Abstract
The generation of induced tissue-specific stem cells has been hampered by the lack of well-established methods for the maintenance of pure tissue-specific stem cells like the ones we have for embryonic stem (ES) cell cultures. Using a cocktail of cytokines and small molecules, we demonstrate that primitive neural stem (NS) cells derived from mouse ES cells and rat embryos can be maintained. Furthermore, using the same set of cytokines and small molecules, we show that induced NS (iNS) cells can be generated from rat fibroblasts by forced expression of the transcriptional factors Oct4, Sox2 and c-Myc. The generation and long-term maintenance of iNS cells could have wide and momentous implications.
Keywords: Induced neural stem cells, Stem cell self-renewal, Fibroblasts, Rat, Induced pluripotent stem cells
Introduction
Neural stem (NS) cells can be expanded in culture while retaining the ability to differentiate into the three main cell types of the central nervous system: neurons, astrocytes and oligodendrocytes [1]. NS cells hold great promise for treating neurodegenerative diseases and spinal cord injury. If the full potential of NS cells in cell replacement therapies is to be realized, non-cerebrocerebellar sources of patient-specific NS cells must be found because the brain tissue is not readily accessible.
Induced pluripotent stem (iPS) cells [2] as well as neurons [3–6], cardiomyocytes [7–9], blood [10] and hepatocytes [11,12] have been generated from fibroblasts through reprogramming by defined transcription factors. These breakthroughs in cellular reprogramming have provided a new avenue for generating patient-specific NS cells from readily accessible tissues such as dermal fibroblasts. Successful generation of induced NS cells will require the identification of NS cell fate-inducing factors as well as application of carefully-contrived culture conditions that can capture the NS cell state and allow its long-term self-renewal. Several recent studies have demonstrated the feasibility of converting mouse fibroblasts directly into neural progenitors/NS cells by forced expression of different sets of transcription factors [13–16]. In the present study, we developed a culture condition that allows long-term expansion of Sox1-positive primitive NS cells. Using this culture condition, we efficiently generated iNS cells from rat fibroblasts by combined expression of the transcriptional factors Oct4, Sox2 and c-Myc.
Results
Maintenance of Sox1-GFP positive NS cells by leukemia inhibitory factor, CHIR99021 and Y27632
Sox1 is one of the earliest specific markers of neuroectoderm [17]. By taking advantage of the Sox1-GFP knock-in reporter mouse embryonic stem (ES) cell line we developed [18], we generated pure Sox1-GFP positive NS cells and tested different combinations of cytokines and small molecules for propagation of Sox1-positive NS cells. We found that concurrent treatment with leukemia inhibitory factor (LIF), CHIR99021 and Y27632 (“LIF/CHIR/Y” hereafter) allowed long-term expansion of Sox1-GFP positive NS cells, even from single cells (Figure 1A). CHIR99021 and Y27632 are small molecules that specifically inhibit glycogen synthase kinase-3 (GSK3) and Rho-associated kinase (ROCK), respectively [19,20]. Similar NS cell lines from rat fetal brains and human ES cells were also derived and maintained in the presence of LIF/CHIR/Y (Figure 1B–E). As expected, Sox1-positive NS cells failed to be maintained in the presence of fibroblast growth factor 2 (FGF2) and epidermal growth factor (EGF) (Figure 1F), the conventional culture condition for the propagation of neural progenitors [21,22].
Figure 1.
