Abstract
Purpose:
The aim of this study was to demonstrate that modern data mining tools can be used as one step in reducing the labor necessary to produce and maintain systematic reviews.
Methods:
We used four continuously updated, manually curated resources that summarize MEDLINE-indexed articles in entire fields using systematic review methods (PDGene, AlzGene, and SzGene for genetic determinants of Parkinson disease, Alzheimer disease, and schizophrenia, respectively; and the Tufts Cost-Effectiveness Analysis (CEA) Registry for cost-effectiveness analyses). In each data set, we trained a classification model on citations screened up until 2009. We then evaluated the ability of the model to classify citations published in 2010 as “relevant” or “irrelevant” using human screening as the gold standard.
Results:
Classification models did not miss any of the 104, 65, and 179 eligible citations in PDGene, AlzGene, and SzGene, respectively, and missed only 1 of 79 in the CEA Registry (100% sensitivity for the first three and 99% for the fourth). The respective specificities were 90, 93, 90, and 73%. Had the semiautomated system been used in 2010, a human would have needed to read only 605/5,616 citations to update the PDGene registry (11%) and 555/7,298 (8%), 717/5,381 (13%), and 334/1,015 (33%) for the other three databases.
Conclusion:
Data mining methodologies can reduce the burden of updating systematic reviews, without missing more papers than humans.
Keywords: citation screening, machine learning, meta-analysis, support vector machine, text classification
Systematic reviews, meta-analyses, and field synopses (i.e., systematically curated compendia summarizing entire fields) have gained acceptance as a practical way to provide reliable and comprehensive syntheses of the exponentially expanding medical evidence base. MEDLINE indexes more than 20,000 new randomized trials and over 9,000 genetic association studies from 2010 alone, and the increasing trajectory of publication rates shows no signs of slowing.1 It is difficult to keep up with new information for both performing new reviews and updating existing reviews.1
Indeed, it is estimated that more than half of the systematic reviews in the Cochrane Library have not been updated for at least two years.2 Similarly, a recent survey of organizations that produce and maintain systematic reviews suggests that at least half of existing reviews are already out of date, limiting their utility.3 Exacerbating the challenge of information overload, the standards for systematic reviews and meta-analyses are more demanding now than they were only 10 years ago. The scenario is probably worse in the scientific fields with rapid data turnaround, such as genetic/-omic research. In fact, field synopses in genetics are attempts to cope with data and information overload.4
The time required to complete a systematic review and meta-analysis has not decreased over the past three decades. Allen and Olkin5 calculated that a well-performed systematic review with meta-analysis can take between 1,000 and 2,000 person hours; part of this time appears to be related to topic refinement and set-up and the rest depends on the number of included papers. The time to complete systematic reviews has increased despite improvements in software tools that facilitate the process (e.g., reference management software), likely because the efficiencies of such tools are no match for the exponential growth of the literature. Based on our own experience, the US Agency for Healthcare Research and Quality's comparative effectiveness reviews take at least 13 months to complete, an amount of time that has grown consistently during the past 15 years.
Strict adherence to the US Institute of Medicine's 21 standards and 82 elements of performance,6 the 100 Methodological Expectations of Cochrane Intervention Reviews (MECIR), and the detailed guidance from internationally acknowledged entities7,8,9,10,11,12,13 could further prolong the completion of systematic reviews. Long timelines generate high costs; increased fiscal constraints thus necessitate modernizing all stages of the systematic review pipeline to increase efficiency. Some improvements will refine processes to remove unnecessary redundancies, some will necessitate the development of new, publicly available resources,14,15 and others will demand the development and application of novel methodologies and tools.16,17
Here we describe a tool to expedite the process of updating systematic reviews, meta-analyses, or field synopses by semiautomating the step of screening citations published after the latest update. Widespread use of the tool would eliminate a substantial amount of the work involved in updating reviews, thereby saving time and human resources and increasing the likelihood that reviews are kept current.
Materials and Methods
Overview
We aim to semiautomate the updating of systematic reviews, meta-analyses, and field synopses, thereby reducing reviewers' efforts and the associated costs. As an example, we describe the application of data mining methods from computer science to facilitate updating four field synopses. Field synopses are continuously updated and manually curated collections of publications that synthesize a broad field. The steps involved in identifying eligible studies for field synopses are as follows. First, one searches databases, such as MEDLINE, to retrieve potentially relevant studies. Second, she or he screens the titles and abstracts of all retrieved citations to identify those that meet predefined eligibility criteria. These are exactly the same steps one would follow to update a systematic review or meta-analysis. Therefore, to avoid confusion, from here on we refer to all literature databases that are updated following the above protocol as systematic reviews.
