Abstract
In mammary tumors, intravital imaging techniques have uncovered an essential role for macrophages during tumor cell invasion and metastasis mediated by an epidermal growth factor (EGF)/colony stimulating factor-1 (CSF-1) paracrine loop. It was previously demonstrated that mammary tumors in mice derived from rat carcinoma cells (MTLn3) exhibited high velocity migration on extracellular matrix (ECM) fibers. These cells form paracrine loop-dependent linear assemblies of alternating host macrophages and tumor cells known as “streams.” Here, we confirm by intravital imaging that similar streams form in close association with ECM fibers in a highly metastatic patient-derived orthotopic mammary tumor (TN1). To understand the in vivo cell motility behaviors observed in streams, an in vitro model of fibrillar tumor ECM utilizing adhesive 1D micropatterned substrates was developed. MTLn3 cells on 1D fibronectin or type I collagen substrates migrated with higher velocity than on 2D substrates and displayed enhanced lamellipodial protrusion and increased motility upon local interaction and pairing with bone marrow-derived macrophages (BMMs). Inhibitors of EGF or CSF-1 signaling disrupted this interaction and reduced tumor cell velocity and protrusion, validating the requirement for an intact paracrine loop. Both TN1 and MTLn3 cells in the presence of BMMs were capable of co-assembling into linear arrays of alternating tumor cells and BMMs that resembled streams in vivo, suggesting the stream assembly is cell autonomous and can be reconstituted on 1D substrates. Our results validate the use of 1D micropatterned substrates as a simple and defined approach to study fibrillar ECM-dependent cell pairing, migration and relay chemotaxis as a complementary tool to intravital imaging.
Keywords: mammary tumor, macrophage, metastasis, cell motility, ECM
Introduction
Cell migration is a signature aspect of various disease processes including the invasion of tumor cells into the surrounding tissue stroma, across the vascular endothelium and dissemination and seeding at distant sites via the circulation. Coordinated cell migration is an important component of cell motility leading to tumor metastasis. Thus, investigating the mechanisms that underlie coordinated cell migration is essential in understanding metastatic spread.1–3
Intravital imaging of tumors in live rodents has been used successfully to characterize the highly complex and detailed carcinoma and stromal cell behaviors within intact primary tumors.4–9 We have used both transgenic mouse models engineered to express fluorescent proteins specifically in the mammary gland (MMTV-PyMT) as well as rat breast carcinoma MTLn3 cells grown in immunodeficient mice or immunocompetent rats, respectively, in conjunction with multi-photon microscopy. Such imaging approaches allow for observations at single-cell resolution and permit quantification of parameters of individual cell motility, as well as interactions between tumor and stromal cells that occur during tumor invasion, intravasation and extravasation. In mammary tumors, intravital imaging techniques have been utilized to define the essential role of macrophages in these behaviors (reviewed in refs. 4 and 10–12). Specifically, tumor cell chemo-taxis toward macrophages is essential for invasion within primary mammary tumors and in vitro,13,14 whereas chemotaxis of tumor cells toward perivascular macrophages is required during intravasation in vivo.15 Both of these events are mediated by a paracrine interaction involving reciprocal signaling between carcinoma cells and macrophages involving EGF receptor ligands and the macrophage growth factor, CSF-1. This paracrine loop sustains a relay chemotaxis signal between the two cell types, enabling them to travel long distances within the primary tumor13,14,16,17 and promote the migration of tumor cells to blood vessels, giving rise to increased intravasation.3,10,13,16
Utilizing second harmonic light scattering, intravital multi-photon microscopy can be used for the simultaneous imaging of the extracellular matrix (ECM) components and the tumor-associated cells that reside within the tumor microenvironment. Imaging of fluorescent protein (FP)-expressing rat MTLn3 carcinoma orthotopic mammary tumors in mice revealed enhanced tumor cell velocity when migrating on ECM fibers.18,19 Two distinct patterns of cell movement were consistently observed: streaming movement, defined as coordinated cell motility whereby the tumor cells align and move linearly in an ordered single file manner, and random cellular movement, defined as uncoordinated carcinoma cell motility independently of other carcinoma cells.3 Furthermore, macrophages were found to co-migrate with carcinoma cells during streaming movement in a coordinated manner.3 We have previously found that streaming migration is overall more efficient than random cell movement in vivo, with cells in streams moving significantly faster and with an increased path length and directionality compared with randomly moving single cells.13,16,19 Interestingly, streaming movement was often found to occur along ECM fibers aligned perpendicular to nearby blood vessels.16 These observations suggest that ECM fibers play an important role in organizing and facilitating streaming migration of carcinoma cells and macrophages in vivo.
In order to further study the minimum requirements for ECM-associated macrophage/tumor cell pairing and streaming as observed in vivo by intravital imaging, an in vitro 1D model of the tumor ECM utilizing adhesive linear micropatterned substrates was evaluated. Contrasting fluorescently labeled tumor cells, macrophages and 1D matrix were imaged by wide-field microscopy in vitro and their motile behavior analyzed by time lapsed animation. This approach was used to determine if various motility parameters including cell protrusion, enhanced tumor cell migration on ECM fibers due to carcinoma cell/macrophage pairing and streaming migration is the result of cell-autonomous behaviors.