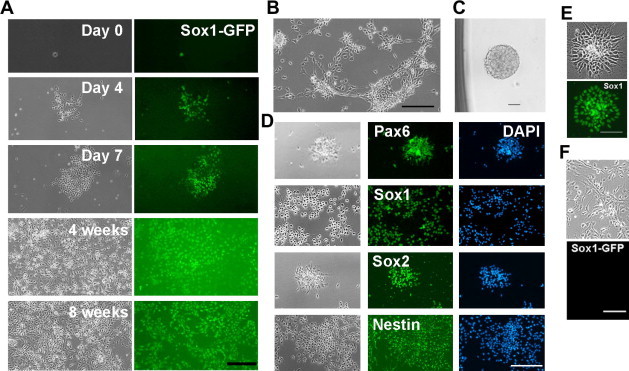
Expansion of Sox1-positive NS cells in the presence of LIF/CHIR/Y A. Clonal expansion of Sox1-GFP NS cells in the presence of LIF/CHIR/Y. Sox1-GFP positive NS cells derived from 46C ES cells were deposited into 0.1% gelatin-coated 96-well plates at 1 cell/well and cultured in N2B27 medium supplemented with LIF/CHIR/Y. Scale bar, 50 μm. B. Phase contrast image of NS cells derived from an E11.5 rat embryo and maintained in the presence of LIF/CHIR/Y at Passage 7. Scale bar, 50 μm. C. A neurosphere generated from a single primary rat NS cell in the presence of LIF/CHIR/Y. D. Immunostaining of primary rat NS cells maintained in the presence of LIF/CHIR/Y for 5 passages. Scale bar, 50 μm. E. Sox1 immunostaining of NS cells derived from H9 human ES cells and maintained in the presence of LIF/CHIR/Y. Scale bar, 50 μm. F. NS cells derived from 46C ES cells and maintained in the presence of FGF2/EGF for 2 passages. Scale bar, 50 μm.
iNS cells derived from rat fibroblasts
Next, we investigated whether iNS cells could be generated from fibroblasts in the presence of LIF/CHIR/Y. Initially, we transduced mouse and rat embryonic fibroblasts with retroviral vectors encoding the reprogramming factors Oct4, Sox2, Klf4 and c-Myc, and cultured the cells in the LIF/CHIR/Y condition. Three weeks after transduction, approximately 50 and 20 colonies emerged from 1 × 105 plated mouse and rat fibroblasts, respectively. Colonies formed from mouse fibroblasts showed the appearance of iPS cells (Figure S1). Surprisingly, most of the colonies formed from rat fibroblasts morphologically resembled the NS cells maintained in the presence of LIF/CHIR/Y (Figure 2A). We picked 11 NS cell-like colonies and found that all of them were expanded in the presence of LIF/CHIR/Y to establish stable cell lines (Figure 2B and C). These cells expressed NS cell markers (Figure 2D–H) and differentiated exclusively into different subtypes of neurons, astrocytes and oligodendrocytes upon the removal of LIF/CHIR/Y, even after 60 passages (Figure 3). Moreover, neurons derived from these rat iNS cells exhibited typical functional membrane properties of neurons (Figure S2). Similar iNS cells were also efficiently generated from rat tail-tip fibroblasts using both retroviral and PiggyBac transposon systems (Figure S3).
Figure 2.
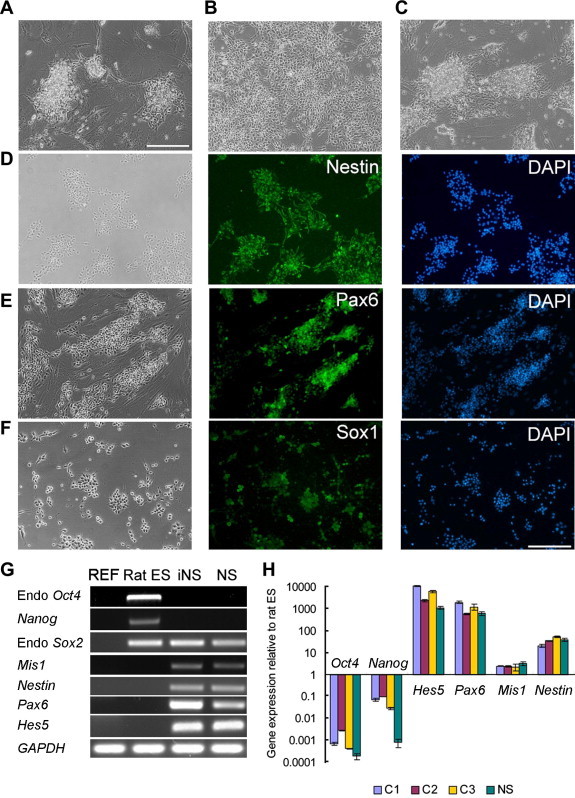
iNS cells derived from rat fibroblasts A. Morphology of rat iNS cell colonies 21 days after transfection of Oct4, Sox2, Klf4 and c-Myc into rat embryonic fibroblasts. Scale bar, 50 μm. B. Rat iNS cells cultured on feeders in the presence of LIF/CHIR/Y at Passage 21. C. Rat iNS cells cultured on 0.1% gelatin in the presence of LIF/CHIR/Y at Passage 15. D–F. Immunostaining of rat iNS cells maintained in the presence of LIF/CHIR/Y. Scale bar, 50 μm. G. RT-PCR analysis of gene expression in rat embryonic fibroblasts (REF), rat ES cells, rat iNS cells and primary rat NS cells derived from E11.5 rat fetal brain and maintained in the presence of LIF/CHIR/Y. GAPDH was used as a loading control. H. qRT-PCR analysis of gene expression. C1, C2 and C3 were three rat iNS cell clones. NS: primary NS cells derived from E11.5 rat fetal brain. Data are presented as mean ± standard deviation (SD) of three biological replicates.
