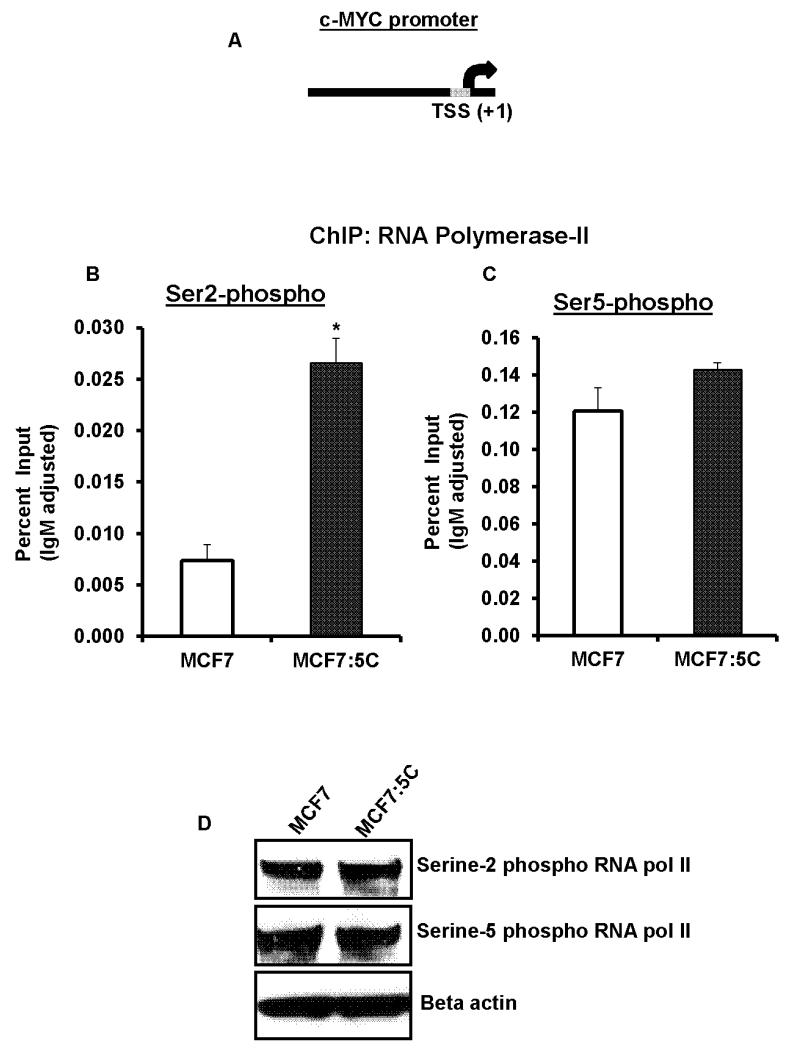
To further determine the mechanism of steady-state transcriptional over-expression of the cMYC mRNA in MCF7:5C cells we probed the proximal promoter of the cMYC gene (Figure 4A) in terms of recruitment of phosphorylated serine-5 and phosphorylated serine-2 RNA polymerase II, which is responsible for the initiation and the elongation of the transcription of RNA, respectively. ChIP assay using phospho-specific RNA polymerase II antibodies revealed that in MCF7:5C cells the recruitment of serine-2 phosphorylated RNA polymerase II was more than 3 fold higher than parental MCF7 cells (Figure 4B). However, no difference was observed in the recruitment of serine-5 phosphorylated RNA polymerase II at the cMYC promoter in MCF7:5C and MCF7 cells (Figure 4C). We further confirmed that the total levels of phosphorylated serine-2 or serine-5 RNA polymerase was not different in MCF7:5C cells as compared to MCF7 cells (Figure 4D).
Figure 4. Recruitment of serine-5 and serine-2 -phosphorylated RNA polymerase II at the cMYC promoter.