We simulated a prospective update of the four systematic review databases during 2010. We used citations published through 2009 to train a classification model to distinguish “relevant” from “irrelevant” citations (i.e., to discern citations likely to be ultimately included in the systematic review from those likely to be excluded). We then used this trained classification model (classifier) to automatically screen articles published in 2010 (i.e., the articles that would be considered when updating the systematic review). A human would manually review only the citations deemed relevant by the classifier. All articles designated as irrelevant would be excluded from further consideration, thereby saving human effort. We assessed the performance of the system with respect to the studies that were ultimately included by the researchers in the 2010 update of the systematic reviews.
Databases
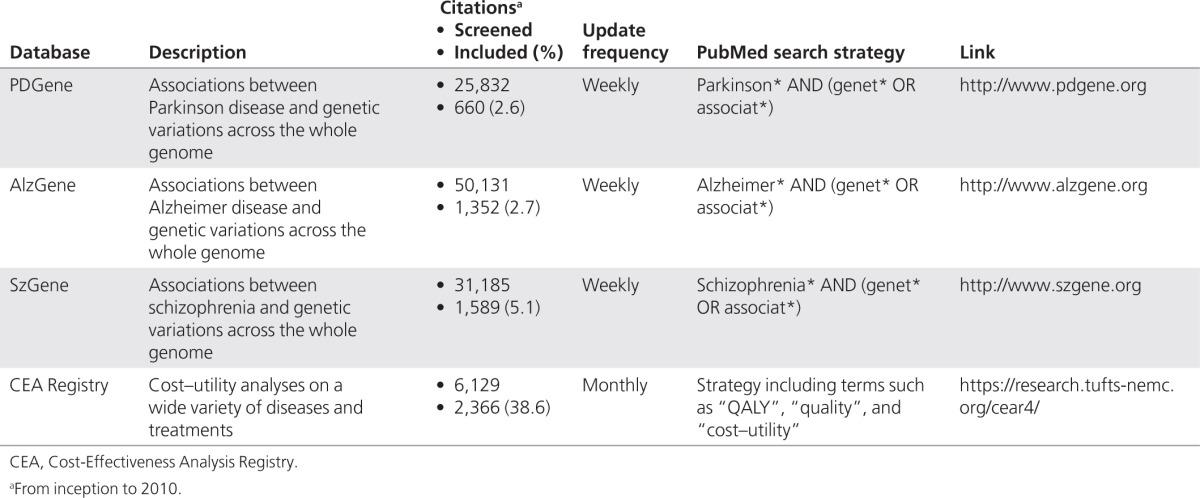
We used four systematic reviews to validate our approach. Three synthesize genetic association studies, investigating Parkinson disease (PDGene),18 Alzheimer disease (AlzGene),19 and schizophrenia (SzGene),20 respectively (Table 1). The fourth is the Tufts Cost-Effectiveness Analysis Registry (CEA Registry), which summarizes information from published cost-effectiveness analyses. We added this fourth field synopsis to gain insights on the generalizability of our approach to nongenetic synopses. The protocol and methods for our four data sets are available on their respective websites (Table 1). In contrast to typical systematic reviews, these address much broader questions and are updated on a weekly or monthly basis. For example, the AlzGene review evaluates the strength of the association between Alzheimer disease and genetic variations across the whole genome, whereas a typical systematic review would probably evaluate only a subset of such genetic variations (e.g., in the APOE gene). Note that attaining perfect (100%) sensitivity with semiautomated updating is much more difficult when all reported variations across thousands of genes are of interest rather than only APOE variations.
Table 1. Characteristics of the four systematic reviews (field synopses).
To simulate a prospective test of our semiautomated system, we segmented each of the data sets into a training set comprising all citations published through 31 December 2009 and an update (validation) set composed of citations published between 1 January 2010 and 31 December 2010 (Supplementary Figure S1 online). This is equivalent to a prospective evaluation of our semiautomated system throughout 2010.
Classification of citations
For each of the four databases, we aim to train a classifier to discriminate relevant from irrelevant citations. There are two components to operationalizing this: a method to encode text in an analyzable format and a classification method.
Text encoding. We created computer-friendly representations of the titles, abstracts (when available), and medical subject heading terms (MeSH; when available) of citations using a standard encoding scheme. Specifically, we created a long list that includes all words present in at least three citations in the set of abstracts screened for the original review. The length of the list (which corresponds to the number of words encoded) depends on the data set, but is typically in the tens of thousands. Each document was then represented simply as a vector with elements 1 and 0, denoting whether the citation contained the corresponding word in this long list. This encoding scheme is known as the “bag-of-words” representation.
