Abstract
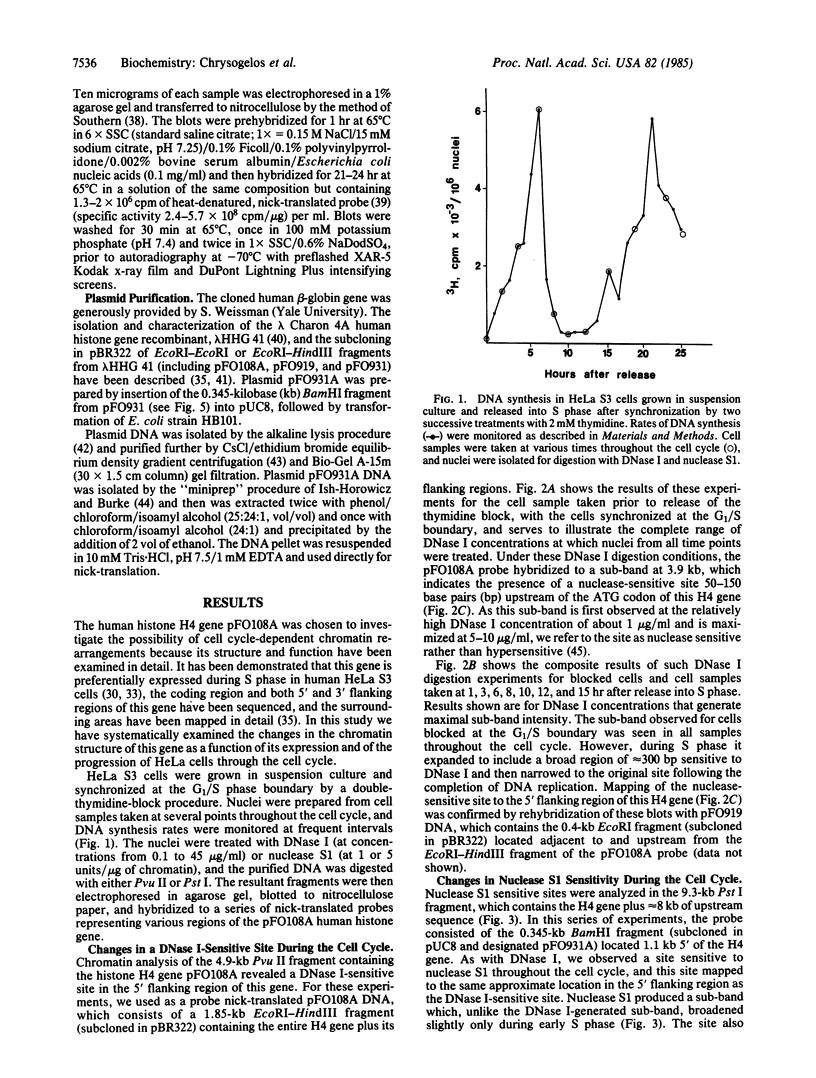
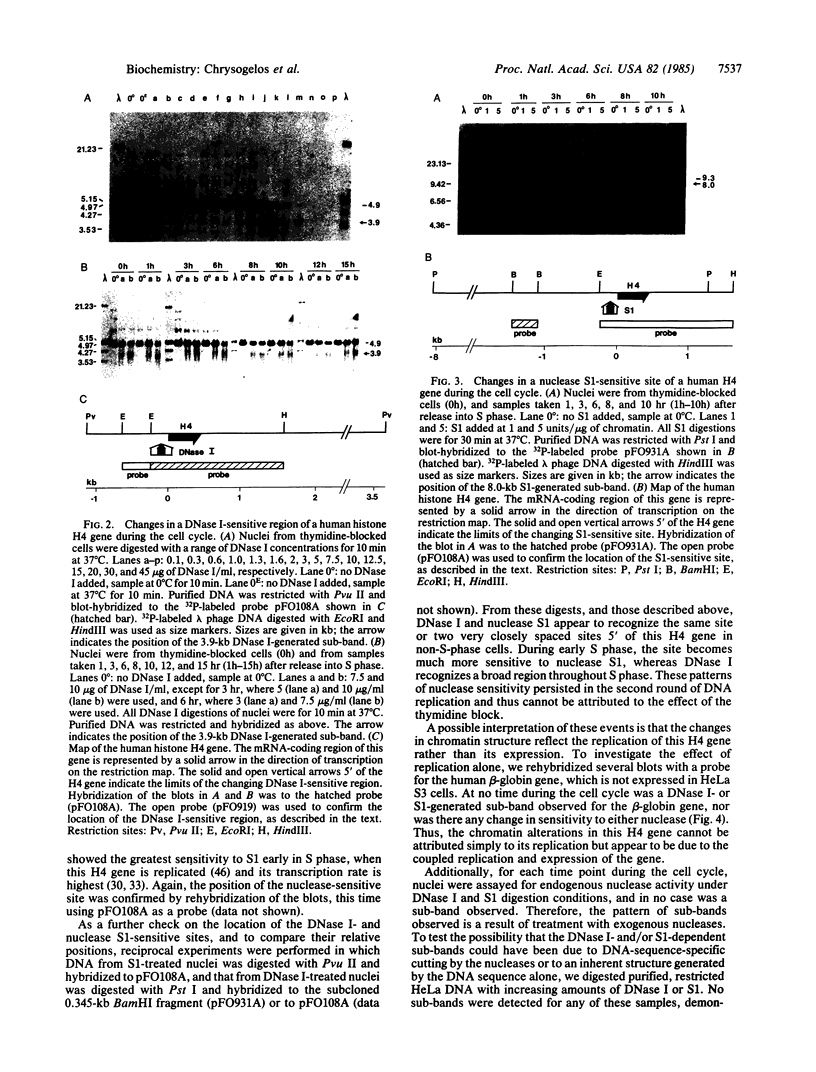
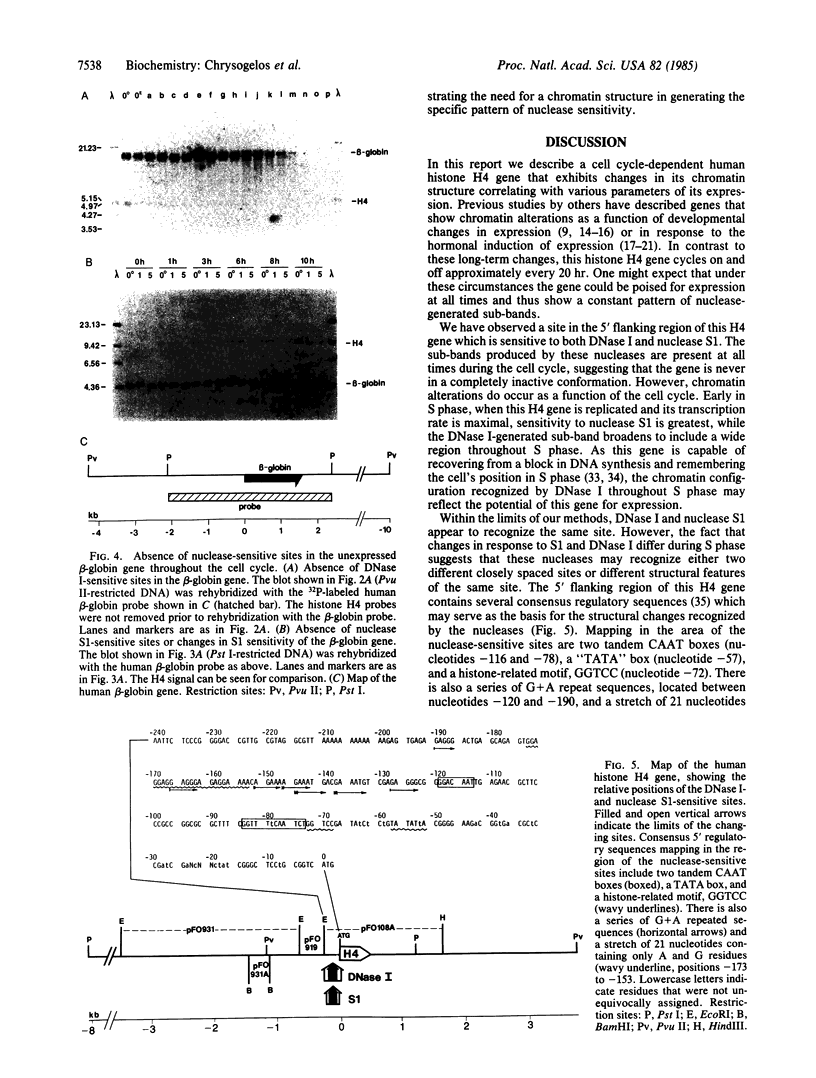
By use of synchronized human HeLa S3 cells, a site sensitive to both DNase I and nuclease S1 was identified 50-150 base pairs upstream of the ATG codon of a cell cycle-dependent histone H4 gene. This site expanded to include a broad region of approximately equal to 300 base pairs sensitive to DNase I throughout S phase and then narrowed again to the original site after the completion of DNA replication. The level of nuclease S1 sensitivity was greatest during early S phase, when the gene is replicated and its transcription rate is maximal. The chromatin structure of the human beta-globin gene, which is not expressed in HeLa cells, was also analyzed throughout the cell cycle, and in no case was a sub-band seen as a result of DNase I or nuclease S1 digestion, nor were there any changes in nuclease sensitivity correlated with its replication. Thus the cell cycle-dependent chromatin alterations in this histone H4 gene appear to be due to the coupled replication and expression of this gene rather than simply its replication. These results suggest that histone genes, as compared with developmentally regulated genes, exhibit an "intermediate" level of regulation whereby the gene is never in a completely inactive conformation, but changes in chromatin structure occur as a function of the cell cycle and expression.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumbach L. L., Marashi F., Plumb M., Stein G., Stein J. Inhibition of DNA replication coordinately reduces cellular levels of core and H1 histone mRNAs: requirement for protein synthesis. Biochemistry. 1984 Apr 10;23(8):1618–1625. doi: 10.1021/bi00303a006. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Olah J., Birnstiel M. L. Major changes in the 5' and 3' chromatin structure of sea urchin histone genes accompany their activation and inactivation in development. Cell. 1983 Jul;33(3):843–848. doi: 10.1016/0092-8674(83)90026-0. [DOI] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Gurley L. R., Tobey R. A. Syntheses and modulations in the chromatin contents of histones H1 degrees and H1 during G1 and S phases in Chinese hamster cells. Biochemistry. 