(A) Schematic presentation of cMYC promoter showing the transcription start site (TSS). The grey box represents the region (~150bp upstream of TSS) probed using real-time PCR following ChIP assay. (B) Recruitment of serine-2 phosphorylated RNA polymerase II and (C) serine-5 phosphorylated RNA polymerase II was assessed by ChIP assay followed by real-time PCR in MCF7: and MCF7:5C cells. Values are represented as percent input of the starting chromatin, adjusted for control IgM recruitment for each sample. (* p<.05 versus MCF7 cells) (D) Total protein levels of serine-2 and serine-5 phosphorylated RNA polymerase II in MCF7 and MCF7:5C cells.
Levels of cyclin dependent kinase 9 (CDK9) and its role in estrogen-independent growth of endocrine-therapy resistant cells
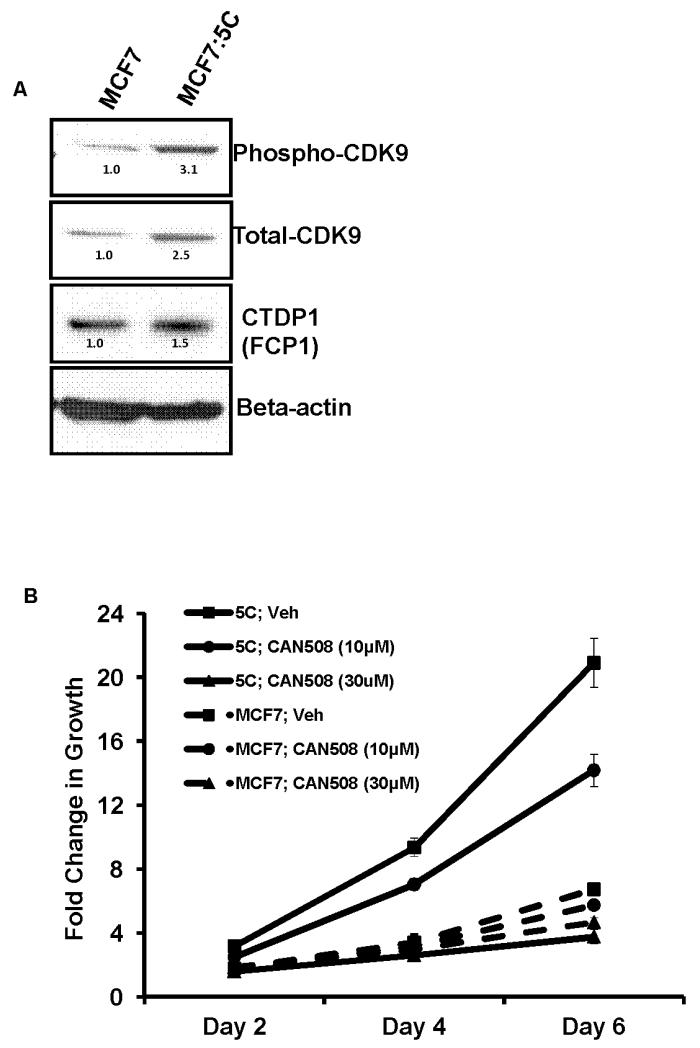
CDK9 is a major kinase which is responsible for the phosphorylation of serine-2 RNA polymerase II [32,33] and the elongation of RNA transcripts [34]. We therefore examined the total CDK9 levels in the endocrine-therapy resistant cells and observed an over-expression in all the cells as compared to the MCF7 cells (Supplementary Figure 4SA). In MCF7:5C cells, the total as well as the phosphorylated CDK9 levels were elevated by 2.5 and 3.1 fold respectively (Figure 5A). We also observed a slight increase in the levels of CTDP/ FCP1 protein in MCF7:5C cells, which is known to dephosphorylate CDK9 [32] (Figure 5A). Interestingly, FCP1 has also been reported to stimulate transcription elongation [35]. Next, we used a specific, potent, competitive inhibitor of CDK9, known as CAN 508 [36] to study the role of CDK9 in estrogen-independent growth of MCF7:5C cells and compared it with the parental MCF7 cells. A dose dependent effect was observed in MCF7:5C cells where 30μM of CAN 508 compound completely inhibited its growth over a six day period (Figure 5B). Furthermore, 30 μM of CAN 508 drastically blocked the growth of all endocrine-therapy resistant breast cancer cells used in this study (supplementary figure 4SB) whereas it had minimal growth inhibitory effect on the parental MCF7 cells.
