Abstract
α-synuclein, a presynaptic protein of poorly defined function, constitutes the main component of Parkinson disease-associated Lewy bodies. Extensive biophysical investigations have provided evidence that isolated α-synuclein is an intrinsically disordered protein (IDP) in vitro. Subsequently serving as a model IDP in numerous studies, α-synuclein has aided in the development of many technologies used to characterize IDPs and arguably represents the most thoroughly analyzed IDP to date. Recent reports, however, have challenged the disordered nature of α-synuclein inside cells and have instead proposed a physiologically relevant helical tetramer. Despite α-synuclein’s rich biophysical history, a single coherent picture has not yet emerged concerning its in vivo structure, dynamics, and physiological role(s). We present herein a review of the biophysical discoveries, developments, and models pertinent to the characterization of α-synuclein’s structure and analysis of the native tetramer controversy.
Keywords: Parkinson disease, in-cell NMR, molecular biophysics, protein structure, synucleopathies, α-synuclein
Historical Perspective and Biomedical Relevance
Initially identified in cholinergic synaptic vesicles, a 140-amino acid protein that localized specifically to the presynaptic terminals and nuclei of neuronal cells was aptly termed, “synuclein.”1 Five years after this discovery, the isolation of α-synuclein from amyloid deposits in human patients suffering from Alzheimer disease (AD) sparked an intense research movement toward characterizing the protein’s structure and physiological function(s).2 Protease digestion of the purified AD amyloid samples yielded 2 co-purifying α-synuclein peptides, of which sequence analyses and secondary structure predictions indicated high propensities to form fibrilizing β-sheets.2 These peptides, referred to as the non-Aβ component (NAC, residues 61–95) of AD, fibrilize in a time- and concentration-dependent manner in vitro3 and have since been shown to be necessary to facilitate fibrilization of wild-type (WT) α-synuclein.4 Because of α-synuclein’s seminal association with AD amyloid deposits, in the early literature the protein was occasionally referred to as the NAC precursor (NACP)5 or, rather confusingly, by its other alias, synelfin.6 Additionally, it should be noted that the presence of α-synuclein in AD plaques remains controversial: 2 research groups attempted to reproduce the results from Ueda et al.2 and were unable to find evidence of NAC peptides in AD deposits.7,8
Fluorescence in situ hybridization (FISH) identified the human gene encoding α-synuclein, SNCA, and its cytogenetic address was mapped to the region 4q21.3-q22.9,10 Shortly thereafter, genetic analysis of an Italian family predisposed to early-onset Parkinson disease (PD)11 reported a correlation between the chromosomal region 4q21-q23 and inherited PD.12 These results, together with the previously reported location of SNCA and the toxicity of NAC peptides, prompted a thorough mutational analysis of SNCA in individuals with early-onset PD. A single base pair change at position 209 in SNCA, encoding an A53T mutation in the α-synuclein sequence, was reported in all but 1 affected family member.13 Utilizing secondary structure prediction, the authors postulated that Ala53 resided in a β-sheet-encapsulated portion of an α-helix. It was proposed that the A53T substitution might disrupt local α-helical conformation and result in extension of the surrounding β-sheet structure, leading to protein aggregation and potentially to amyloid formation.13 (Indeed, the A53T variant aggregated more rapidly than WT α-synuclein under oxidizing conditions in vitro).14 The following year, another familial genetic study revealed a correlation between inherited PD and an A30P mutation encoded in SNCA.15 Soon afterward, a newly identified point mutation (E46K)16 and duplication17 and triplication18 of the SNCA locus were reported in individuals with early-onset autosomal dominant PD. Moreover, immunohistochemistry identified α-synuclein as the major constituent of PD-associated cytoplasmic Lewy bodies, the hallmark insoluble aggregates localized to regions of neurodegeneration in both familial and sporadic PD.14 Within the last year, 4 additional pathogenic PD-related missense mutations in SNCA have been reported: A18T,19 A29S,19 and H50Q,20 which were identified in sporadic PD cases, and G51D21 associated with early-onset hereditary PD. Intriguingly, the close sequence proximity of a subset of PD-associated mutational sites (Glu46, His50, Gly51, and Ala53) suggested a critical physiological role for this region of α-synuclein,22 whereas mutations that introduced potential phosphorylation sites (A18T and A29S) pointed to a possible pathological role for certain post-translational modifications (PTMs).19