Results
Carcinoma cell/macrophage co-migration on ECM fibers within orthotopic human breast tumors in vivo
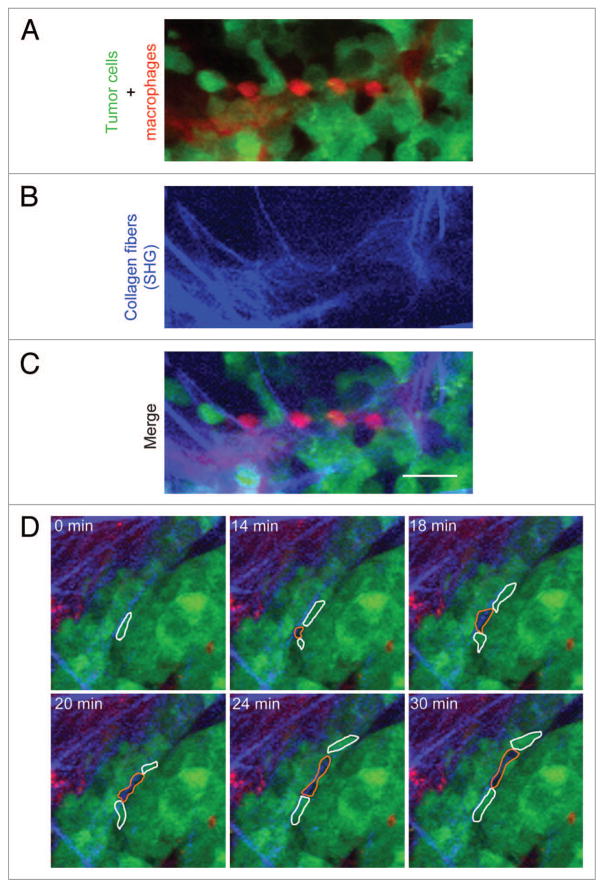
In order to determine whether the in vivo motility behaviors described in rat and mouse tumors would be present in a mammary tumor derived from primary human breast cancer cells, we used the TN1 breast tumor generated as described previously20 for our study. TN1 human breast tumor cells were isolated from a pleural effusion of a patient with breast cancer, labeled by stable expression of GFP and injected in the mammary fat pad of immunodeficient mice. TN1 orthotopic tumors in mice are invasive and metastatic,20 and therefore an appropriate model to study invasion of human breast cancer cells in vivo. When TN1 tumors were imaged by intravital multiphoton microscopy, we observed pairing of tumor cells with macrophages on collagen fibers (Fig. 1A–C), similarly to what has been observed before for mouse and rat mammary tumors. We also observed that TN1 tumor cells exhibited high velocity amoeboid migration in association with collagen fibers as unattached cells that followed each other, frequently retaining a space between cells of approximately 10–20 μm (Fig. 1D; Vid. S1). When individual TN1 cells migrated over the tumor, it was obvious that the space observed between the moving TN1 cells was indeed a host cell, identified here as a moving shadow (Fig. 1D; Vid. S1). We have previously identified these shadow host cells as macrophages.15,16 This data, combined with the visualization of tumor cell-macrophage pairs (Fig. 1A–C), suggest that cell migration in the TN1 tumors occurs in streams with tumor-associated macrophages (i.e., single file of unattached tumor cells and alternating macrophages following one another). This type of motility is similar to what has been reported previously for mouse and rat mammary tumors16,19 demonstrating that the tumor cell migration behaviors observed in rodent mammary tumors are also evident in patient-derived mammary tumors. This finding raises the interesting question of whether the ability of tumor cells to pair and stream with macrophages are autonomous properties of the interacting macrophages and tumor cells.
Figure 1.
Streaming migration in a patient primary tumor cell-derived mammary tumor in vivo. (A–C) A representative image from intravital multiphoton microscopy of the human mammary tumor (TN1) in a living mouse. Tumor cells stably express GFP (green), mouse macrophages were labeled by tail-vein injection of Texas-red dextran (red), and collagen fibers were visualized by second harmonic generation (SHG) as blue fibers (blue). (A) The image shows the pattern of alternating tumor cells (green) and macrophages (red). (B) SHG channel showing the collagen fibers. (C) Merge image of A and B, showing the alternating tumor cells and macrophages aligned on the collagen fibers. The stream consists of the alignment of macrophage-tumor cell pairs. Scale bar = 25 μm. (D) Stills from Video S1 showing TN1 cells (green cells outlined in white) moving coordinately in a stream with host macrophages (black shadow outlined in orange). Note that the stream in Video S1 is appearing at the 14 min time point, because the streaming cells are moving up from the optical section immediately below. Optical sections were taken at 5 μm apart.