Figure 5. Total CDK9 levels and effect of its inhibition on estrogen-independent growth.
(A) Protein levels of phospho and total CDK9 and CTDP1 was assessed using western blotting in MCF7 and MCF7:5C cells. The numbers above each band correspond to the fold change in protein levels versus MCF7 cells adjusted for beta actin levels for each sample. (B) Total DNA was measured to assess the growth of MCF7 and MCF7:5C cells after 2, 4 and 6 days of treatment with indicated doses of the CDK9 inhibitor, CAN508.
CDK9 inhibition blocks transcription of cMYC RNA and levels of cMYC protein in MCF7:5C cells
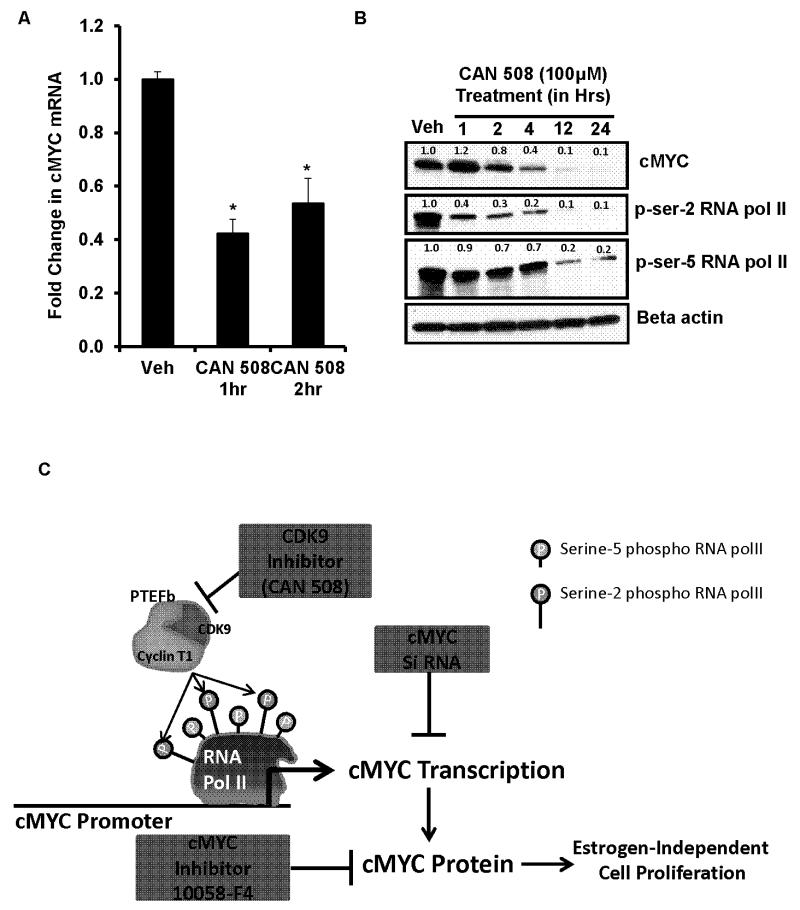
Inhibition of CDK9 in MCF7:5C cells by using CAN 508, resulted in approximately 60% decrease in cMYC mRNA within one hour of treatment (Figure 6A). This was followed by time dependent decline in cMYC protein levels (Figure 6B). Concomitant inhibition of serine-2 phosphorylated RNA polymerase II CTD was also observed within an hour of treatment (Figure 6B) indicating its role in cMYC transcription. As evident, serine-5 phosphorylation of RNA polymerase II CTD was not much altered within 4 hours of CDK9 inhibition. Although later time points showed marked reduction in serine-5 phosphorylation, along with serine-2 phosphorylation which was most likely due to secondary effects of CDK9 inhibition. Inhibition of CDK9 also completely blocked the phosphorylation of retinoblastoma (Rb) protein within twelve hours of treatment (Supplementary Figure S5) in the MCF7:5C cells.