Figure 3.
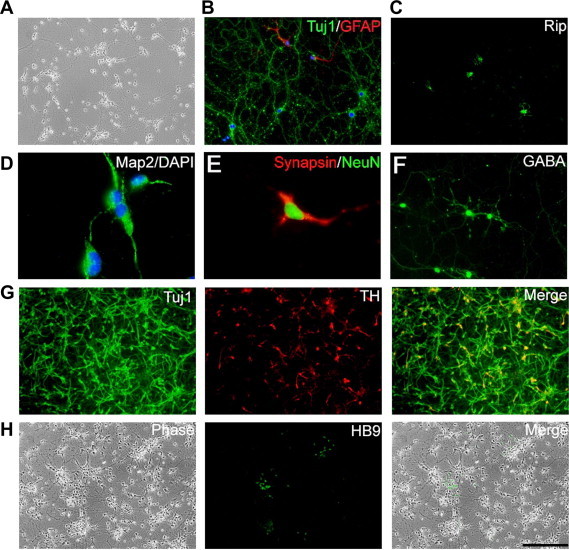
Neuronal and glial differentiation of rat iNS cells A. Phase contrast image of neurons spontaneously differentiated from rat iNS cells after the removal of LIF/CHIR/Y. B. Tuj1 and GFAP immunostaining of cells generated from rat iNS cells after exposure to EGF and FGF2 for 10 days followed by culturing in N2B27 medium plus 1% serum for another 7 days. C. Exposure to PDGF-AA and T3 (triiodothyronine) induced differentiation of rat iNS cells toward Rip-positive oligodendrocytes. D–H. Different subtypes of neurons derived from rat iNS cells. Scale bar, 50 μm.
Regional specification of rat iNS cells derived and cultured in the presence of LIF/CHIR/Y
Early stage NS cells possess the capability of differentiating toward region-specific neuronal fates in response to patterning cues but NS cells maintained in the presence of FGF2/EGF lose this capability [23]. Rat iNS cells generated and maintained in the presence of LIF/CHIR/Y expressed Pax6 and Sox1 (Figure 2E and F), indicating an early stage NS cell identity. To further confirm the developmental stage of these rat iNS cells, we examined their gene expression pattern by RT-PCR. As shown in Figure 4A and B, rat iNS cells and E11.5 rat fetal brain tissue expressed DACH1, PLZF, LIX1 and FAM70A, genes that are uniquely expressed in early stage NS cells, whereas the expression of S100B, PMP2 and AQP4, specific markers for FGF2/EGF NS cells [23], was not detected. In contrast with rat iNS cells maintained in the presence of LIF/CHIR/Y, NS cells maintained in the presence of FGF2/EGF expressed S100B, PMP2 and AQP4, but not DACH1, PLZF, LIX1 and FAM70A (Figure 4A).
Figure 4.
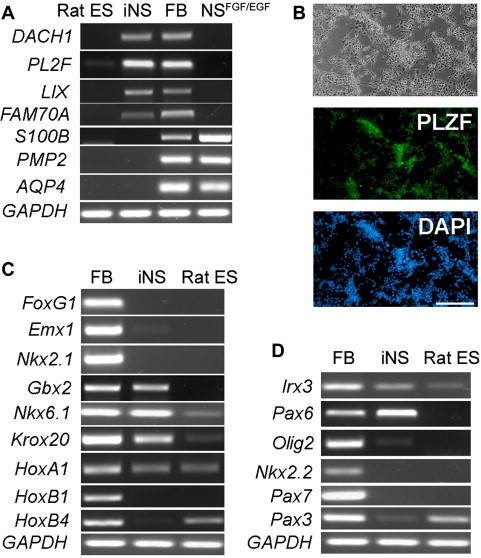
Regional identity of rat iNS cells A. RT–PCR analysis of the expression of genes unique to rosette NS cells (DACH1, PLZF, LIX1 and FAM70A) and genes specifically expressed in FGF2/EGF NS cells (S100B, PMP2 and AQP4). FB: E11.5 rat fetal brain. B. PLZF immunostaining of rat iNS cells. C. RT-PCR analysis of the expression of markers along the anterior–posterior axis. D. RT-PCR analysis of the expression of markers along the dorsal–ventral axis.