Modeling the tumor ECM on 1D adhesive substrates
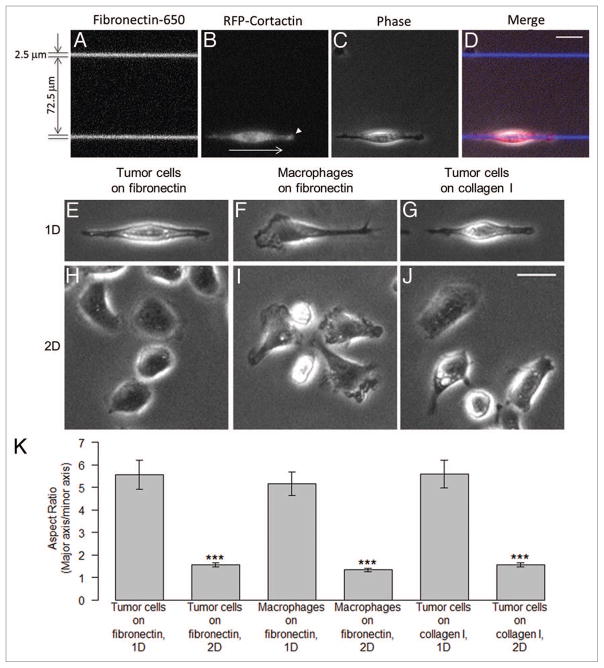
The in vivo motility rates of MTLn3 tumor cells measured by intra-vital microscopy were significantly higher on linear ECM fibers than on 2D substrates in vitro.13,16,19,21 Our analysis of mouse and rat tumors revealed that breast tumor cells move in vivo as unattached amoeboid cells at high velocities (~3 μm/min), often using the collagen fibers of the tumor ECM as “highways.”16,18,19 We set out to investigate this effect in a simplified model system utilizing micropatterned 1D adhesive substrates to mimic the linear ECM fibers of the tumor microenvironment. In vivo, these fibers are believed to be composed of collagen type I and fibronectin based on their ability to generate a second harmonic signal and the requirement of β1 integrin for tumor cell migration.22 Since ECM fibers imaged by second harmonic within orthotopic or transgenic tumor models ranged from 2–5 μm in width,18 we chose a CYTOOchip™ Motility coverslip micropatterned with linear 2.5 or 5 μm wide adhesive stripes from CYTOO Inc. for all time lapse experiments. MTLn3 cells stably expressing TagRFP-cortactin, a marker for actin polymerization-dependent protrusions such as lamellipodia and invadopodia, were plated on 650-labeled fibronectin micropatterned stripes in the presence of 5% FBS, (Fig. 2A–D). MTLn3 cells spread equally well on 2.5 and 5 μm stripes composed of either fibronectin or type I collagen within 3 to 4 h and adopted a similar highly elongated and polarized morphology, regardless of the substrate (Fig. 2E and I). This observation is similar to the “uni-axial” phenotype of NIH-3T3 fibroblasts migrating on 1D adhesive micropatterened substrates.23 During migration on the fibronectin stripe, active ruffling and protrusion occurred at one axis of the cell, as marked by enhanced TagRFP-cortactin accumulation at the leading edge (Fig. 2B, white arrowhead), while the extension of lateral lamel-lipodia were suppressed. For comparison, MTLn3 cells plated on the 2D regions of the coverslip coated with fibronectin or type I collagen had a flattened, non-polarized morphology as quantified by aspect ratio (Fig. 2K) and extended protrusions randomly around the cell surface (Fig. 2F and J).
Figure 2.
Modeling the fibrillar ECM of the tumor microenvironment using in vitro 1D micropatterned adhesive substrates. (A) An area of CYTOOchip™ Motility coverslip showing 650 nm labeled fibronectin 1D stripes with 2.5 μm width and the gap between the stripes of 72.5 μm. (B) A TagRFP-cortactin expressing MTLn3 cell on a 1D stripe in RFP channel and (C) the corresponding phase contrast image. A white arrow and an arrowhead in (B) indicate the direction of movement of the cell and cortactin accumulation at the leading edge, respectively. (D) Merge image of A, B and C. (E–J) Representative phase contrast images of the morphology of MTLn3 cells and BMMs on 1D and 2D substrates. (E) MTLn3 cell on 1D fibronectin stripe, (F) MTLn3 cell on 2D fibronectin, (G) BMM on 1D fibronectin stripe, (H) BMMs on 2D fibronectin, (I) MTLn3 cell on 1D type I collagen stripe and (J) MTLn3 cells on 2D type I collagen. (K) Quantification of cell morphologies on 1D and 2D substrates are shown as aspect ratio (major axis/minor axis). Statistical significance was calculated using unpaired, two-tailed Student’s t test. ***p value < 0.001. Error bars represent the SEM. Scale bars = 25 μm.
On 1D adhesive stripes, MTLn3 cells were highly contact-inhibited and displayed rapid changes in cell polarity and directionality, especially when encountering an adjacent cell on the stripe (Vid. S2). The motility of MTLn3 cells remained restricted to the adhesive stripe, with no spreading or migration onto the adjacent non-adhesive regions throughout the duration of the assay (Vid. S2). Cell velocities on 2.5 μm-wide fibronectin or type I collagen stripes averaged 1 μm/min (Fig. 3B; Fig. S1) and persistent unidirectional motility of up to 100 μm or more was routinely observed while maintaining a highly polarized morphology. In contrast, MTLn3 cells plated on the 2D regions of the coverslip and displayed relatively low rates of motility (< 0.1 μm/min). These results suggest that the increased motility measured in vivo relative to the speeds using traditional 2D substrates may be due to the 1D geometry of the micropatterned substrate mimicking the geometry of fibers observed in vivo.
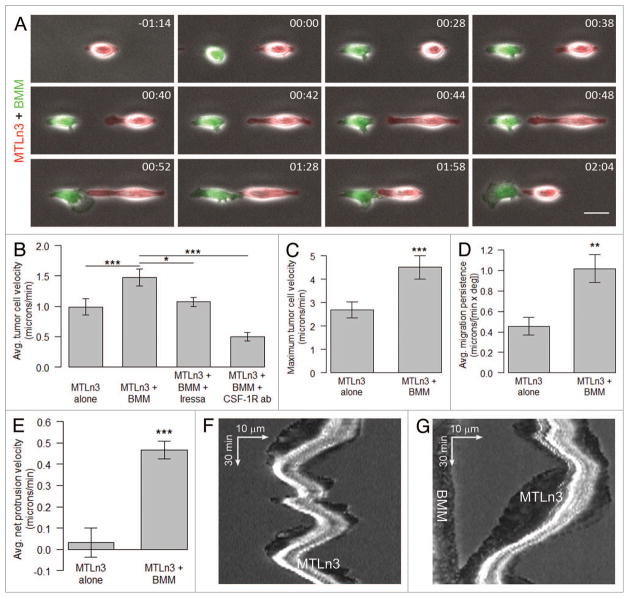
Figure 3.