Classification method. A detailed exposition of the methods is provided in the Supplementary data online. Briefly, we used Support Vector Machines (SVMs),21 which are state-of-the-science methods for text classification.22 SVMs in their canonical form perform poorly when there are far fewer examples (in our case, citations) from one class (relevant citations) than the other (irrelevant citations). This is known as class imbalance, and it is the norm in literature searches conducted for systematic reviews. Further, in systematic reviews false negatives (relevant citations misclassified as irrelevant) are much more costly than false positives (irrelevant citations misclassified as relevant). We explicitly accounted for these asymmetric misclassification costs in two ways. First, we modified the SVMs to emphasize sensitivity (we penalized more severely for false negatives compared with false positives). Second, we did not train SVMs on the entire training set, but instead on a balanced sample comprising an equal number of relevant and irrelevant examples. This was accomplished by using all relevant examples and a randomly selected sample of the irrelevant examples and is analogous to performing a nested case–control study in a cohort with sparse index cases.
Although undersampling discards information, empirical evidence shows that it yields classifiers with higher sensitivity.23,24,25,26,27 However, undersampling introduces randomness (the random sampling of the irrelevant citations in the training set). To reduce random variation in the model's predictions, we used an ensemble of 11 sets of SVM classifiers trained independently (an approach called bagging).28,29 The final classification decision was taken as a majority vote over this ensemble (see Figure 1). We used an odd number of classifiers in the ensemble to avoid ties (the exact choice of 11 was motivated by our previous experimental work, in which this value worked well across many data sets).24,25
Figure 1.
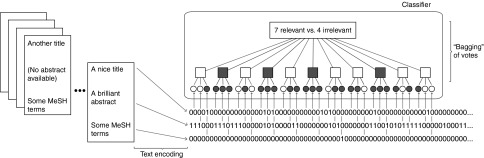
Outline of the classification method. The title, abstract, and MeSH term components of citation documents are encoded as series of 0's and 1's (i.e., as separate “bag-of-words” representations; see Methods section). We used an ensemble of 11 base classifiers (squares) comprising three Support Vector Machines (SVMs, circles), one per encoded component. Open circles and black circles stand for SVMs that classify their encoded components as relevant and irrelevant, respectively. If at least one of the SVMs suggests that the citation is relevant, the corresponding base classifier casts a relevant vote (white squares); otherwise, it casts a vote for irrelevant (black squares). The overall disposition is given according to the majority vote of the ensemble of 11 base classifiers (here, relevant with 7 vs. 4 votes)—this is called “bagging”. The proportion of votes for the “winning” disposition is a proxy for the confidence of the classifier in its ultimate vote (here 7/11, or 0.64). MeSH, medical subject heading.
Analyses
For each data set, we calculated the sensitivity and specificity of the classifiers on the update set. The reference standard was whether a citation was ultimately included in the systematic review (based on manual screening). We report the number of citations that reviewers would have needed to screen, had they been using the proposed semiautomated system to update reviews in 2010, versus the number of citations they actually screened. We assessed the variability of overall results by repeating all analyses 20 times using different random number seeds. We arbitrarily considered the first run the main analysis and report minimum and maximum results from the other 19.
Analyses of misclassifications: Our classification model does not provide rationales for its predictions. To provide some intuition as to whether the model and the human experts tend to make the same “mistakes,” we performed the following analysis. For each data set, we asked our experts to manually rescreen “false-positive” citations (i.e., citations not included in the final database but predicted to be relevant by the classifier based only on titles and abstracts and not on the full text). The experts then categorized the rescreened citations as “clearly irrelevant”, “unclear,” or “clearly relevant” (scoring them as 0, 1, or 2, respectively). For these same “false-positive” citations, we measured the confidence of the classification model in its (mistaken) predictions, as described below. An association between higher scores in human rescreening and higher confidence in the classification models predictions suggests that model and human experts tend to make the same mistakes.
We took the proportion of votes in agreement with the majority vote as a measure of the model's confidence in its predictions. For example, in Figure 1, 7 of the 11 base classifiers predict that the citation is relevant, and the confidence of the classification is 7/11 (≈0.64). Thus, confidence ranges from 6/11 (≈0.54) to 11/11 (1.0).