Next, we analyzed the expression of regional identity markers along the anterior–posterior and dorsal–ventral axes of the brain. The expression of telencephalic markers FoxG1, Emx1 and Nkx2.1 was not detected by RT-PCR in rat iNS cells; instead, rat iNS cells highly expressed anterior hindbrain markers Gbx2, Nkx6.1 and Krox20. The posterior hindbrain markers HoxB1 and HoxB4, however, were not expressed in rat iNS cells (Figure 4C). Along the dorsal–ventral axis, rat iNS cells expressed ventral hindbrain markers Irx3 and Pax6, but not the extreme ventral markers Olig2 and Nkx2.2 or the dorsal markers Pax7 and Pax3 (Figure 4D). Taken together, these results suggest that rat iNS cells generated and maintained in the presence of LIF/CHIR/Y represent early stage primitive NS cells and have an anterior-ventral hindbrain character.
Oct4, Sox2 and c-Myc are sufficient to reprogram rat fibroblasts into a NS cell fate
Loss of function in the tumor-suppressive p53 pathway has been shown to dramatically accelerate the reprogramming process [24–26]. Indeed, when fibroblasts derived from p53−/− rat embryos were subjected to reprogramming [27], iNS cell-like colonies emerged as early as 4 days after transduction (data not shown). To determine which of the four factors are required to generate iNS cells, we transduced rat fibroblasts with different combinations of the four factors. The results, as summarized in Figure S4, showed that Oct4/Sox2/c-Myc and Sox2/c-Myc were sufficient to generate iNS cells from wild-type and p53−/− rat fibroblasts, respectively. No iNS cell-like colonies emerged in any of the combinations without either Sox2 or c-Myc, suggesting that both Sox2 and c-Myc were required for the conversion of rat fibroblasts into iNS cells. Finally, we investigated whether the induction of iNS cells entails a passage through the iPS cell stage. We derived fibroblasts from Oct4-GFP transgenic rat embryos in which the GFP transgene was driven by the Oct4 promoter. GFP-positive cells were never observed during the reprogramming of fibroblasts to iNS cells (Figure S5), suggesting that iNS cells were most likely converted directly from fibroblasts without passing through an intermediate iPS state.
Discussion
Here we show that self-renewal of Sox1-positive primitive NS cells derived from mouse, rat and human can be efficiently maintained in serum-free N2B27 medium supplemented with LIF/CHIR/Y. Moreover, by forced expression of Oct4/Sox2/c-Myc, rat fibroblasts can be directly converted into self-renewing iNS cells under the LIF/CHIR/Y condition. These rat iNS cells have all the key characteristics of primary NS cells including expression of the NS cell markers and the potential to differentiate into astrocytes, oligodendrocytes, and mature neurons with functional membrane properties. Intriguingly, combined expression of the same factors failed to convert mouse fibroblasts to iNS cells. We reason that additional factor(s) or different combinations of factors are required for the generation of mouse iNS cells.
Rat iNS cells require LIF for self-renewal and predominately differentiate into neurons upon the removal of self-renewal factors, suggesting that they may be equivalent to in vivo primitive NS cells [28]. Interestingly, rat iNS cells maintained in the presence of LIF/CHIR/Y have an anterior-ventral hindbrain character, distinguishing them from mouse iNS cells maintained in the presence of FGF2/EGF [16] or rosette human NS cells [23]. Further studies will be needed to determine whether these rat iNS cells are capable of differentiation toward different region-specific neuronal fates in response to patterning cues.
Rat iNS cells could not be generated from rat fibroblasts in the absence of LIF and CHIR99021, further supporting the notion that culture environments could dictate the fate of reprogrammed cells [29]. We hypothesize that iNS cells can also be generated from human fibroblasts using an approach similar to that described here. Generation of patient-specific iNS cells will not only enable us to develop new tools for the diagnosis of neurological diseases, but also provide a pure and renewable source of patient-specific neural cells for use in cell replacement therapies and drug discovery.