However, mutations in SNCA account for only a small portion of PD, with spontaneous oligomerization and aggregation of WT α-synuclein encompassing 95% of all PD cases.23 Early diagnostic symptoms of PD, which include rigidity, resting tremors, and difficulty completing basic motor tasks, are precluded by massive dopaminergic neuronal loss in the substantia nigra and other brain regions.24 Although LBs alone cannot account for such neuronal death: studies in animal models and postmortem human brain samples have demonstrated the occurrence of LB-independent and α-synuclein-associated neurodegeneration through other toxic forms of the protein, e.g., soluble oligomers and prion-like propagation (reviewed in refs. 25 and 26). Toxic α-synuclein deposits have also been identified in other neurodegenerative diseases such as dementia with Lewy bodies, PD with dementia, multiple system atrophy, and a host of other Lewy body diseases (LBDs), collectively referred to as “synucleopathies” (reviewed in ref. 27). Conformational plasticity within the native state of α-synuclein, which, depending on local environmental conditions,28 can initiate nucleation of toxic structures (i.e., soluble oligomers and/or insoluble aggregates), has led to its designation as a “protein chameleon.”29-31
Despite heavily focused research, the physiological function(s) of α-synuclein still remain unclear. A plethora of roles have been proposed, including the orchestration of vesicle trafficking;32,33 control of the neuronal apoptotic response;34,35 maintenance of dopamine homeostasis36 and mitochondrial membrane integrity;37 inhibition of mitochondrial fusion,38 vesicle fusion,39,40 complex I activity,41,42 and free radical production;43 formation of transmembrane ion channels;44,45 general chaperone activity;46 and regulation of synaptic plasticity47 and cell signaling.48,49 In addition, recent data suggested that α-synuclein promotes soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-complex assembly by directly binding to vesicle-associated membrane protein 2 (VAMP2).50 This study, along with 2 additional publications, reported that α-synuclein did not affect the kinetics or efficiency of vesicle fusion, but rather enhanced clustering of vesicle-mimics in a VAMP2- and phospholipid-dependent manner.51,52
Historically characterized as an intrinsically disordered protein (IDP), numerous studies have utilized α-synuclein as a model protein to develop technologies to investigate the biophysical parameters of IDPs.53-59 Recent reports, however, have disputed the longstanding notion that α-synuclein is a disordered monomer, and have instead suggested the formation of a helical tetramer under physiological conditions.60,61 In either case, it is hoped that structural analyses of WT α-synuclein will provide insight into its function and also the onset and progression of neurodegenerative diseases. This review recapitulates the historical discoveries, developments, and models that have contributed to our structural understanding of α-synuclein thus far, culminating in a discussion of the recently proposed native tetramer theory and studies on the functional impact of Nα-acetylation.
Low-Resolution Biophysical Analyses of α-Synuclein
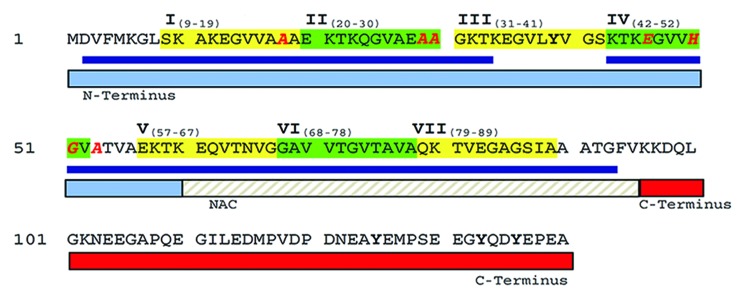
The peptide sequence of α-synuclein comprises 3 distinct regions (Fig. 1): N-terminal residues 1–60 that contain 4 11-residue imperfect repeats with a conserved KTKEGV motif; residues 61–95 that harbor 3 additional KTKEGV repeats and include the hydrophobic and amyloidogenic NAC region; and C-terminal residues 96–140 that are enriched in acidic and proline residues. This C-terminal region facilitates interactions with more than 30 different proteins.62
Figure 1. Amino acid sequence of α-synuclein and its organization. The 7 imperfect 11-residue repeats are highlighted and designated with Roman numerals. Regions found to adopt helical conformation in the presence of micelles are underlined in blue. Mutational sites that are known to, or believed to cause early-onset PD are indicated by red letters. Reprinted with permission from reference 51. Copyright 2012, Springer Science + Business Media.