To test for an association between human rescreening and model confidence, we compared the distribution of human scores across the extreme quartiles of the distribution of confidence scores over false-positive citations using Fisher's exact test. In the case of the smaller CEA Registry, we performed this comparison across the top and bottom halves of the confidence score distributions. To also quantify the direction of the association, we used a proportional odds ordinal logistic regression to calculate the odds ratio for a human assigning a higher score (from 0 to 1 or from 1 to 2). The predictor was the percentile of classifier confidence (higher vs. lower).
Software
We used the LIBSVM30 Support Vector Machine implementation and its Python interface. All of our code is open source and is available from the authors. Statistical analyses were performed in Stata SE, version 11 (Stata, College Station, TX). All P values are two tailed and considered significant at the 0.05 level.
Results
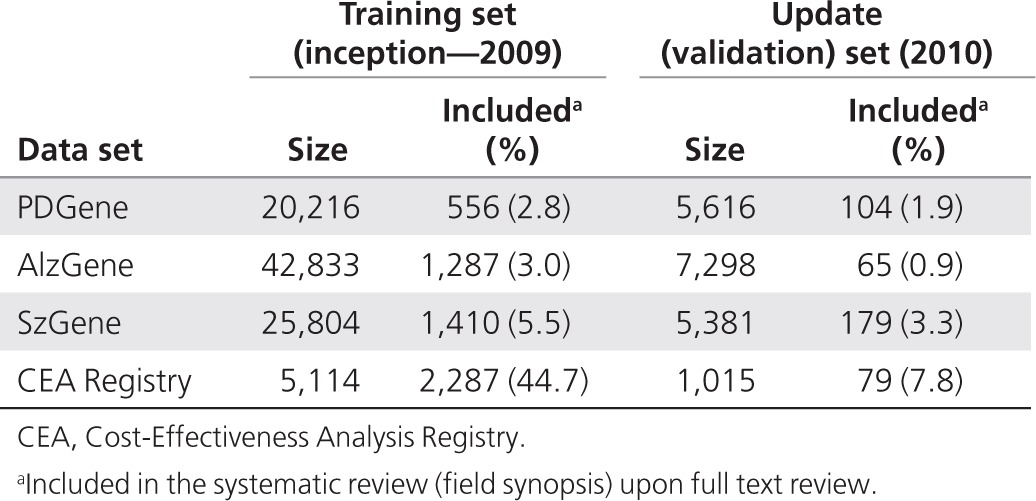
Table 2 lists the size of the initial (training) and update (validation) sets in the four databases, as well as the proportion of citations that were finally included upon full-text review. Researchers screened between ~25,000 and 50,000 citations for the three genetic synopses and a total of 6,129 for the CEA Registry (from inception through 2010; these comprise the training sets). Of these, only a minority was finally included in the systematic reviews. For example, during 2010, 0.9 to 3.3% of citations reviewed were included in the genetic databases, and 7.8% were included in the CEA Registry. Semiautomation could therefore save considerable resources that are otherwise spent reading irrelevant citations.
Table 2. Training and update (validation) sets in the four systematic reviews.
Performance in the update set. In all three genetic topics, the proposed semiautomated strategy correctly identified all citations that were included in the systematic reviews in 2010 (100% sensitivity) and considered relevant only ~10% of the papers that were excluded by the human experts (specificity of about 90%). Had the semiautomated system been used in 2010, the human experts would have needed to screen only 605 (PDGene), 555 (AlzGene), and 717 (SzGene) titles and abstracts compared with the 5,616, 7,298, and 5,381 citations they manually screened for the three data sets (Table 3). This translates to reductions in labor of ~81, 92, and 87%, respectively.
Table 3. Empirical results of classifier performance over the four systematic reviews in the 2010 update.
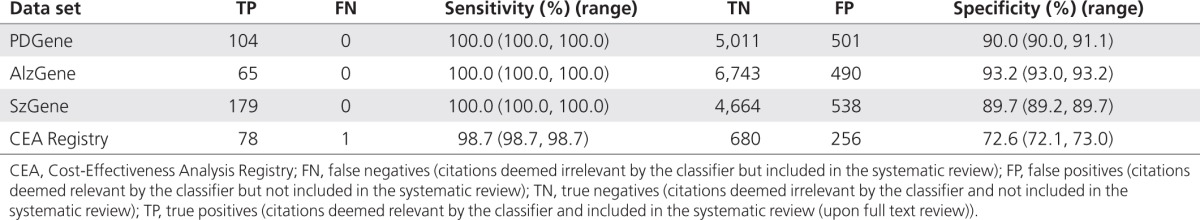
In the case of the CEA Registry, the classifier missed only 1 eligible article (sensitivity about 99%), and incorrectly considered relevant ~28% of the papers that were excluded by human reviewers in 2010 (specificity around 73%). Relying on the semiautomated system throughout 2010, researchers would have needed to screen only 334 of 1,015 citations (a reduction in labor of ~67%).