Figure 6. Reduction of cMYC mRNA and protein by CDK9 inhibition and the proposed model of cMYC transcriptional regulation in MCF7:5C cells.
(A) Levels of cMYC mRNA was measured by quantitative RT-PCR in MCF7:5C cells after one and two hrs of CDK9 inhibition by 100μM of CAN508. (* p<.05 versus vehicle (Veh) treatment). (B) Protein levels of cMYC, phospho-serine-2 and serine-5 RNA polymerase II after inhibition of CDK9 by 100μM of CAN508 for indicated time points. The numbers above each band correspond to the fold change in protein levels versus vehicle (Veh) treatment adjusted for beta actin levels for each sample. (C) The cartoon depicts our findings on the CDK9 mediated cMYC transcriptional regulation and its role in estrogen-independent growth of the MCF7:5C cells.
Discussion
Accumulative evidence indicates that cMYC overexpression and subsequent genes up-regulated in breast cancers are associated with resistance to AIs [8] and antiestrogens [9,7]. This study establishes the role and mechanism of cMYC regulation in the estrogen-independent growth of ERα+, endocrine-resistant breast cancer cells. All the resistant cell models used in this study are MCF7-derived cell lines. Importantly, MCF7 cells retain the ERα protein after acquiring endocrine therapy resistance which mimics the clinical scenario as 80% of the endocrine-therapy resistant breast cancer patients are ERα positive [37]. Interestingly, despite the limited availability of cell lines, significant translational advances have occurred [24]. Based on our results, we decipher a novel mechanism of transcriptional over-expression of cMYC in resistant breast cancer cells (Figure 6C) which involves CDK9 mediated hyper-phosphorylation of serine-2 RNA polymerase-II CTD at the promoter of cMYC gene. This, in turn, is responsible for the transcriptional elongation and overexpression of cMYC. Our analysis of the annotated breast cancer patient’s database (Figure 3) suggested that over-expression of cMYC correlates with the failure of endocrine therapy (but not chemotherapy) and eventual relapse of the disease.
Ectopic overexpression of cMYC in MCF7 cells is reported to be sufficient to confer resistance to endocrine therapy [7,10]. We observed elevated cMYC levels in the ERα+, endocrine therapy -resistant breast cancer cells (Figure 1A and B) which proliferated in the absence of estrogen. A previous study has also reported high cMYC levels in long-term estrogen deprived cells [38]. Inhibition of cMYC or its depletion blocked the proliferation of the cells (Figure 2A and Supplementary Figure S2A) demonstrating the critical role of cMYC overexpression in estrogen-independent growth of these resistant breast cancer cells. The reduction in ‘S’ phase cells (Figure 2C and D) was achieved by de- phosphorylation of tumor suppressor retinoblastoma (Rb) protein (Supplementary Figure S2B) which is known to arrest the cells in G1 phase of the cell-cycle [39].
Further, using a pulse chase assay, we ascertained that the high basal level of cMYC mRNA in the MCF7:5C cells was due to the high rate of transcription and not enhanced stability of the transcripts (Figure S3). Since therapeutic targeting of cMYC is not feasible, we studied the upstream factors responsible for cMYC transcriptional over-expression from its natural proximal promoter in the MCF7:5C cells. Transcription of cMYC gene is regulated at the elongation step by promoter-proximal pausing of RNA polymerase II in eukaryotes [40,41]. Importantly, cMYC is a well-defined estrogen-regulated gene [42] and the estrogen-induced growth of the hormone responsive breast cancer cells is contingent upon the expression of cMYC gene in these cells as majority of growth related genes which are estrogen regulated are cMYC target [43]. In MCF7 cells, studies have demonstrated [44] that the proximal promoter of the cMYC gene is pre-loaded with RNA polymerase II which is phosphorylated at serine 5 of its CTD, in the absence of estrogen. However, phosphorylation of serine-2 of CTD of RNA polymerase II is needed to overcome the elongation block of the transcripts which is achieved after estrogen stimulation. Our findings are consistent. In MCF7 cells we observed high levels of serine-5 phosphorylation, and low serine-2 phosphorylation of RNA polymerase II CTD at the cMYC promoter under basal conditions (Figure 4B and C). In contrast, under identical condition, the phosphorylation of serine-2 of CTD of RNA polymerase II is markedly elevated in MCF7:5C cells (Figure 4A and B) which drives the higher transcriptional elongation of cMYC.