Materials and methods
Primitive NS cells culture
Prospective head regions from E9.5 to E11.5 rat embryos at neural-fold stage were excised and treated with dissociation buffer containing 1 mM EDTA and 0.25% trypsin in phosphate-buffered saline (PBS) for 4 min. Neuroepithelium was detached from the underlying tissue and mechanically dissected into single cells. The cells were cultured in poly-l-lysine/laminin-coated dishes with N2B27 medium [30] containing LIF (1000 units/ml), CHIR99021 (3 μM) and Y27632 (10 μM). CHIR99021 and Y27632 were synthesized in the Division of Signal Transduction Therapy, University of Dundee, UK.
ES cell culture and differentiation
Rat ES cells were routinely maintained in the 2i condition as described [31–33]. 46C mouse ES cells were routinely cultured and converted to Sox1-GFP positive NS cells as described [18]. Transient selection with puromycin (0.5 μg/ml) and fluorescence activated cell sorting (FACS) were used to eliminate non-neural cells and obtain homogeneous population of Sox1-GFP positive NS cells. Sox1-GFP NS cells were then re-plated onto 0.1% gelatin- or poly-l-lysine/laminin-coated plates and cultured in N2B27 medium supplemented with LIF (1000 units/ml), CHIR99021 (3 μM) and Y27632 (10 μM). For the derivation of clonal lines, individual Sox1-GFP NS cells were deposited into 96-well plates and cultured in the presence of LIF/CHIR/Y. The presence of one cell per well was confirmed under microscopy 1 h after plating. Established Sox1-GFP NS cell lines were trypsinized and expanded every 3–4 days at a subculture ratio of 1:2–3.
Immunostaining
Immunostaining was performed according to a standard protocol. Alexa Fluor 488 and Alexa Fluor 546 were used as secondary conjugates and nuclear counterstaining was performed with DAPI. Primary antibodies used include the following: Nestin (1:400; Santa Cruz Biotechnology); Pax6 (1:50; DSHB, Iowa); Sox1 (1:100; BD Biosciences); Sox2 (1:200; Santa Cruz Biotechnology); Tuj1 (1:2000, Sigma); GFAP (1:300; Santa Cruz Biotechnology); GABA (1:1000; Sigma); tyrosine hydroxylase (1:1000; Sigma); Rip (1:50; DSHB, Iowa); MAP2 (1:500; Millipore); HB9 (1:50; DSHB, Iowa); PLZF (1:100; Calbiochem, Iowa); synapsin (1:500; Millipore); NeuN (1:500; Millipore); Nanog (1:200; Santa Cruz Biotechnology) and SSEA1 (1:200; Santa Cruz Biotechnology).
Differentiation of NS cells
For astrocyte differentiation, iNS cells were cultured as free-floating neurospheres in N2B27 medium supplemented with 20 ng/ml of EGF and 20 ng/ml of FGF2 for 7–10 days. Spheres were trypsinized into single cells and replated onto poly-l-lysine/laminin-coated 4-well plates at 5 × 104 cells/cm2 in N2B27 medium supplemented with 1% fetal calf serum. For oligodendrocyte differentiation, iNS cells were seeded at 5 × 104 cells/cm2 and cultured in N2B27 medium supplemented with FGF2 (20 ng/ml), EGF (20 ng/ml) and SHH (200 ng/ml) (all from PeproTech) for 10 days. At day 11, cells were dissociated with trypsin and replated onto poly-l-lysine/laminin-coated dishes in the presence of PDGF-AA (10 ng/ml) and T3 (triiodothyronine, Sigma, 40 ng/ml) to promote differentiation toward oligodendrocyte lineage [34]. For neuronal differentiation, cells were harvested by trypsinization and transferred to petri dishes in N2B27 medium. After 4 days, aggregated cells were replated onto poly-l-lysine/laminin-coated 4-well plates at a density of 3–4 × 104 cells/cm2 in N2B27 medium and cultured for another 8–10 days. For induction of midbrain precursors, iNS cells were cultured in N2B27 medium supplemented with 200 ng/ml SHH, 100 ng/ml FGF8 (PeproTech) and 160 μM ascorbic acid (Sigma–Aldrich) for 8 days. The cells were subsequently differentiated in N2B27 medium with SHH (200 ng/ml), FGF8 (100 ng/ml) and BDNF (20 ng/ml) for 10 days. Motor neurons were generated by exposing iNS cells to SHH (500 ng/ml) and RA (0.05 μM) for 7 days, followed by culturing in the presence of 20 ng/ml BDNF and 200 ng/ml SHH in N2B27 medium.