Sensitivity analyses. All results were robust when we repeated the entire analysis an additional 19 times using different random number seeds. No eligible papers were missed in the three genetic topics, and the same eligible paper was always missed in the CEA Registry. The specificity of the classifiers was practically identical to that of the main analyses (Table 2).
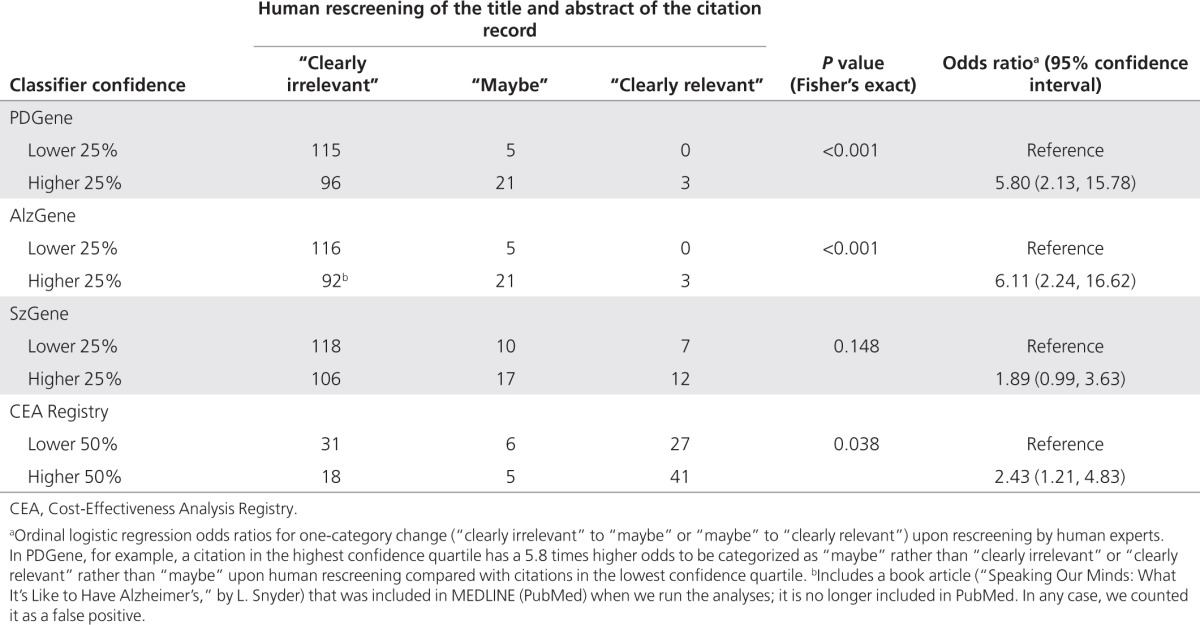
Analysis of misclassifications. As mentioned above, only a single citation31 in one data set (CEA Registry) would have been a false negative (i.e., missed if the semiautomated system was actually in use during 2010). The confidence of the classification model in that prediction was high because 10 of 11 base classifiers categorized the citation as irrelevant. Upon re-review, human experts deemed that this citation might also have been missed by an inexperienced reviewer: only a single sentence in the abstract suggests that a cost-effectiveness (or cost-utility) analysis might have been performed.
Table 4 describes the results of human rescreening of citations that were not included in the systematic reviews, but were suggested as relevant by the classifier (false positives) for different strata of classifier confidence. Classifier confidence appears to be associated with human experts' dispositions upon re-review. Based on ordinal logistic regressions, false positives in the highest 25% (50% for the CEA Registry) had between 1.9 and 6.1 higher odds of being deemed “clearly relevant” rather than “maybe” or “maybe” rather than “clearly irrelevant” upon re-review by human experts. Confidence intervals included 1 only in the SzGene data set.
Table 4. Human expert rescreening of classifier “false positives” stratified by classifier confidence.
Discussion
If we are ever to keep up with the information overload while adhering to the current demanding standards, we must modernize the methodology and conduct of systematic reviews, meta-analyses, and field synopses without compromising their scientific validity. This is particularly relevant to genetics and genomics, in which data accumulate rapidly. To this end, we demonstrated that data mining methodologies can reduce the burden of updating of systematic reviews without sacrificing their comprehensiveness.