The kinase complex responsible for the phosphorylation of serine-2 of RNA polymerase II CTD and inducing transcriptional elongation is known as positive transcriptional elongation factor-b (PTEF-b) which is composed of CDK9 and cyclin T1 [45-47]. Our observation of higher levels of CDK9 in MCF7:5C cells (Figure 5A) and in other resistant breast cancer cells (Supplementary figure S4A) strongly suggested that it is responsible for elevated serine-2 phosphorylation of RNA polymerase II CTD at the cMYC promoter of MCF7:5C cells. Indeed, using a pharmacological agent, CAN 508, which specifically inhibits CDK9 activity [48,36] the growth of MCF7:5C cells (Figure 5B) as well as other endocrine therapy resistant MCF7 derived ERα+ breast cancer cells (supplementary figure S4B) were selectively inhibited. This demonstrated that the estrogen-independent growth of the endocrine therapy resistant breast cancer cells was driven by CDK9. We further confirmed that inhibition of CDK9 led to the reduction of cMYC mRNA levels within one hour of treatment in MCF7:5C cells followed by the protein level (Figures 6A and B). The concurrent decrease in global serine-2 (but not serine-5) phosphorylation of RNA polymerase II CTD (Figures 6B) -suggested that CDK9 was responsible for cMYC transcriptional over-expression in the resistant cells. In addition, we confirmed that CDK9 inhibition reduced the level of phospho-Rb protein (supplementary figure S5) in a similar manner as cMYC depletion in the MCF7:5C cells. This supports our hypothesis that the growth suppressive effect of CDK9 inhibition reduces cMYC levels in the endocrine-therapy resistant breast cancer cells. Furthermore, we found that CDK9 or cMYC inhibition was not deleterious to the immortalized human epithelial cells (MCF10A) (supplementary figure S6) indicating that CDK9 can be a potential novel therapeutic target.
Since we did not detect any difference in the global level of serine-2 phosphorylated RNA polymerase II CTD between MCF7:5C and its parental MCF7 cells (Figure 4D); further studies are required to establish the chromatin modifications at the cMYC promoter which ensue in the process of acquiring resistance. These changes are crucial as it allows the RNA polymerase II CTD to be hyper-permissive for serine-2 phosphorylation, thus ensuring elongation of the cMYC transcripts in the MCF7:5C cells. Intriguingly, recent reports have indicated that in hematological malignancies bromo-domain containing protein 4 (BRD4), which has been known to recruit CDK9 and regulate serine-2 phosphorylation of RNA polymerase II [49,50], is involved in cMYC overexpression [18,51].
In this study, we have delineated the transcriptional mechanism of cMYC over-expression, endocrine-therapy resistant, ERα+ breast cancer cells, and propose that recruitment of hyper-phosphorylated serine-2 RNA polymerase II at the cMYC promoter which is mediated by CDK9, is responsible for the estrogen independent proliferation of these cells. We therefore suggest that there will be a potential clinical benefit by using CDK9 inhibitors in the treatment of endocrine therapy resistant breast cancers.
Supplementary Material
Acknowledgements
This work (VCJ) was supported by the Department of Defense Breast Program under Award number W81XWH-06-1-0590 Center of Excellence; subcontract under the SU2C (AACR) Grant number SU2C-AACR-DT0409; the ‘Susan G Komen For The Cure Foundation’ under Award number SAC100009 and the Lombardi Comprehensive Cancer Center Support Grant (CCSG) Core Grant NIH P30 CA051008. The views and opinions of the author(s) do not reflect those of the US Army or the Department of Defense.