Retrovirus packaging
The day before transfection, GP2–293 cells were seeded onto 10 cm dishes. On the next day, pMXs-based retroviral vectors (pMXs-mOct4, pMXs-mSox2, pMXs-mKlf4 and pMXs-mc-Myc, Addgene) and pCMV-VSVG (Addgene) were co-transfected into GP2–293 cells using Lipofectamine LTX Transfection Reagent (Invitrogen) according to the manufacturer’s protocols. 24 h after transfection, the medium was replaced and the virus-containing supernatants were collected 48 and 72 h after transfection. After centrifugation to remove the cell debris, the viruses were filtered through a 0.45-mm cellulose acetate filter and stored at −80 °C.
Generation of rat iNS cells
Oct4-GFP transgenic rats were obtained from the Rat Resource and Research Center. Timed pregnant Sprague–Dawley (SD) rats were purchased from Harlan. Animal experiments were performed according to the investigator’s protocols approved by the USC Institutional Animal Care and Use Committee. For rat embryonic fibroblast (REF) isolation, embryos from E14.5 timed-pregnant rats were washed with PBS. The head and intestinal organs were removed from isolated embryos. The remaining tissue was manually dissociated and incubated in 0.25% trypsin for 10–15 min. After trypsinization was halted with serum medium and centrifugation, the cells were plated into 10-cm tissue culture dishes with MEF medium (GMEM containing 10% fetal bovine serum, 2 mM l-glutamine and 100 IU/ml penicillin/streptomycin). To establish postnatal tail-tip fibroblasts (TTFs), the tails from one-week-old p53−/−, p53−/+ and p53+/+ rat pups were peeled, minced into 1-cm pieces, placed in 35-mm culture dishes, and incubated in MEF medium until confluent. For iNS reprogramming, the REFs or TTFs were seeded at 1 × 105 cells per 35-mm dish, and the four-factor viral cocktail was added every other day in the presence of polybrene (4 μg/μl). After the second virus infection, cells were transferred onto γ-irradiated CF1-MEF feeders and cultured in neural reprogramming medium (mouse ES medium and N2B27 medium mixed 1:1 and supplemented with 3 μM CHIR99021 and 1000 units/ml LIF). iNS cells were generated and maintained in N2B27 medium supplemented with LIF/CHIR/Y.
Electrophysiology analysis
Cells cultured on glass coverslips were transferred to a submersion recording chamber and visualized under IR-DIC optics (Olympus BX51 WI). The recording chamber was perfused with artificial cerebrospinal fluid (ACSF) containing (in mM) 126 NaCl, 3 KCl, 1.25 NaH2PO4, 2 MgSO4, 26 NaHCO3, 2 CaCl2 and 10 dextrose, saturated with 95% O2/5% CO2. Whole-cell recording electrode solution contained (in mM): 130 K-gluconate, 4 KCl, 2 NaCl, 0.2 EGTA, 0.3 GTP-Tris, 4 ATP-Mg, 10 HEPES and 14 phosphocreatine-Tris (pH 7.25; ∼290 mOsm). Resting membrane potentials (RMPs) were measured upon establishing whole cell configuration following gigaohm seal break-ins. After current steps were injected to elicit action potentials, neurons were tested with a series of voltage commands to record the action currents. Neurons were then voltage clamped at RMP in the presence of TTX (1 μM) and bicuculline (20 μM) to record miniature excitatory postsynaptic currents (mEPSCs). The AMPA receptor antagonist CNQX (20 μM) was washed in at the end of experiments to verify the mEPSCs were indeed glutamatergic. Electrical signals were amplified with a Multiclamp 700B amplifier, digitized at 10 kHz with Digidata 1440A and acquired under control of pClamp 10.2 software (all from Molecular Devices).