Tumor cell-macrophage pairing and stimulated motility on 1D adhesive substrates is dependent on EGF/CSF-1 paracrine signaling. Stills from Video S3 showing TagRFP-cortactin expressing MTLn3 cells plated for 3–4 h on 2.5 μm-wide unlabeled fibronectin stripes and time-lapse imaged with a 20×objective for 1.5–2 h to establish baseline motility rates. Following addition of an equal number of CellTracker™ green-labeled BMMs, the cells were imaged for 6 h, treated with either EGFR inhibitor Iressa (1 μM) or the CSF-1 receptor function blocking antibody AFS98 (50 μg/μl) and imaged for an additional 8 h. Fluorescence and phase channels were merged. (A) An MTLn3 cell (red) and a BMM (green) interacting with each other on the 1D stripe. Time is in hr:min. Scale bar = 25 μm. (B) Average tumor cell velocity (μm/min) for MTLn3 cells alone, MTLn3+BMM, MTLn3+BMM+Iressa and MTLn3+BMM+CSF-1R antibody. (C–E) The maximum tumor cells velocity (μm/min), average migration persistence [μm/(min × deg)] and average net protrusion velocity (μm/min) for MTLn3 cells alone and MTLn3+BMMs. (F) Kymograph plot showing random MTLn3 cell movement in the absence of BMMs. (G) Kymograph plot showing directed MTLn3 cell protrusion and motility toward the BMM. Statistical significance was calculated using paired, two-tailed Student’s t-test. *p value < 0.05, **p value < 0.01 and ***p value < 0.001. Error bars represent the SEM.
Since previous studies employing intravital imaging of both xenograph and transgenic mammary tumors identified macrophages as the primary host cell co-migrating with tumor cells13,19,21 we set out to determine if we could reconstitute macrophage/tumor cell pairing and co-migration on the micropatterned 1D substrates in vitro. In our 1D motility assay, BMMs expressing GFP or labeled with CellTracker™ green plated on 2.5 or 5 μm wide adhesive stripes, rapidly attached and began to migrate within 15 min. BMMs displayed a highly polarized shape on 1D compared with 2D substrates (Fig. 2G, H and K) and extended broad, fan-shaped lamellipodia that were not limited to the adhesive stripe during migration (Vids. S4 and S6). This is in contrast to the leading lamellipodia of MTLn3 cells that were considerably smaller and confined to the adhesive stripe (Vid. S2). Unlike MTLn3 cells that are highly contact-inhibited for motility, BMMs were often observed migrating over the surface of the tumor cells (Vids. S4 and S5), a behavior highly reminiscent of ECM fiber-independent motility exhibited by tumor macrophages in vivo.18
To investigate the interaction of tumor cells and macrophages in our 1D motility assay, TagRFP-cortactin labeled MTLn3 cells and GFP-expressing BMMs were plated onto 1D adhesive stripes and their behavior was observed by time lapsed imaging. Within 30 min of a BMM attaching and spreading in the vicinity of an MTLn3 cell (< 2 cell diameters), the tumor cell extended a protrusion toward the BMM. This was also accompanied by a rapid reciprocal protrusion response by the BMM toward the tumor cell (Fig. 3A). Average net MTLn3 cell protrusion velocity increased approximately 15-fold from 0.03 μm/min in the absence of BMMs to 0.47 μm/min in the presence of BMMs (Fig. 3E). Kymographic analysis confirmed this increase in MTLn3 cell protrusion following the addition of BMMs (Fig. 3F and G). Occasionally, a fraction of BMMs migrated off the adhesive stripe, across a non-adhesive area and onto an adjacent adhesive stripe containing a tumor cell (Vid. S4 and Fig. S2). These findings support the concept that macrophages actively migrate toward tumor cells on any available substrate. The increase in MTLn3 cell protrusion was accompanied by a 1.5 fold increase in average tumor cell velocity after addition of BMMs (Fig. 3B). Maximum tumor cell velocity in 1D increased from 2.69 ± 0.35 μm/min (MTLn3 alone) to 4.52 ± 0.50 μm/min (MTLn3 plus BMMs), a range in agreement with the high velocity migration values of tumor cells on ECM fibers (3.4 μm/min) observed in vivo.19,21 In addition, the average MTLn3 cell migration persistence on 1D substrates increased 2.2 fold in the presence of BMMs (Fig. 3D).
Stimulation of tumor cell motility by macrophages on 1D adhesive substrates is dependent on an EGF/CSF-1 paracrine signaling
Previous studies have demonstrated a paracrine interaction between carcinoma cells and macrophages that depends on EGF secretion by macrophages and binding to the EGF receptor (EGFR) on tumor cells inducing CSF-1 secretion by tumor cells in xenograft and transgenic mammary tumors13 and MDA-MB-231 cells.17 Since this paracrine interaction is essential for tumor cell migration in mammary tumors,13 we examined whether the enhancement in tumor cell motility in the presence of macrophages we observe in our 1D model is dependent on this EGF/CSF-1 paracrine loop. Treatment with Iressa, an inhibitor of EGFR signaling, reduced MTLn3 cell protrusion and decreased average cell motility to levels equivalent to MTLn3 cells alone (Fig. 3B). Similar results were obtained with the EGFR inhibitor AG-1478 (data not shown). In addition, inhibition of CSF-1 receptor signaling with the function-blocking antibody AFS-98 reduced average tumor cell velocity by 66% of MTLn3 + BMM. (Fig. 3B) supporting a role for the EGF/CSF-1 paracrine loop during macrophage-enhanced tumor cell motility in our 1D model of the tumor ECM.