Abbreviations
- AI
aromatase inhibitor
- ChIP
chromatin-immuno precipitation assay
- CTD
carboxy-terminal domain
- E2
17β-estradiol
- ERα
estrogen receptor alpha
- RT-PCR
real time polymerase chain reaction
Footnotes
Disclosures: None.
REFERENCES
- 1.Sengupta S, Jordan VC. Selective estrogen modulators as an anticancer tool: mechanisms of efficiency and resistance. Adv Exp Med Biol. 2008;630:206–219. doi: 10.1007/978-0-387-78818-0_13. [DOI] [PubMed] [Google Scholar]
- 2.Obiorah I, Jordan VC. Progress in endocrine approaches to the treatment and prevention of breast cancer. Maturitas. 2011;70(4):315–321. doi: 10.1016/j.maturitas.2011.09.006. doi:10.1016/j.maturitas.2011.09.006 S0378-5122(11)00325-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol. 2007;25(36):5815–5824. doi: 10.1200/JCO.2007.11.3886. doi:JCO.2007.11.3886 [pii] 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 4.Chia S, Gradishar W, Mauriac L, Bines J, Amant F, Federico M, Fein L, Romieu G, Buzdar A, Robertson JF, Brufsky A, Possinger K, Rennie P, Sapunar F, Lowe E, Piccart M. Double-blind, randomized placebo controlled trial of fulvestrant compared with exemestane after prior nonsteroidal aromatase inhibitor therapy in postmenopausal women with hormone receptor-positive, advanced breast cancer: results from EFECT. J Clin Oncol. 2008;26(10):1664–1670. doi: 10.1200/JCO.2007.13.5822. doi:JCO.2007.13.5822 [pii] 10.1200/JCO.2007.13.5822. [DOI] [PubMed] [Google Scholar]
- 5.Nabholtz JM, Buzdar A, Pollak M, Harwin W, Burton G, Mangalik A, Steinberg M, Webster A, von Euler M, Arimidex Study Group Anastrozole is superior to tamoxifen as first-line therapy for advanced breast cancer in postmenopausal women: results of a North American multicenter randomized trial. J Clin Oncol. 2000;18(22):3758–3767. doi: 10.1200/JCO.2000.18.22.3758. [DOI] [PubMed] [Google Scholar]
- 6.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. doi:10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNeil CM, Sergio CM, Anderson LR, Inman CK, Eggleton SA, Murphy NC, Millar EK, Crea P, Kench JG, Alles MC, Gardiner-Garden M, Ormandy CJ, Butt AJ, Henshall SM, Musgrove EA, Sutherland RL. c-Myc overexpression and endocrine resistance in breast cancer. J Steroid Biochem Mol Biol. 2006;102(1-5):147–155. doi: 10.1016/j.jsbmb.2006.09.028. doi:S0960-0760(06)00265-2 [pii] 10.1016/j.jsbmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. doi:10.1038/nature11143 nature11143 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller TW, Balko JM, Ghazoui Z, Dunbier A, Anderson H, Dowsett M, Gonzalez-Angulo AM, Mills GB, Miller WR, Wu H, Shyr Y, Arteaga CL. A gene expression signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clin Cancer Res. 2011;17(7):2024–2034. doi: 10.1158/1078-0432.CCR-10-2567. doi:1078-0432.CCR-10-2567 [pii] 10.1158/1078-0432.CCR-10-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venditti M, Iwasiow B, Orr FW, Shiu RP. C-myc gene expression alone is sufficient to confer resistance to antiestrogen in human breast cancer cells. Int J Cancer. 2002;99(1):35–42. doi: 10.1002/ijc.10269. doi:10.1002/ijc.10269 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, Liu XS, Shioda T, Toner M, Loda M, Brown M, Brugge JS, Ramaswamy S. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107(8):3698–3703. doi: 10.1073/pnas.0914203107. doi:0914203107 [pii] 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, Prindiville SA, Calzone KA, Soballe PW, Hu Y, Chowdhury SA, Schwartz R, Schaffer AA, Ried T. Single-Cell Genetic Analysis of Ductal Carcinoma in Situ and Invasive Breast Cancer Reveals Enormous Tumor Heterogeneity yet Conserved Genomic Imbalances and Gain of MYC during Progression. Am J Pathol. 2012;181(5):1807–1822. doi: 10.1016/j.ajpath.2012.07.012. doi:10.1016/j.ajpath.2012.07.012 S0002-9440(12)00577-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83(12):1688–1695. doi: 10.1054/bjoc.2000.1522. doi:10.1054/bjoc.2000.1522 S0007092000915222 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JE., Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2(10):740–749. doi: 10.1038/nrc906. doi:10.1038/nrc906 nrc906 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. doi:10.1038/nature07260 nature07260 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414(6865):768–773. doi: 10.1038/414768a. doi:10.1038/414768a 414768a [pii] [DOI] [PubMed] [Google Scholar]