RT-PCR and qRT-PCR
Total RNA was extracted using Trizol reagent kit (Invitrogen) according to the manufacture’s instruction. cDNA was synthesized with 1 μg of total RNA using Cloned AMV First-Strand cDNA Synthesis Kit. PCR reaction mixtures were prepared with Taq DNA polymerase (Invitrogen). qRT-PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) according to manufacturer’s instructions. Signals were detected with an ABI7900HT Real-Time PCR System (Applied Biosystems). The relative expression level was determined by the 2-dCT method and normalized against GAPDH. Primers used are listed in Tables S1 and S2.
Authors’ contributions
GX and PH derived and characterized Sox1-GFP mouse NS cells and rat iNS cells. CQ developed conditions for the maintenance of NS cells and the conversion of rat fibroblasts into iNS cells. SQ contributed to the characterization of rat iNS cells. CT contributed to the conversion of iNS cells from p53 mutant rat fibroblasts. QLY conceived the study, designed the experiments, and wrote the manuscript with assistance from GX and PH. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Acknowledgements
We thank members of the Ying lab and the USC Stem Cell Core for technical assistance; Charles Ashton for critical reading of the manuscript. This work was supported by USC startup fund to QLY and in part by NIH (Grant No. R01OD010926) to QLY.
Footnotes

Appendix Supplementary. data
Figure S1.
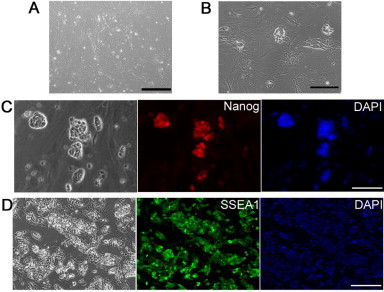
Reprogramming of mouse embryonic fibroblasts by Oct4/Sox2/Klf4/c-Myc under the LIF/CHIR/Y condition (A) Phase contrast image of mouse embryonic fibroblasts (MEFs) derived from E13.5 CF1 mice. (B) Morphology of iPS cell-like colonies 21 days after transfection with Oct4/Sox2/Klf4/c-Myc. (C and D) Immunostaining of mouse iPS cells with ES markers Nanog (C) and SSEA1 (D). Scale bars, 50 μm.
Figure S2.
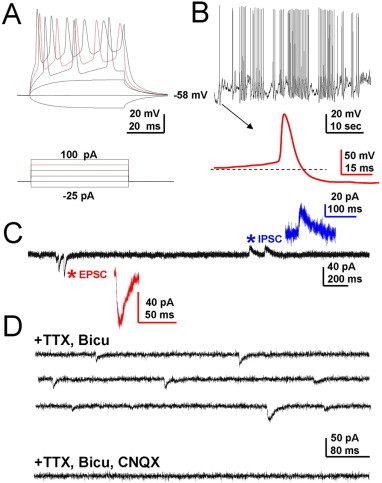
Characterization of membrane properties and neurotransmitter phenotype of neurons derived from rat iNS cells (A) Injection of current steps into iPS-derived neurons elicits action potentials (APs). (B) Spontaneous AP firing of an iPS-derived neuron. Inset, expanded view of a single AP. (C) Some cells show both spontaneous excitatory postsynaptic currents (EPSCs) and spontaneous inhibitory postsynaptic currents (IPSCs). (D) Isolated miniature EPSC (top panel) in the presence of tetrodotoxin (TTX) and bicuculline (Bicu) can be further blocked by AMPA receptor antagonist CNQX (lower panel), indicating that the neuron is glutamatergic in nature.
Figure S3.
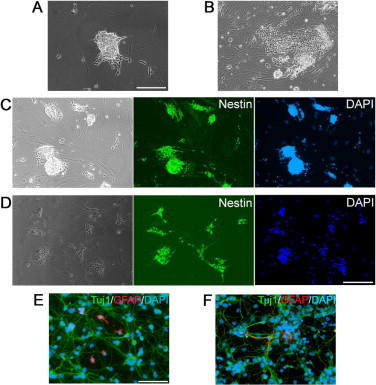
Rat iNS cells derived from rat tail-tip fibroblasts (A and B) Morphology of iNS colonies generated from rat tail-tip fibroblasts using retrovirus carrying the four factors (A) or PiggyBac transposon carrying the four factors (B). C-F. Rat iNS cells derived by retrovirus carrying the four factors (C and E) or PiggyBac transposon carrying the four factors (D and F) expressed NS marker Nestin (C and D) and were able to differentiate into Tuj1positve neurons and GFAP positive astrocytes (E and F). Scale bars, 50 μm.