Early characterization of α-synuclein’s secondary structure utilized circular dichroism (CD) spectroscopy.63 Analysis of the CD spectrum of free α-synuclein led to estimates of less than 2% α-helical and nearly 70% random coil content in solution. The Stokes radius was consistent with a globular protein of 57 kDa rather than 14.5 kDa expected for a protein of 140 amino acids.63 By utilizing the Guiner approximation, small-angle X-ray scattering (SAXS) can be used to determine the radius of gyration (Rg) for a native globular protein of n amino acids by means of the expression, RgN = 2.9 • n1/3.64 Guiner analysis of the X-ray scattering curves for α-synuclein produced a Rg of 41 Å, which was larger than the theoretical Rg of a 140-residue globular protein (15.1 Å) but smaller than that of a completely unfolded polypeptide of an equivalent number of residues (52 Å).65 Thus, α-synuclein was found to lack significant secondary structure and to adopt a conformation that was slightly more compact than the extended random coil state. Corroborating these data, Fourier-transform infrared (FTIR) spectroscopy displayed a broad peak at 1650 cm−1, supporting the notion of a high degree of structural disorder.65
Lipid binding and the 5-helix model
The sequence similarity between the 11-residue repeats of α-synuclein and the amphipathic α-helices of apolipoproteins led Davidson et al.66 to hypothesize that α-synuclein interacted with phospholipid bilayers in a similar manner. Intriguingly, α-synuclein was found to bind exclusively to acidic vesicles.66 As monitored with CD spectroscopy, lipid binding induced a coil-to-helix transition wherein the α-helical content of α-synuclein increased from 2 to 71%. Collectively, these data led to a secondary structure predication-based hypothesis that α-synuclein interacted with lipids in a 5-helix manner,66 hereafter referred to as the 5-helix model. The model required 5 separate helices to optimize the charge distribution over certain stretches of the polypeptide sequence. Reminiscent of class A2 helices, the 4 helices located in residues 1–60 were believed to facilitate lipid binding while the fifth helix located in residues 61–94, presumed to constitute a class G helix, was thought to modulate protein-protein interactions.66
NMR Spectroscopy-Based Analyses of Free and SDS Micelle-Bound α-Synuclein
Residues 1–100 bind SDS micelles in an α-helical conformation
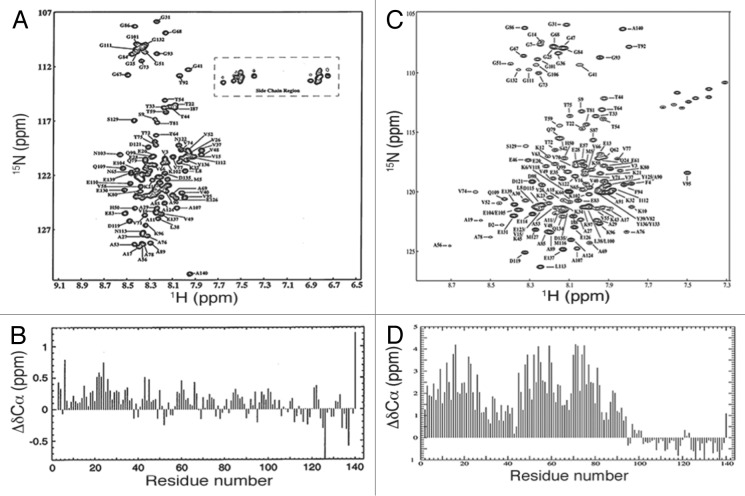
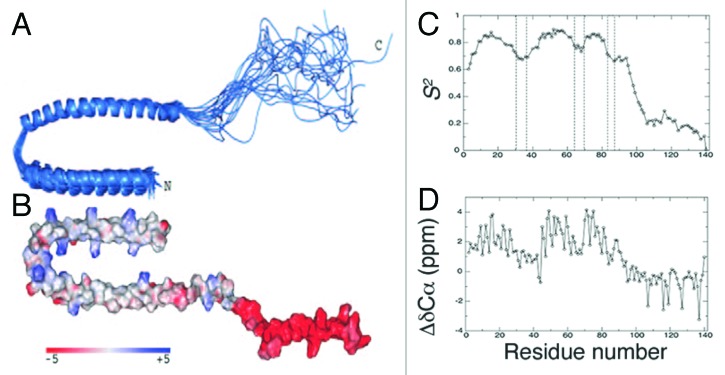
Eliezer et al.67 used multi-dimensional nuclear magnetic (NMR) spectroscopy to further characterize the lipid binding interactions of α-synuclein and accompanying conformational changes. Two-dimensional (2D) 1H-15N heteronuclear single quantum coherence (1H-15N HSQC) spectra, which correlate the chemical shifts of each amide/amino nitrogen to the chemical shifts of its corresponding amide/amino proton(s), provide a “fingerprint” of a protein that is uniformly ([U-]) enriched with 15N, a NMR-active stable isotope. 