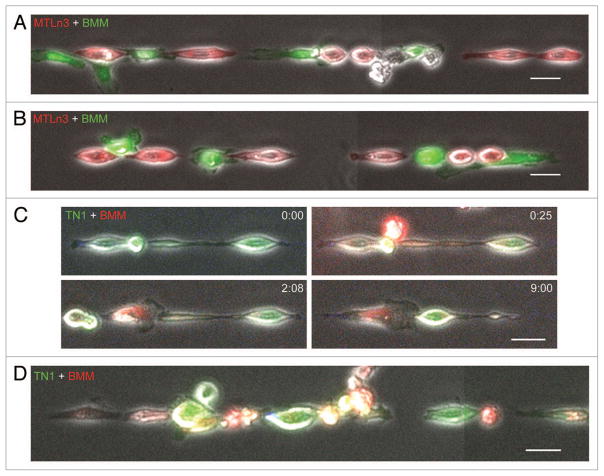
Patient-derived TN1 tumors, imaged here for the first time, in addition to mouse and rat mammary tumors imaged previously by intravital multiphoton microscopy,16,19 have revealed that tumor-associated macrophages can co-assemble into linear streams with alternating tumor cells along the ECM fiber (Fig. 1A–C). Intriguingly, we observed similar co-assembly of alternating MTLn3 cells and BMMs in our 1D model (Fig. 4A and B). These co-assemblies occurred more often in regions along the 1D substrate where roughly equal ratios of MTLn3 cells and BMMs were bound. Changing the ratio of MTLn3 cells to BMMs (2:1 or 1:2, respectively) during plating on the 1D substrate did not significantly alter the occurrence of steams. Furthermore, these co-assemblies of alternating MTLn3 cells and BMMs persisted for several hours (Vid. S6). To test the validity of the 1D assay more directly, we investigated whether this co-assembly on 1D substrates could occur with human patient-derived TN1 cells. We found that TN1 cells demonstrated pairing behavior (Fig. 4C) and formed streams with BMMs (Fig. 4D) similar to MTLn3 cells (Fig. 3A, 4A and B). Taken together, these observations suggest that in vivo cell migration behaviors observed in rodent and human tumor models can be successfully reconstituted on linear micropatterned 1D substrates and their co-assembly is cell autonomous and dependent on an intact EGF/CSF-1 paracrine interaction.
Figure 4.
Spontaneous assembly of streams of tumor cells and BMMs on 1D adhesive substrates. MTLn3 cells stably expressing TagRFP-cortactin were plated for 3–4 h on 2.5 μm-wide unlabeled fibronectin stripes. After time lapse imaging for 1.5–2 h, CellTracker™ green-labeled BMMs were added and imaged for an additional 6 h. Regions of the stripe that contained tumor cells and BMMs (A and B) were capable of forming streams of alternating tumor cells and macrophages like those observed in Figure 1 by intravital imaging of patient TN1 tumors. Refer to Video S6 for an example of MTLn3+BMM stream formation on 1D stripes. (C) GFP expressing TN1 cell pairing with CellTracker™ red-labeled BMM. Time is in hr:min. Panel labeled with time 0:00 shows TN1 cells on 1D stripes before the addition of BMMs. Panel labeled with time 0:25 shows the beginning of TN1 and BMM cell pairing. (D) A representative image showing a stream of alternating TN1 cells (green) and BMMs (red) on 1D substrates. Scale bars = 25 μm.
Discussion
The tumor microenvironment is composed of a complex array of cells including macrophages, fibroblasts, adipocytes, immune cells and tumor-associated vasculature and supporting ECM fibers. The ECM is an important determinant of the tumor microenvironment and acts as the primary scaffold for cell migration within the tumor.10 Tumor-associated cells can adhere and interact with various ECM substrates within the tumor microenvironment such as collagen, fibronectin and laminin through their integrin receptors. Regulation of integrin binding is critical for mediating tumor-associated cell attachment and de-adhesion that occurs during invasion and migration on various ECM substrates. In addition to using ECM fibers as scaffolds, tumor-associated cells can facilitate invasion by degrading ECM proteins by secretion of matrix metalloproteases (MMPs) at specialized structures called invadopodia (carcinoma cells) or podosomes (macrophages).24,25
Investigations into the tumor microenvironment have been facilitated by multiphoton-based intravital microscopy of living tumors. The ability to simultaneously image fluorescently tagged tumor-associated cells and α-helical containing ECM proteins, such as type I collagen, by second-harmonic generation, without the need for a fluorescent tag, has proven to be a powerful tool in gathering detailed observations of the complex cell interactions with the ECM during invasion and metastasis.4,19 Mammary tumor cells derived from transgenic mouse models (MMTV-PyMT) as well as rat MTLn3 tumors grown in immunodeficient mice, display a highly efficient form of migration called streaming that involves the coordinated motility of amoeboid cells that are not in direct contact with one another.13,16,18,19 For the first time, we show by intravital imaging that streaming migration occurs on fibrillar collagen in a patient-derived primary mammary tumor.