- 17.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, Cleary MA, Park JR, Grandori C. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109(24):9545–9550. doi: 10.1073/pnas.1121119109. doi:10.1073/pnas.1121119109 1121119109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. doi:10.1016/j.cell.2011.08.017 S0092-8674(11)00943-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. An estrogen receptor positive MCF-7 clone that is resistant to antiestrogens and estradiol. Mol Cell Endocrinol. 1992;90(1):77–86. doi: 10.1016/0303-7207(92)90104-e. doi:0303-7207(92)90104-E [pii] [DOI] [PubMed] [Google Scholar]
- 20.Pink JJ, Jiang SY, Fritsch M, Jordan VC. An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer Res. 1995;55(12):2583–2590. [PubMed] [Google Scholar]
- 21.Brunner N, Boulay V, Fojo A, Freter CE, Lippman ME, Clarke R. Acquisition of hormone-independent growth in MCF-7 cells is accompanied by increased expression of estrogen-regulated genes but without detectable DNA amplifications. Cancer Res. 1993;53(2):283–290. [PubMed] [Google Scholar]
- 22.Brunner N, Frandsen TL, Holst-Hansen C, Bei M, Thompson EW, Wakeling AE, Lippman ME, Clarke R. MCF7/LCC2: a 4-hydroxytamoxifen resistant human breast cancer variant that retains sensitivity to the steroidal antiestrogen ICI 182,780. Cancer Res. 1993;53(14):3229–3232. [PubMed] [Google Scholar]
- 23.Brunner N, Boysen B, Jirus S, Skaar TC, Holst-Hansen C, Lippman J, Frandsen T, Spang-Thomsen M, Fuqua SA, Clarke R. MCF7/LCC9: an antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen. Cancer Res. 1997;57(16):3486–3493. [PubMed] [Google Scholar]
- 24.Sweeney EE, McDaniel RE, Maximov PY, Fan P, Jordan VC. Models and Mechanisms of Acquired Antihormone Resistance in Breast Cancer: Significant Clinical Progress Despite Limitations. Horm Mol Biol Clin Investig. 2012;9(2):143–163. doi: 10.1515/hmbci-2011-0004. doi:10.1515/hmbci-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ariazi EA, Cunliffe HE, Lewis-Wambi JS, Slifker MJ, Willis AL, Ramos P, Tapia C, Kim HR, Yerrum S, Sharma CG, Nicolas E, Balagurunathan Y, Ross EA, Jordan VC. Estrogen induces apoptosis in estrogen deprivation-resistant breast cancer through stress responses as identified by global gene expression across time. Proc Natl Acad Sci U S A. 2011;108(47):18879–18886. doi: 10.1073/pnas.1115188108. doi:10.1073/pnas.1115188108 1115188108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis JS, Meeke K, Osipo C, Ross EA, Kidawi N, Li T, Bell E, Chandel NS, Jordan VC. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst. 2005;97(23):1746–1759. doi: 10.1093/jnci/dji400. doi:97/23/1746 [pii] 10.1093/jnci/dji400. [DOI] [PubMed] [Google Scholar]
- 27.Ellis MJ, Gao F, Dehdashti F, Jeffe DB, Marcom PK, Carey LA, Dickler MN, Silverman P, Fleming GF, Kommareddy A, Jamalabadi-Majidi S, Crowder R, Siegel BA. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: a phase 2 randomized study. JAMA. 2009;302(7):774–780. doi: 10.1001/jama.2009.1204. doi:10.1001/jama.2009.1204 302/7/774 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S, Obiorah I, Maximov P, Curpan R, Jordan VC. Molecular mechanism of action of bisphenol and bisphenol A mediated by oestrogen receptor alpha in growth and apoptosis of breast cancer cells. Br J Pharmacol. 2013;169(1):167–178. doi: 10.1111/bph.12122. doi:10.1111/bph.