2D 1H-15N HSQC spectra of [U-15N]-labeled α-synuclein (Fig. 2A) displayed NMR signals with narrow line widths, which generally indicate high backbone mobility, and limited dispersion of 1H chemical shifts, which are characteristic for unfolded proteins. Sequence-specific assignment of these resonances—necessary for the calculation of secondary structure propensities—required the production of [U-13C,15N]-labeled α-synuclein and the acquisition of various 3D NMR spectra. Differences in the experimentally observed 13Cα chemical shift- and the random coil 13Cα chemical shift-values for any given residue type, typically referred to as the secondary 13Cα chemical shift (ΔδCα), report on secondary structure propensities, with α-helical and β-sheet ΔδCα values around +2.6 ppm and –1.4 ppm, respectively.68 Lack of significant ΔδCα values confirmed the absence of pronounced secondary structure in free α-synuclein (Fig. 2B). However, it should be noted that the first 100 residues displayed a slight propensity for α-helical torsion angles, particularly for the region between residues 18–31, which was predicted to populate α-helical conformations about 10% of the time. The C-terminal residues 101–141 appeared to preferentially adopt transient β-sheet conformations.67 Measurements of 15N relaxation rates provided insights into the backbone dynamics of free α-synuclein. The rates with which 15N transverse magnetization decayed (R2) exhibited a maximum around residue 122 of the acidic C-terminus, indicative of diminished relative flexibility that was consistent with a role in modulating protein-protein interactions.69 To investigate the conformational changes within α-synuclein upon lipid binding, the authors chose sodium dodecyl sulfate (SDS) detergent micelles as a lipid-mimetic. Surprisingly, addition of SDS micelles led to the disappearance of 105 resonances from the 2D 1H-15N HSQC spectra of α-synuclein acquired at 10°C.69 The remaining 30 resonances corresponded to those previously assigned to the C-terminal region, suggesting that these residues remained disordered and did not interact with micelles. It was found that NMR signals of N-terminal α-synuclein residues could be observed at 40°C, under conditions that enhanced the tumbling rates of SDS-micelle bound-α-synuclein.69 Comparison of 1H-15N HSQC and HNCA (1HN and 13Cα correlation) spectra between micelle-bound and free α-synuclein revealed that, in the presence of micelles, 1H chemical shifts had dispersed (Fig. 2C) and ΔδCα values of N-terminal residues 1–100 had increased by ~2 ppm (Fig. 2D), diagnostic of helical conformations. These results led to the conclusion that binding of α-synuclein to SDS micelles elicited formation of an extended α-helix encompassing residues 1–100.67,69
Figure 2. 2D 1H-15N HSQC spectra and ΔδCα values of uniformly labeled 15N α-synuclein were indicative of a disordered protein that adopted helical conformations in the presence of SDS micelles. (A) Backbone 1H resonances encompassed small chemical shift ranges, which were suggestive of enhanced backbone mobility (B) Lack of significant ΔδCα values of free α-synuclein suggested a lack of secondary structure. Reprinted from reference 67 with permission from Elsevier. (C) Although signals from the ~100 N-terminal residues were not observed in the 1H-15N HSQC spectrum of this sample at 10°C, they are observed here at elevated temperatures under conditions in which micelles tumble more rapidly. Reprinted from reference 70 with permission from Elsevier. (D) Upon addition of micelles, ΔδCα values in residues 1–100 increased by ~2 ppm, which indicated that α-synuclein had adopted helical conformations. Reprinted from reference 70 with permission from Elsevier.