Indeed, only a single citation of the many dozens that were included in each topic's 2010 update would have been missed by the semiautomated method. This is directly comparable to the performance of individual human screeners: in empirical explorations, human experts missed on average 8% of eligible citations (ranging from 0 to 24%).32 To minimize the likelihood of overlooking eligible studies, current recommendations suggest using two independent screeners.33,34,35,36 Thus, computer-assisted screening could replace full manual screening for both screeners, replace one screener, or be used in addition to both screeners to further increase the sensitivity of the overall process. To refine the role of the semiautomated process in real-life settings, we must evaluate their utility using study designs similar to those used for evaluating medical tests.37,38,39,40 The most robust design would entail a randomized comparison of using versus not using the semiautomated system during the conduct of systematic reviews. Apart from quantifying the utility of the semiautomated system, such an experiment would potentially allow one to detect unforeseen detrimental consequences (e.g., sloppiness during manual screening as a result of overreliance on the computer).
We note that semiautomating the updating of systematic reviews is different than semiautomating the screening of citations when performing a new systematic review.17 The main difference is that when updating an existing review, one has access to the set of citations that were manually screened for the original review. These citations can be used to train a classifier; the larger this set is, the better the classifier will likely perform, because it has more data from which to learn. By contrast, when performing a new review, no such training data set is available at the outset, and the problem is more challenging. Methods for new systematic reviews are still in development24,25,26,27 and remain to be empirically validated on a large scale.
Allowing for some caveats, our results are applicable to the general case of updating typical systematic reviews and meta-analyses. First, the scope of key questions in the updates should be identical to (or at worst a subset of) the scope in the original review. If the update is on one part of the original review, we would expect a decrease in the specificity of the predictions (some citations that would be eligible for the original review would be out of scope for the update), but no effect on the sensitivity. Further, there is a tacit assumption that there is no abrupt shift in the typical vocabulary of the field, as may happen, for example, in the case of an experimental drug or a disease or condition being referred to with a new name. Finally, the initial review should have a large enough number of citations to train a robust model. Ultimately, systematic reviewers must make a decision regarding the applicability of a model trained on the original review to the citations retrieved for the update; no hard and fast rules currently exist.
The latter observation is perhaps the most important concern when assessing the applicability of our results to updating typical systematic reviews. Specifically, in the three genetic topics, the initial set was an order of magnitude larger than most systematic reviews. However, in our previous work on semiautomating citation screening in new systematic reviews, we have consistently attained 100% sensitivity with training sets of 1,700–2,500 citations in all examined examples.25 Larger reviews are very common; literature searches for typical reviews often return 2,500–5,000 citations. High performance should therefore be attainable when semiautomating review updates. If anything, the systematic reviews (field synopses) in our examples have broader inclusion criteria than most systematic reviews and thus likely represent a more difficult problem: there are more eligible papers and thus more opportunities to miss them. Furthermore, there is greater diversity among the eligible papers.
Moreover, in practice one can assess the likely performance that a classification model will achieve when semiautomating a review update by performing an analysis called cross-fold validation on the citations manually screened for the original review. This analysis involves splitting the original database into, for example, 10 parts and sequentially training a model on 9/10 of the data and then assessing its performance on the remaining 1/10. This is repeated 10 times, and an estimate of performance is taken as average of these 10 tests. If the estimated performance is deemed acceptable (i.e., very high sensitivity is consistently achieved), then the researchers undertaking the update may opt to use the semiautomated approach.
Finally, another limitation is that the computer is a “black box” because no readily interpretable explanation is provided for the classifications. This challenge is inherent to modern text classification methods. However, a detailed rationale for excluding the citations during screening is not necessarily critical. For example, we do not really provide rationales for the millions of citations that are excluded by the search strategy in the first place. In the end, trusting computer algorithms to semiautomate abstract screening in routine settings is contingent on their empirical performance: if there are consistently acceptable empirical results, this criticism is less important.
We find it intuitively agreeable that upon re-reviewing false positives (i.e., citations ultimately not included in the respective synopses but denoted relevant by the model), experts tended to have trouble with the same citations that our model did. This is an indirect indication that the models rely at least on similar patterns of terms as humans do, of course lacking a human's semantic comprehension.
The semiautomated system reduced the number of citations that would have needed to be screened by a human expert by 70–90%, a substantial reduction in workload, without sacrificing comprehensiveness. Regularly updated systematic reviews and meta-analyses will play increasingly crucial roles in translating scientific knowledge into evidence-based medicine. The novel approach developed here is an important and practical step toward reaching that goal. In addition to making the process of updating systematic reviews substantially more efficient, application of our method may also improve the quality of evidence-based medicine.