In mammary tumors, streaming occurs in association with fibrillar ECM and requires macrophages. In vivo, tumor cell motility rates during streaming are far greater than observed on 2D substrates in vitro. This high velocity streaming motility is due in part to the linear fibers of ECM which can act as “highways” for cell migration inside the tumor and ultimately guide tumor cells toward blood vessels.4,21 Host cells such as tumor-associated macrophages are also capable of ECM fiber-directed motility while co-migrating with tumor cells in vivo.18
In order to study the minimum requirements for tumor cell motility on fibrillar ECM in vivo, we developed an in vitro assay utilizing linear, fluorescently labeled stripes of adhesive ECM micropatterned onto glass coverslips (CYTOO Inc.). We found that either fibronectin or type I collagen stripes of 2.5 or 5 μm supported high velocity tumor cell motility (Fig. S1) at rates comparable to in vivo values measured on collagen fibers of similar width imaged by second harmonic generation.21 Tumor cell or BMM spreading on 2D areas failed to generate the highly-polarized, uni-axial phenotype associated with high velocity tumor cell motility (Fig. 2) in agreement with NIH-3T3 fibroblasts migrating on similar micropatterned substrates.23 Initial studies demonstrated that MTLn3 cells in suspension or attached to 2D substrates showed transient protrusive activity over their entire cell surface upon stimulation with EGF.26 However, when MTLn3 cells were plated on micropatterned 1D adhesive vitronectin substrates, lamellipodial protrusion was confined to the distal ends along the axis of the linear stripe, while the extension of lateral lamellipodia over adjacent nonadhesive surfaces were shorter and highly unstable, demonstrating that adhesion to the underlying substrate, dispensable for actin-dependent protrusion, is required for stabilization and final extent of lamellipodial extension.26 Our high-resolution time lapsed analysis (Fig. 2; Vids S3–5) suggests that suppression of lateral lamellipod extension plays a significant role in generating persistent, high velocity tumor cell motility that we observe in our 1D model as well as tumor imaged by multiphoton microscopy in vivo. We propose that cell migration on 1D substrates accurately reflects the tumor cell behavior and morphology observed within the context of a highly fibrillar in vivo tumor microenvironment.
In the metastatic tumor microenvironment, both streaming tumor cell motility and intravasation require an interaction with macrophages through a paracrine loop,5,15 whereby chemotaxis to EGF secreted by macrophages stimulates CSF-1 release by tumor cells. Since macrophages chemotax toward CSF-1, this can promote pairing of the two cell types as well as the formation of higher order interactions between the two cells by relay chemotaxis, resulting in paracrine-dependent streaming.16 Based on intravital images of patient-derived TN1 breast tumor cells grown as orthotopic xenografts in mice shown here as well as previous data collected from rodent breast carcinoma models,16 we conclude that streaming motility is a conserved phenomenon during metastasis of breast carcinoma.
Using 1D micropatterned adhesive stripes to study the fibrillar nature of the tumor ECM, we observed that MTLn3 cells in the presence of BMMs could significantly increase their protrusion and velocity upon close interaction, and this cell pairing depended on an intact EGF/CSF-1 paracrine loop. Moreover, in the presence of BMMs, MTLn3 cells as well as human patient-derived TN1 cells were capable of forming assemblies on 1D substrates that resembled linear arrays of alternating tumor cells and macrophages identified as streams in vivo. These results indicate that the assembly of streams is cell autonomous and can be reconstituted on 1D adhesive substrates without any additional co-factors present.
Despite differences in cell migration and morphology on 2D substrata vs. 3D matrices, the use of high-throughput 2D assays for evaluating cell motility are still widely used. Recent studies have concluded that growth factor-elicited membrane protrusion but not 2D motility correlated with enhanced migration of human breast carcinoma lines in 3D matrices.27 Our data support this finding as evidenced by the enhanced cell polarization and motility of MTLn3 cells in 1D vs. 2D. Since the uni-axial phenotype and enhanced motility of carcinoma cells migrating in 1D are highly representative of 3D motility, our 1D micropatterned substrate model more closely approximates the fibrillar nature of the in vivo tumor microenvironment and offers a simple and more appropriate substrate for detailed analyses of cell protrusion, cell-cell pairing and migration than conventional 2D substrates.
The data presented here validates the use of micropatterned 1D adhesive substrates to study the fibrillar ECM found within the tumor microenvironment. The composition and geometry of the 1D micropatterned substrate can be easily customized in order to study the role of the ECM during high velocity tumor cell migration, cell-cell pairing and streaming. Cell motility parameters such as velocity, persistence, lamellipodial protrusion and retraction can be analyzed with high spatial and temporal resolution by time-lapsed, wide-field fluorescence microscopy. Furthermore, this 1D model of the tumor ECM can facilitate screening for novel gene and drug targets involved in the regulation of EGF/CSF-1 paracrine signaling and applied to monitor relay chemo-taxis and assembly of tumor cell and macrophage streams as a high-throughput assay complementary to imaging of tumors in the living animal by multi-photon microscopy.
Materials and Methods
Cell lines and reagents
All cells were maintained at 37°C in a 5% CO2 incubator. MTLn3 cells derived from the 13762NF rat mammary adenocarcinoma, were cultured in α-MEM (Life Technologies) supplemented with 5% FBS (Gemini Bio-products) and 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies). An MTLn3 cell line stably expressing TagRFP-labeled murine cortactin was generated by geneticin (Life Technologies) selection. For purposes of the 1D assay, TN1 cells stably expressing GFP were briefly cultured in DMEM-F12 (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals) and 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies).
Murine bone marrow-derived macrophages (BMMs) were isolated from wild type or cfms-GFP mice and prepared according to a previously published protocol,28 and grown in α-MEM media with 15% FBS (Sigma), 360 ng/ml recombinant human CSF-1 (Chiron), and 100 U/ml penicillin and 100 ug/ml streptomycin. GFP expressing- or unlabeled BMMs were CSF-1 deprived overnight prior to the experiment.
Paracrine loop interactions between MTLn3 cells and BMMs were inhibited by addition of the EGFR inhibitor AG-1478 (Tyrphostin; Cell Signaling) at 5 μM in DMSO or Iressa (AstraZeneca) at 1μM in DMSO or the rat monoclonal function-blocking antibody against mouse CSF-1 receptor at 50 μg/μl (AFS-98) (NOVUS Biologicals).