α-synuclein binds SDS micelles as 2 α11/3-helices
A more detailed analysis of SDS micelle-bound α-synuclein that utilized ΔδCα’s and sequential (i to i+1) 1HN–1HN nuclear Overhauser effects (NOEs), which generally report on through-space interactions between pairs of hydrogen atoms closer than 5 Å, provided the first evidence to disprove the 5-helix lipid-binding model. If the 5-helix model were correct, one would expect to observe relatively small positive or negative ΔδCα changes upon micelle binding in 4 linker regions separating the 5 predicted helices.66,70 Contrary to this notion, data from ΔδCα values and i to i+1 NOE signals indicated the presence of only 1 helical interruption, located between residues Ser42 and Thr44. Sequential 1HN NOE cross-peak intensities for all other residues in the N-terminus of α-synuclein (1–94) were suggestive of helical structure (Fig. 3), and, based on these data, Bussell and Eliezer70 proposed that α-synuclein bound to SDS micelles via 2 helices. Moreover, the HELNET software program, a predictor of lipid binding affinities of amphipathic helices, indicated that the first helix (residues 1–41) could be either an α-helix (18 residues per 5 full turns) or a non-canonical α11/3-helix (11 residues per 3 full turns), whereas the second helix (residues 45–94) would predominately exist as an α11/3-helix.70 The authors postulated that the energetic cost of assuming an α11/3-helix could be alleviated through a combination of hydrophobic interactions with lipids and further stabilized by the amphipathic nature of the helical surface. Additionally, it was noted that the inter-helix break could serve as a hinge to enable interactions with membranes less curved than SDS micelles (e.g., synaptic vesicles).70
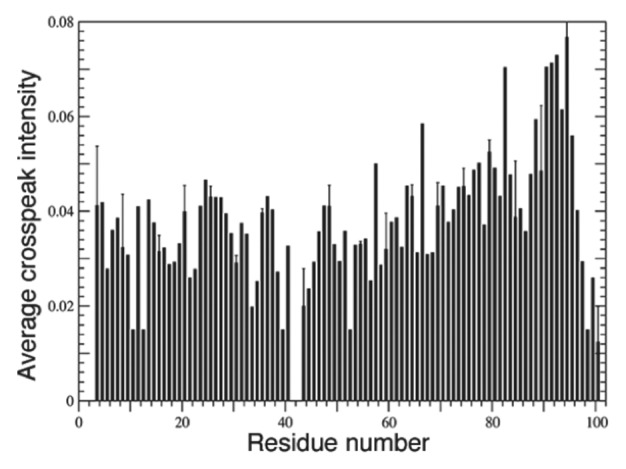
Figure 3. Sequential (i to i + 1) 1HN–1HN NOEs in α-synuclein bound to SDS micelles indicated the formation of 2 broken helices. Only residues 1–100 are depicted here. The lack of NOEs between residue 42 and its neighbors (41 and 43) revealed a break in the helix at this position. All other residues displayed strong NOEs typical of helical conformation. Reprinted from reference 70 with permission from Elsevier.