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22(40):6151–6159. doi: 10.1038/sj.onc.1206641. doi:10.1038/sj.onc.1206641 1206641 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008;15(11):1149–1155. doi: 10.1016/j.chembiol.2008.09.011. doi:10.1016/j.chembiol.2008.09.011 S1074-5521(08)00370-0 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. doi:10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 32.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7(8):557–567. doi: 10.1038/nrm1981. doi:nrm1981 [pii] 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 33.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16(16):2135–2146. doi: 10.1101/gad.999002. doi:10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19(11):1108–1115. doi: 10.1038/nsmb.2399. doi:10.1038/nsmb.2399 nsmb.2399 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18(20):2437–2468. doi: 10.1101/gad.1235904. doi:18/20/2437 [pii] 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 36.Krystof V, Cankar P, Frysova I, Slouka J, Kontopidis G, Dzubak P, Hajduch M, Srovnal J, de Azevedo WF, Jr., Orsag M, Paprskarova M, Rolcik J, Latr A, Fischer PM, Strnad M. 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J Med Chem. 2006;49(22):6500–6509. doi: 10.1021/jm0605740. doi:10.1021/jm0605740. [DOI] [PubMed] [Google Scholar]
- 37.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. doi:10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeng MH, Shupnik MA, Bender TP, Westin EH, Bandyopadhyay D, Kumar R, Masamura S, Santen RJ. Estrogen receptor expression and function in long-term estrogen-deprived human breast cancer cells. Endocrinology. 1998;139(10):4164–4174. doi: 10.1210/endo.139.10.6229. [DOI] [PubMed] [Google Scholar]
- 39.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. doi:0092-8674(95)90385-2 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321(6071):702–706. doi: 10.1038/321702a0. doi:10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 41.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6(11):2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 42.Wang C, Mayer JA, Mazumdar A, Fertuck K, Kim H, Brown M, Brown PH. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol. 2011;25(9):1527–1538. doi: 10.1210/me.2011-1037. doi:10.1210/me.2011-1037 me.2011-1037 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musgrove EA, Sergio CM, Loi S, Inman CK, Anderson LR, Alles MC, Pinese M, Caldon CE, Schutte J, Gardiner-Garden M, Ormandy CJ, McArthur G, Butt AJ, Sutherland RL. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One. 2008;3(8):e2987. doi: 10.1371/journal.pone.0002987. doi:10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29(5):1123–1133. doi: 10.1128/MCB.00841-08. doi:10.1128/MCB.00841-08 MCB.00841-08 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809(1):34–45. doi: 10.1016/j.bbagrm.2010.11.001. doi:10.1016/j.bbagrm.2010.11.001 S1874-9399(10)00139-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. doi:10.1016/j.molcel.2009.10.019 S1097-2765(09)00784-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. doi:S1097-2765(06)00429-1 [pii] 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Baumli S, Hole AJ, Noble ME, Endicott JA. The CDK9 C-helix exhibits conformational plasticity that may explain the selectivity of CAN508. ACS Chem Biol. 2012;7(5):811–816. doi: 10.1021/cb2004516. doi:10.1021/cb2004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, Yang H, Chin KC. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287(51):43137–43155. doi: 10.1074/jbc.M112.413047. doi:10.1074/jbc.M112.413047 M112.413047 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. doi:S1097-2765(05)01432-2 [pii] 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 51.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. doi:10.1038/nature10334 nature10334 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.