Intravital imaging
Human-in-mouse breast cancer orthotopic mammary tumors (TN1) derived from patient tumor cells labeled with optical reporter fusion genes were imaged by time-lapse multiphoton microscopy. The TN1 human-in-mouse tumor model has been described in detail elsewhere.20 Briefly, pleural effusion breast tumor cells were isolated from a patient with triple negative breast cancer (ER-/PR-/Her2-), transduced with lentiviral particles to stably express GFP and then injected in the mammary fat pad of a NOD.SCID mouse (Jackson Laboratories). The TN1 tumor cells were never passaged in culture and only passaged in vivo orthotopically in NOD.SCID mice.
Intravital multiphoton imaging in the TN1 patient-derived tumors was performed with methods similar to previous studies16 using an Olympus FV1000-MPE microscope with a 25×, NA = 1.05 water immersion objective with correction lens. The laser-light source consists of a standard femtosecond-pulsed laser system (Tsunami-Millennia, Newport/Spectra-Physics) used for excitation of fluorophores in the range of 740–950 nm (e.g., CFP, GFP and YFP). The fluorescence and second-harmonic signals generated were collected via a dichroic mirror and sent to two photomultiplier-tube (PMT) detectors to allow detection of CFP (and second harmonic generation), GFP and yellow fluorescent protein (YFP). A portion of the emission of 70 kDa Texas Red 2 dextran can be detected in the YFP channel. Random fields were imaged of 512 × 512 μm at 512 × 512 pixels for total 100 μm depth (21 slices) beginning at the edge of the tumor. To visualize tumor cell motility and streaming behavior, images were taken at 2 min intervals for a total of 30 min. Time-lapse animations were then assembled and analyzed in 3D and through time using ImageJ. A cell movement event was defined as a translocation of > 1 cell diameter (20 μm) observed within a visual field as defined above. In the TN1 tumors imaged in this report, tumor cells stably expressed GFP. Host macrophages were visualized either by labeling them by tail vein injection of 70 kDa Texas Red dextran 2 h before the start of intravital imaging, or by their appearance as shadows on the GFP-expressing tumor due to light scattering, as described previously.15 The extracellular matrix fibers (ECM) within the tumor were imaged by second harmonic light scattering. The second harmonic signal was excited at 760–960 nm and imaged through a filter with a 450–480 nm cutoff.
In vitro 1D tumor ECM motility assay
Tumor cell and BMM motility was analyzed in vitro on a 20 × 20 mm2, 170 μm (#1.5) thick, coverslip micropatterened by microphotolithography with parallel linear stripes of fibronectin labeled with 650 nm dye. (CYTOO chips™ Motility; CYTOO Inc.). CYTOO chips™ Motility containing parallel 2.5 and 5.0 μm wide adhesive stripes, separated by 72.5 and 70 μm of non-adhesive area, respectively, were selected for time lapse imaging. For experiments on 2.5 and 5.0 μm wide type I collagen stripes, CYTOO chips™ Motility, pre-activated coverslips were incubated with 40 μg/ml FITC-conjugated type I collagen derived from bovine skin (Sigma) for 2 h and rinsed 3 times with d-PBS according to manufacturer’s instructions. Approximately 1 × 105 TagRFP-cortactin MTLn3 cells or GFP-TN1 cells were plated on CYTOO chips™ Motility fitted with a CYTOO live cell imaging chamber for 3 to 4 h (16 incubator. The cells h for GFP-TN1 cells) at 37°C in a 5% CO2 were briefly rinsed in imaging media (L-15 + 5% FBS) to remove unattached cells before imaging.
1D imaging
Time lapse images were acquired with a Photometrics CoolSnap HQ2 CCD camera on an integrated wide-field DeltaVision microscope equipped with a high precision X and Y NanoMotion III stage for capturing images at multiple positions (Applied Precision, LLC). Imaging was done at 37°C with a 20× air objective, NA = 0.4. Up to 10 different fields along an adhesive stripe were imaged in the FITC, TRITC, Cy5 and phase channels every 2 min. TagRFP-cortactin-MTLn3 cells or GFP-TN1 cells were imaged for 1.5–2 h. For tumor cell and BMM co-migration experiments, 1 × 105 BMMs (either GFP-BMMs or CellTracker labeled BMMs) were added to the imaging chamber on the microscope stage and imaging was continued for the next 6 h. For EGFR inhibition (with Iressa or AG-1478) or CSF-1R inhibition (with function blocking antibody, AFS-98), inhibitors or the antibody was added to the imaging chamber on the microscope stage and imaging was continued for 8 h. All imaging was done in imaging media (L-15 + 5% FBS) and the time-lapse conditions did not have any significant effect on cell viability as evidenced by mitosis which occurred during the course of the imaging.
Image analysis
Time-lapse movies consisting of 3 fluorescent channels and a phase contrast channel were assembled by using the “Combine…” command in ImageJ. A montage of 6–7 adjacent fields of totaling approximately 2 mm of continuous stripe length was analyzed. Images were thresholded, cell centroid and protrusions were measured in each frame by custom written macros in ImageJ. Average migration persistence was calculated as speed/[1+(100/360) × angle], where angle is the directional change in degrees.29 On 1D stripes angle value is either 0 or 180, depending on whether the cell persists in the same direction or changes direction. For MTLn3 alone or MTLn3+BMM conditions, reported average tumor cell velocities and persistence (Fig. 3B and D; Fig. S1) are the maximum 1 h average values. For EGFR inhibition (with Iressa) or CSF-1R inhibition (with function blocking, AFS-98), we averaged tumor cell velocities during each hour of the 3–8 h period after inhibitor or antibody treatment and calculated the maximum 1 h average velocity. Net protrusion velocities were calculated as the maximum average values over a 20 min period. Cell morphologies on 1D and 2D substrates were quantified as aspect ratio (major axis/minor axis) in ImageJ. Kymograph plugin in ImageJ was used to make kymograph plots. Tumor cell velocity and persistence calculations were done in Microsoft Excel, data were imported into R software for statistical significance calculation using Student’s t-test. Statistical significance was defined as p value < 0.05. All bar plots were made in R.