1982 Aug 17;21(17):3991–4001. doi: 10.1021/bi00260a014. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Mueller G. C. Histone synthesis in vitro on HeLa cell microsomes. The nature of the coupling to deoxyribonucleic acid synthesis. J Biol Chem. 1969 Nov 10;244(21):5947–5952. [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L., Stein G., Stein J. Histone gene expression in human diploid fibroblasts: analysis of histone mRNA levels using cloned human histone genes. Mol Cell Biochem. 1984;60(2):123–130. doi: 10.1007/BF00222482. [DOI] [PubMed] [Google Scholar]
- Hanly S. M., Bleecker G. C., Heintz N. Identification of promoter elements necessary for transcriptional regulation of a human histone H4 gene in vitro. Mol Cell Biol. 1985 Feb;5(2):380–389. doi: 10.1128/mcb.5.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Iqbal M. A., Plumb M., Stein J., Stein G., Schildkraut C. L. Coordinate replication of members of the multigene family of core and H1 human histone genes. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7723–7727. doi: 10.1073/pnas.81.24.7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. S., Bellard M., Dretzen G., Bellard F., Chambon P. A close association between sites of DNase I hypersensitivity and sites of enhanced cleavage by micrococcal nuclease in the 5'-flanking region of the actively transcribed ovalbumin gene. EMBO J. 1984 May;3(5):1137–1144. doi: 10.1002/j.1460-2075.1984.tb01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marashi F., Green L., Briggs R., Duhl D., Hnilica L., Stein J. L., Stein G. S. Variations in the organization and expression of human histone genes in normal diploid and tumor cell lines. Anticancer Res. 1984 Jan-Apr;4(1-2):69–74. [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G. Influence of DNA synthesis inhibition on the coordinate expression of core human histone genes during S phase. Nucleic Acids Res. 1983 Nov 25;11(22):7927–7945. doi: 10.1093/nar/11.22.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Rickles R., Marashi F., Sierra F., Clark S., Wells J., Stein J., Stein G. Analysis of histone gene expression during the cell cycle in HeLa cells by using cloned human histone genes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):749–753. doi: 10.1073/pnas.79.