Disclosure
The authors declare no conflict of interest.
Acknowledgments
T.A.T., B.C.W., C.H.S., J.L., C.E.B., and K.S. were funded by R01 HS018494 (US Agency for Healthcare Research and Quality). No one other than the authors had any role in study design, collection, analysis, or interpretation of data, in the writing of the report, or in the decision to submit the paper for publication. T.A.T. conceived the initial idea and designed the study. B.C.W. wrote code and trained and applied classifiers. L.B., C.M.L., and J.T.C. provided the content of the databases and re-reviewed citations after classification by the semiautomated system. B.C.W. and T.A.T. analyzed results and wrote the first draft of the paper. T.A.T. takes responsibility for the accuracy of the data, analysis, and interpretation. All authors contributed to the interpretation of the results and provided critical revisions to the paper. No other person, including medical editors, contributed to the manuscript. The genetics databases used for this project have been made possible by the kind support of the Cure Alzheimer's Fund (CAF), the Michael J. Fox Foundation (MJFF) for Parkinson's Research, the National Alliance for Research on Schizophrenia and Depression (NARSAD), Prize4Life, and EMD Serono. C.M.L. is supported by the Fidelity Biosciences Research Initiative. L.B. is supported by the German Ministry for Education and Research (BMBF).
Supplementary Material
References
- Bastian H, Glasziou P, Chalmers I. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up. PLoS Med. 2010;7:e1000326. doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch GG.No improvement—still less than half of the Cochrane reviews are up to dateXIV Cochrane Colloquium (Ireland)2006
- Garritty C, Tsertsvadze A, Tricco AC, Sampson M, Moher D. Updating systematic reviews: an international survey. PLoS ONE. 2010;5:e9914. doi: 10.1371/journal.pone.0009914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodsham AJ, Higgins JP. Online genetic databases informing human genome epidemiology. BMC Med Res Methodol. 2007;7:31. doi: 10.1186/1471-2288-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IE, Olkin I. Estimating time to conduct a meta-analysis from number of citations retrieved. JAMA. 1999;282:634–635. doi: 10.1001/jama.282.7.634. [DOI] [PubMed] [Google Scholar]
- Institute of Medicine of the National Academies . National Academies Press: Washington, DC; 2011. Finding What Works in Healthcare. Standards for Systematic Reviews. [PubMed] [Google Scholar]
- Chou R, Aronson N, Atkins D.et al. AHRQ series paper 4: assessing harms when comparing medical interventions: AHRQ and the effective health-care program J Clin Epidemiol 201063502–512. [DOI] [PubMed] [Google Scholar]
- Fu R, Gartlehner G, Grant M.et al. Conducting quantitative synthesis when comparing medical interventions: AHRQ and the Effective Health Care Program J Clin Epidemiol 2011641187–1197. [DOI] [PubMed] [Google Scholar]
- Harris RP, Helfand M, Woolf SH, Methods Work Group, Third US Preventive Services Task Force et al. Current methods of the US Preventive Services Task Force: a review of the process. Am J Prev Med. 2001;20 3 Suppl:21–35. doi: 10.1016/s0749-3797(01)00261-6. [DOI] [PubMed] [Google Scholar]
- Helfand M, Balshem H. AHRQ series paper 2: principles for developing guidance: AHRQ and the effective health-care program. J Clin Epidemiol. 2010;63:484–490. doi: 10.1016/j.jclinepi.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Owens DK, Lohr KN, Atkins D.et al. AHRQ series paper 5: grading the strength of a body of evidence when comparing medical interventions–agency for healthcare research and quality and the effective health-care program J Clin Epidemiol 201063513–523. [DOI] [PubMed] [Google Scholar]
- Slutsky J, Atkins D, Chang S, Sharp BA. AHRQ series paper 1: comparing medical interventions: AHRQ and the effective health-care program. J Clin Epidemiol. 2010;63:481–483. doi: 10.1016/j.jclinepi.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Whitlock EP, Lopez SA, Chang S, Helfand M, Eder M, Floyd N. AHRQ series paper 3: identifying, selecting, and refining topics for comparative effectiveness systematic reviews: AHRQ and the effective health-care program. J Clin Epidemiol. 2010;63:491–501. doi: 10.1016/j.jclinepi.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Lin BK, Clyne M, Walsh M.et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database Am J Epidemiol 20061641–4. [DOI] [PubMed] [Google Scholar]
- Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- Yu W, Clyne M, Dolan SM.et al. GAPscreener: an automatic tool for screening human genetic association literature in PubMed using the support vector machine technique BMC Bioinformatics 20089205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BC, Trikalinos TA, Lau J, Brodley C, Schmid CH. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics. 2010;11:55. doi: 10.1186/1471-2105-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill CM, Roehr JT, McQueen MB.et al. Comprehensive research synopsis and systematic meta-analyses in Parklinson's disease: The PDGene database 2012. in press. [DOI] [PMC free article] [PubMed]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen MB.et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database Nat Genet 200840827–834. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. Springer: New York; 2000. The Nature of Statistical Learning Theory. [Google Scholar]
- Joachims T. Springer: Berlin, Germany,; 1998. Text Categorization With Support Vector Machines: Learning With Many Relevant Features. [Google Scholar]
- Van Hulse J, Khoshgoftaar TM, Napolitano A.Experimental perspectives on learning from imbalanced dataInternational Conference on Machine Learning (ICML)2007935–942.