Supplementary Material
Acknowledgments
We would like to thank Analytical Imaging Facility and Gruss Lipper Biophotonics Center at Albert Einstein College of Medicine for technical assistance. We also wish to thank Jeffrey Segall and Esther Arwert for their helpful discussions. Grant support was provided by NIH CA100324 (J.S.C, D.C.), CA150344 (R.J.E.), CA164468 (A.P.), a Komen postdoctoral fellowship KG111405 (V.P.S.), NIH T90 DK070103-05, DOD BCRP W81XWH-09-1-0331, 1K12CA139160-02, Chicago Fellows Program and CTSA (UL1 RR024999) at the University of Chicago (H.L.).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Müller A, Homey B, Soto H, Ge NF, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. http://dx.doi.org/10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 2.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–57. doi: 10.1038/nrm2720. http://dx.doi.org/10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 3.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–87. doi: 10.1038/nrc3078. http://dx.doi.org/10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3:921–30. doi: 10.1038/nrc1231. http://dx.doi.org/10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, et al. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–11. doi: 10.1158/0008-5472.CAN-06-3714. http://dx.doi.org/10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- 6.Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1:155–67. doi: 10.1242/dmm.000596. discussion 165 http://dx.doi.org/10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, et al. Intravital imaging of meta-static behavior through a mammary imaging window. Nat Methods. 2008;5:1019–21. doi: 10.1038/nmeth.1269. http://dx.doi.org/10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andresen V, Alexander S, Heupel W-M, Hirschberg M, Hoffman RM, Friedl P. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr Opin Biotechnol. 2009;20:54–62. doi: 10.1016/j.copbio.2009.02.008. http://dx.doi.org/10.1016/j.copbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Perentes JY, Duda DG, Jain RK. Visualizing anti-tumor immune responses in vivo. Dis Model Mech. 2009;2:107–10. doi: 10.1242/dmm.001842. http://dx.doi.org/10.1242/dmm.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–6. doi: 10.1016/j.cell.2006.01.007. http://dx.doi.org/10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi H, Pixley F, Condeelis J. Invadopodia and podosomes in tumor invasion. Eur J Cell Biol. 2006;85:213–8. doi: 10.1016/j.ejcb.2005.10.004. http://dx.doi.org/10.1016/j.ejcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Kedrin D, Wyckoff J, Sahai E, Condeelis J, Segall JE. Imaging tumor cell movement in vivo. Curr Protoc Cell Biol. 2007;Chapter 19(19):7. doi: 10.1002/0471143030.cb1907s35. [DOI] [PubMed] [Google Scholar]
- 13.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–9. doi: 10.1158/0008-5472.CAN-04-1449. http://dx.doi.org/10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 14.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–83. doi: 10.1158/0008-5472.CAN-04-1853. http://dx.doi.org/10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 15.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823. http://dx.doi.org/10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 16.Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci. 2011;124:2120–31. doi: 10.1242/jcs.086231. http://dx.doi.org/10.1242/jcs.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–506. doi: 10.1158/0008-5472.CAN-09-1868. http://dx.doi.org/10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidani M, Wyckoff J, Xue C, Segall JE, Condeelis J. Probing the microenvironment of mammary tumors using multiphoton microscopy. J Mammary Gland Biol Neoplasia. 2006;11:151–63. doi: 10.1007/s10911-006-9021-5. http://dx.doi.org/10.1007/s10911-006-9021-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Wyckoff JB, Frohlich VC, Oleynikov Y, Hüttelmaier S, Zavadil J, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–88. [PubMed] [Google Scholar]
- 20.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107:18115–20. doi: 10.1073/pnas.1006732107. http://dx.doi.org/10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60:5401–4. [PubMed] [Google Scholar]
- 22.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. http://dx.doi.org/10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–90. doi: 10.1083/jcb.200810041. http://dx.doi.org/10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quaranta V. Motility cues in the tumor microenvironment. Differentiation. 2002;70:590–8. doi: 10.1046/j.1432-0436.2002.700912.x. http://dx.doi.org/10.1046/j.1432-0436.2002.700912.x. [DOI] [PubMed] [Google Scholar]
- 25.Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–57. doi: 10.1038/nrm1436. http://dx.doi.org/10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- 26.Bailly M, Yan L, Whitesides GM, Condeelis JS, Segall JE. Regulation of protrusion shape and adhesion to the substratum during chemotactic responses of mammalian carcinoma cells. Exp Cell Res. 1998;241:285–99. doi: 10.1006/excr.1998.4031. http://dx.doi.org/10.1006/excr.1998.4031. [DOI] [PubMed] [Google Scholar]
- 27.Meyer AS, Hughes-Alford SK, Kay JE, Castillo A, Wells A, Gertler FB, et al. 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J Cell Biol. 2012;197:721–9. doi: 10.1083/jcb.201201003. http://dx.doi.org/10.1083/jcb.201201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley ER. Murine bone marrow-derived macrophages. In: Pollard Jeffrey W, Walker John M., editors. Basic Cell Culture Protocols. 2. Totowa, NJ: Humana Press, Inc; 1997. pp. 301–5. [Google Scholar]
- 29.Farina KL, Wyckoff JB, Rivera J, Lee H, Segall JE, Condeelis JS, et al. Cell motility of tumor cells visualized in living intact primary tumors using green fluorescent protein. Cancer Res. 1998;58:2528–32. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.