These results were independently confirmed by limited tryptic digest experiments on α-synuclein in the presence of SDS micelles.71 Theoretically, α-helical regions stabilized by micelle binding should be protected from proteolysis, whereas unstructured and exposed regions are more likely to be cleaved. Indeed, following digestion with trypsin, 2 protected fragments were recovered and identified as N-terminal peptides of 4 and 6 kDa. Based on the size of these peptides, cleavage was hypothesized to occur at Lys43 or Lys45 (i.e., at the unprotected helical break).71 NMR analysis of the solvent accessibilities of α-synuclein amide protons provided a more detailed definition of the inter-helix break site. Because intensities of exchange cross-peaks in 15N 3D (NOESY)-HSQC and (TOCSY)-HSQC NMR spectra are related to the degrees of solvent exposure of the backbone amides, experimentally determined values for disordered C-terminal α-synuclein residues exhibited strong exchange cross-peaks (as expected for solvent exposed amide protons), whereas values for structured residues within the N-terminal helices were weak, or completely absent. Exchange cross-peaks of intermediate intensities were observed for residues Lys43 and Thr44, which independently confirmed limited proteolysis, ΔδCα, and 1HN NOE results.70,71
Paramagnetic relaxation enhancement (PRE) exploits the altered relaxation rates of nuclei in the presence of paramagnetic probes and enables identification of interatomic (i.e., paramagnetic center-nuclear spin) distances up to 25 Å apart. Bussell et al.72 analyzed α-synuclein helix periodicity and the topology of its SDS micelle-bound conformation via PRE measurements. The addition of paramagnetic Mn2+ led to the broadening of most α-synuclein 1H-15N HSQC resonances; however, signals from residues Lys10, Ser42, and Val74 were much less affected and indicated diminished solvent interactions.72 Further inspection of the PRE data revealed a 3- to 4-residue periodicity suggestive of a helical structure. To investigate helical periodicity, the group incorporated 4-OH-TEMPO spin labels into SDS micelles and analyzed the experimental α-synuclein signal intensities as a function of azimuthal angles (11 possible azimuth angles for an 11/3 helix and 18 possible angles for a canonical α-helix). Theoretically, plotting PRE-mediated α-synuclein peak-broadening data against the correct periodicity should yield a sine wave, owing to the oscillations between successive helix residues and the micelle spin probe.72 Sinusoidal traces were indeed observed upon plotting PRE data acquired with 3 different micelle-incorporated spin probes (4-OH-TEMPO, 5-, and 16-doxylstearate) vs. 11/3 periodicity, which prompted the group to conclude that α-synuclein formed 2 α11/3-helices when bound to SDS-micelles.72
Long-range interactions of free α-synuclein desolvate the NAC region
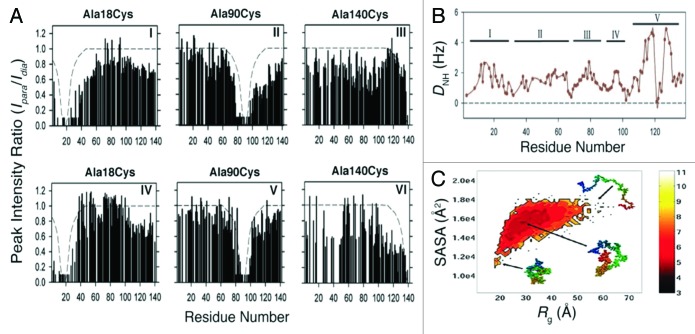
Another group utilized PRE measurements to further characterize the ensemble of structures adopted by free α-synuclein in solution. Bertoncini et al.73 incorporated a cysteine residue at each of three different α-synuclein locations (A18C, A90C, or A140C) and individually labeled each cysteine with (1-oxy-2,2,5,5-tetramethyl-D-pyrroline-3-methyl)-methanethiosulfonate (MTSL). Peak intensity ratios between 2D 1H-15N HSQC spectra of [U-15N], MTSL-labeled α-synuclein with oxidized and reduced spin labels (Ipara/Idia) reported on nearby proton-spin label interactions (Fig. 4A, I –III). Particularly interesting were the long-range interactions observed between spin-labeled A18C and residues 60 and 115–140 (Fig. 4, I) that