- Small KM, Wallace BC, Brodley CE, Trikalinos TA.The constrained weight space SVM: Learning with labeled featuresInternational Conference on Machine Learning (ICML)2011865–872.
- Wallace BC, Small KM, Brodley CE, Trikalinos TA.Active learning for biomedical citation screeningKnowledge Discovery and Data Mining (KDD)2010173–182.
- Wallace BC, Small KM, Brodley CE, Trikalinos TA.Modeling annotation time to reduce workload in comparative effectiveness reviewsProc ACM International Health Informatics Symposium (IHI)201028–35.
- Wallace BC, Small KM, Brodley CE, Trikalinos TA.Who should label what? Instance allocation in multiple expert active learningProc SIAM International Conference on Data Mining2011176–187.
- Breiman L. Bagging predictors. Journal of Machine Learning. 1996. pp. 123–140.
- Kang P,, Cho S. EUS SVMs: ensemble of under-sampled SVMs for data imbalance problems. 2006;;4232:837–846. [Google Scholar]
- Chang C-C, Lin C-J.LIBSVM: A Library for Support Vector Machines . http://www csie ntu edu tw/~cjlin/libsvm .
- Seeman E, Boonen S, Borgström F.et al. Five years treatment with strontium ranelate reduces vertebral and nonvertebral fractures and increases the number and quality of remaining life-years in women over 80 years of age Bone 2010461038–1042. [DOI] [PubMed] [Google Scholar]
- Edwards P, Clarke M, DiGuiseppi C, Pratap S, Roberts I, Wentz R. Identification of randomized controlled trials in systematic reviews: accuracy and reliability of screening records. Stat Med. 2002;21:1635–1640. doi: 10.1002/sim.1190. [DOI] [PubMed] [Google Scholar]
- Centre for Reviews and Dissemination . York Publishing Services: York, UK; 2009. Systematic Reviews: CRD's Guidance for Undertaking Reviews in Healthcare. [Google Scholar]
- Higgins J, Green S.Cochrane Handbook for Systematic Reviews of Interventions version 5.1.0The Cochrane Collaboration. http://www.cochrane-handbook.org .
- Institute of Medicine . National Acedemies Press: Washington, DC,; 2011. Finding What Works in Health Care. Standards for Systematic Reviews. [PubMed] [Google Scholar]
- Relevo R, Balshem H. Finding evidence for comparing medical interventions: AHRQ and the Effective Health Care Program. J Clin Epidemiol. 2011;64:1168–1177. doi: 10.1016/j.jclinepi.2010.11.022. [DOI] [PubMed] [Google Scholar]
- Trikalinos TA, Siebert U, Lau J. Decision-analytic modeling to evaluate benefits and harms of medical tests: uses and limitations. Med Decis Making. 2009;29:E22–E29. doi: 10.1177/0272989X09345022. [DOI] [PubMed] [Google Scholar]
- Hayen A, Macaskill P, Irwig L, Bossuyt P. Appropriate statistical methods are required to assess diagnostic tests for replacement, add-on, and triage. J Clin Epidemiol. 2010;63:883–891. doi: 10.1016/j.jclinepi.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Irwig L, Bossuyt P, Glasziou P, Gatsonis C, Lijmer J. Designing studies to ensure that estimates of test accuracy are transferable. BMJ. 2002;324:669–671. doi: 10.1136/bmj.324.7338.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord SJ, Irwig L, Bossuyt PM. Using the principles of randomized controlled trial design to guide test evaluation. Med Decis Making. 2009;29:E1–E12. doi: 10.1177/0272989X09340584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