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D. E., Canfield T. K., Gartler S. M. Chromatin structure of active and inactive human X chromosomes. Nucleic Acids Res. 1984 Feb 24;12(4):1829–1845. doi: 10.1093/nar/12.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E., Borun T. W. The cytoplasmic synthesis of histones in hela cells and its temporal relationship to DNA replication. Proc Natl Acad Sci U S A. 1967 Feb;57(2):409–416. doi: 10.1073/pnas.57.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Shephard E. A., Phillips I., Davis J., Stein J. L., Stein G. S. Evidence for the resumption of DNA replication prior to histone synthesis in HeLa cells after release from treatment with hydroxyurea. FEBS Lett. 1982 Apr 19;140(2):189–192. doi: 10.1016/0014-5793(82)80891-0. [DOI] [PubMed] [Google Scholar]
- Sierra F., Lichtler A., Marashi F., Rickles R., Van Dyke T., Clark S., Wells J., Stein G., Stein J. Organization of human histone genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra F., Stein G., Stein J. Structure and in vitro transcription of a human H4 histone gene. Nucleic Acids Res. 1983 Oct 25;11(20):7069–7086. doi: 10.1093/nar/11.20.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spelsberg T. C., Steggles A. W., Chytil F., O'Malley B. W. Progesterone-binding components of chick oviduct. V. Exchange of progesterone-binding capacity from target to nontarget tissue chromatins. J Biol Chem. 1972 Mar 10;247(5):1368–1374. [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Stein G. S., Borun T. W. The synthesis of acidic chromosomal proteins during the cell cycle of HeLa S-3 cells. I. The accelerated accumulation of acidic residual nuclear protein before the initiation of DNA replication. J Cell Biol. 1972 Feb;52(2):292–307. doi: 10.1083/jcb.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein G., Park W., Thrall C., Mans R., Stein J. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature. 1975 Oct 30;257(5529):764–767. doi: 10.1038/257764a0. [DOI] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]