suggested transient contacts between N- and C-termini. Interactions with residues 115–140 persisted following treatment with 8 M urea, which otherwise unfolded the protein and restricted peak broadening to 15 residues from the spin-label (Fig. 4A, IV; compare with Fig. 4A, V). Spin-labeled A90C was found to interact with all of the C-terminal residues (Fig. 4A, II), whereas the spin label on A140C broadened residues 80–100 (Fig. 4A, III).73 Extensive PREs in spin-labeled A140C were observed upon 8 M urea treatment (Fig. 4A, VI), presumably due to inherent flexibility in C-terminal residues. Measurements of 1-bond 1H-15N residual dipolar couplings (RDCs) of free α-synuclein aligned in both bacteriophage Pf1 and 5% (w/v) n-octyl-penta(ethylene glycol)/octanol (C8E5) were consistent with a hydrophobic cluster centered around residues 115–119 and 125–129 (Fig. 4B).73 Furthermore, similar RDCs identified 4 other domains within the protein: domain I (residues 1–28), domain II (residues 33–65), domain III (residues 61–95, i.e., the NAC region), domain IV (residues 95–105), and the aforementioned C-terminal domain V (residues 115–119 and 125–129) (Fig. 4B). Bernado et al.74 compared these experimentally-derived α-synuclein RDCs73 to calculated RDCs averaged over the protein’s conformational ensembles. Calculated RDCs of residues 30–110 were in close agreement with prior data,73 but the N- and C-termini exhibited complex behaviors that were not accurately represented by the simulation74 However, after dividing α-synuclein’s amino acid sequence into 7 20-residue domains and only accepting conformers that contained inter-domain Cβ–Cβ residue distances of less than 15 Å, calculations yielded RDCs comparable with experimentally acquired values. Of note, sequence-wide calculated RDC distributions agreed with experimental data only when residues 1–20 and 121–140 established contacts, in accordance with previously observed residual N- and C-terminal interactions.73,74 Further dividing domains 1–20 and 121–140 into 5-residue fragments generated highly reproducible RDCs that were dependent on long-range interactions between residues 6–10 and 136–140, specifically. Collectively, these experimental and calculated RDC data suggested nonrandom, long-range interactions between the N- and C-termini of free α-synuclein.73,74
Figure 4. Both PRE in amide protons and experimental and calculated RDCs of free α-synuclein exhibited signs of nonrandom, long-range contacts. (A) Three alanine residues were individually substituted by cysteine to enable spin label incorporation. Intensity ratios of 1H-15N peaks with oxidized and reduced spin labels (Ipara/Idia) revealed persistent interactions between α-synuclein’s N- and C-termini, as well as C-terminal interactions with NAC residues 80–100. Dashed lines denoted expected paramagnetic effects for an equivalent random coil. (I–III) Ipara/Idia ratios of α-synuclein in buffer and (IV–VI) Ipara/Idia ratios in the same buffer plus 8 M urea. (B) One-bond N-H (1)DNH) residual dipolar couplings (RDCs) measured for α-synuclein in aligned media. Roman numberals indicated specific domains within the protein that displayed similar RDCs. (A and B) Adapted from reference 73 with permission. Copyright 2005 National Academy of Sciences, USA. (C) MD simulations-generated free energy landscape of α-synuclein, defined by the expression, F(Rg, SASA) = –ln p(Rg, SASA), depicted quantitatively with a heat scale. SASA denoted solvent exposed surface area and p(Rg, SASA) represented probability distributions of Rg and SASA. Adapted with permission from reference 79. Copyright 2009 American Chemical Society.