Figure S4.
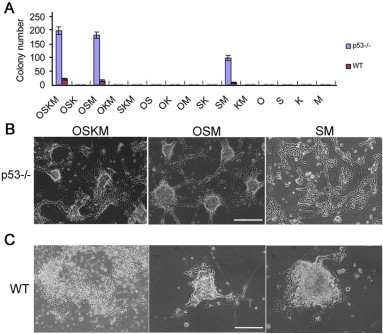
Generation of rat iNS cells by different combination of transcription factors (A) Quantification of iNS colonies derived from p53−/− and wild type (WT) REFs infected with different combinations of the four transcription factors Oct4 (O), Sox2 (S), Klf4 (K) and c-Myc (M). REFs were plated at 1 × 105 cells per 35-mm dish. iNS cell-liked colonies were scored under a microscope. Data are presented as mean±SD of three biological replicates (colony number per 1 × 105 REFs plated). (B and C) Representative images of iNS colonies generated from different transcription factor combinations. Scale bars, 50 μm.
Figure S5.
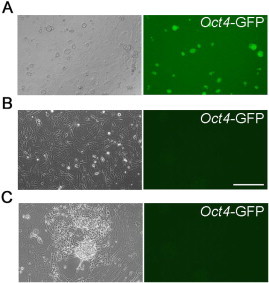
Conversion of Oct4-GFP REFs into iNS cells (A) Phase contrast and fluorescent images of ES cells derived from Oct4-GFP transgenic rat. (B) Phase contrast and fluorescent images of REFs derived from Oct4-GFP transgenic rat. Scale bar, 50 μm. (C) Phase contrast and fluorescent images of iNS cells derived from Oct4-GFP REFs.
RT-PCR primers
Quantitative real-time PCR primers
References
- 1.Gage F.H. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang Z.P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D.R., Yang T.Q. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caiazzo M., Dell’Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 6.Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ieda M., Fu J.D., Delgado-Olguin P., Wedantham V., Hayashi Y., Bruneau B.G. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efe J.A., Hilcove S., Kim Janghwan, Zhou H., Quyang K., Wang G. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 9.Wada R., Muraoka N., Inagawa K., Yamakawa H., Miyamoto K., Sadahiro T. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo E., Rampalli S., Risueno R.M., Schnerch A., Mitchell R., Fiebig-Comyn A. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 11.Huang P., He Z., Ji S., Sun H., Xiang D., Liu C. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 12.Sekiya S., Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 13.Kim J., Efe J.A., Zhu S., Talantova M., Yuan X., Wang S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lujan E., Chanda S., Ahlenius H., Sudhof T.C., Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thier M., Worsdorfer P., Lakes Y.B., Gorris R., Herms S., Opitz T. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Han D.W., Tapia N., Hermann A., Hemmer K., Hoing S., Arauzo-Bravo M.J. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Pevny L.H., Sockanathan S., Placzek M., Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 18.Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 19.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 21.Tropepe V., Hitoshi S., Sirard C., Mak T.W., Rossant J., van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 22.Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N.D., Tabar V., Studer L. Human ESC-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marion R.M., Strati K., Li H., Murga M., Blanco R., Ortega S. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong C., Li P., Wu N.L., Yan Y., Ying Q.L. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hitoshi S., Seaberg R.M., Koscik C., Alexson T., Kusunoki S., Kanazawa I. Primitive neural stem cells from the mammalian epiblast differentiate to definitive neural stem cells under the control of Notch signaling. Genes Dev. 2004;18:1806–1811. doi: 10.1101/gad.1208404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han D.W., Greber B., Wu G., Tapia N., Araúzo-Bravo M.J., Ko K. Direct reprogramming of fibroblasts into epiblast stem cells. Nat Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- 30.Ying Q.L., Smith A.G. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 31.Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong C., Huang G., Ashton C., Li P., Ying Q.L. Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc. 2011;6:827–844. doi: 10.1038/nprot.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billon N., Jolicoeur C., Ying Q.L., Smith A., Raff M. Normal timing of oligodendrocyte development from genetically engineered, lineage-selectable mouse ES cells. J Cell Sci. 2002;115:3657–3665. doi: 10.1242/jcs.00049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR primers
Quantitative real-time PCR primers