Time-resolved tryptophan fluorescence energy-transfer (FET) measurements and electron transfer (ET) reactions independently confirmed the nonrandom contacts observed in free α-synuclein. Lee et al.75 produced α-synuclein analogs with tryptophan substituted at residues Phe4, Tyr39, or Phe94, which functioned as fluorescent donors, and combined these with individually nitrated (NO2) tyrosine residues (A19Y, Tyr39, V74Y, or Tyr136) to act as fluorescent acceptors. FET rates of modified, free α-synuclein variants corresponded to heterogeneous probability distributions of fluorescent donor-acceptor (DA) distances.75 Fluorescent donors and acceptors separated by 15 and 20 residues (e.g., F4W/A19Y-NO2 and V74Y-NO2/F94W) all displayed similar DA distance distributions that contained short (≤ 15 Å, ~10%), intermediate (~20 Å, ~45%), and extended (≥ 30 Å, ~45%) conformations irrespective of sequence location. Intriguingly, ~80% of N- and C-termini (W4/Y136-NO2) distances were ≥ 40 Å, yet 20% fell in the intermediate range near 20 Å. Taken together, these FET data suggested that free α-synuclein rapidly interconverted between a wide range of conformations on the microsecond time-scale.75 These results were in agreement with probability distance distributions derived from photoinduced ET reactions between the triplet excited state of tryptophan (3Trp*) and Tyr-NO2 using the Trp- and Tyr-substituted α-synuclein variants from Lee et al.75,76 Additionally, ET-derived intrachain contact rates of F4W/Tyr136-NO2 significantly deviated from values expected for a random coil conformation, which confirmed that α-synuclein’s N- and C-termini made nonrandom contacts in solution.77
Similar PRE-based measurements and ensemble molecular dynamics (MD) simulations supported the above results. Dedmon et al.78 individually placed MTSL spin-labels at α-synuclein positions Q24C, S42C, Q62C, S87C, or N107C, and 1H-15N HSQC peak-broadening (Ipara/Idia) results from each variant suggested nonrandom, long-range interactions between C-terminal residues and the central NAC region. Ensemble MD simulations, which reported on the likelihood of inter-residue distances shorter than 8.5 Å, included only a subset of experimentally acquired PRE restraints, yet identified highly probable contacts between residues 120–140 and residues ~30–100.78 Using the PRE-data of Dedmon et al.78 and spin-labeled N122C α-synuclein, Allison et al.79 applied ensemble MD simulations with stringent ensemble-averaged restraints and created a free energy landscape of α-synuclein (Fig. 4C). Structures with low Rg but high solvent exposed surface area (SASA) represented the most populated ensembles of α-synuclein conformations, which prompted the authors to hypothesize that such conformations enabled favorable binding interactions in crowded intracellular environments.79
By translating PRE and RDC data into distance restraints, Bertoncini et al.73 used XPLOR-NIH80 to compile the 7 most representative conformations of free α-synuclein in solution (Fig. 5). Torsion angle calculations began with a random coil conformation at 3000 K and followed with simulated annealing and energy minimization to satisfy experimentally derived distance constraints as the temperature was reduced to 20 K. It should be noted that distance calculations derived from PRE measurements often generate large variations, and that averaging over temporal and spatial measurements may combine features that are usually absent in a given molecule.73 Nonetheless, the calculated structures suggested that C-terminal residues 110–130 shielded parts of the hydrophobic NAC region (residues 85–95) and interacted with the N-terminus near Glu20 (Fig. 5).73 These results were consistent with the hypothesis that full-length α-synuclein was less amyloidogenic than truncated α-synuclein (residues 1–102), presumably because C-terminal interactions prevented NAC-mediated aggregation.81 The group concluded that the release of intrinsic long-range interactions would expose hydrophobic NAC patches, such that aggregative β-sheet conformations became accessible and toxic oligomerization and aggregation could occur.73
Figure 5. Native-state conformations of α-synuclein as determined by XPLOR-NIH with structural constraints derived from PRE and RDC measurements. Four clusters in which the 10 lowest-energy structures from each cluster are overlaid. RDCs are shown with a continuous color scale. Note that the C-terminal region contacted NAC residues and N-terminal residues. Adapted from reference 73 with permission. Copyright 2008 National Academy of Sciences, USA.
Solution Structure of α-Synuclein Bound to SDS Micelles
α-synuclein forms 2 canonical α-helices upon binding to SDS micelles
Utilizing RDCs and PRE, Ulmer et al.82 solved the solution structure of SDS micelle-bound α-synuclein. The group measured RDCs in polyacrylamide gels and acquired PRE-derived long-range distance restraints by incorporating a cysteamyl-EDTA tag (complexed with Mn2+) into the α-synuclein variant S89C. Analysis of the structure revealed 2 curved helices, termed helix-N (residues 3–37) and helix-C (residues 45–92), which surprisingly adopted an anti-parallel configuration (Fig. 6A). On average, 1 turn of helix-N contained 3.60 residues with a twist angle of 100.1° and 1 turn of helix-C encompassed 3.56 residues at a twist angle of 101.2°, diagnostic of canonical α-helices. Thus, the SDS micelle binding model with 2 11/3 α-helices was superseded by a high-resolution structure with 2 canonical (18/5) α-helices.82 Intriguingly, the PRE data alluded to variations within inter-helical distances, indicative of enhanced backbone dynamics. General order parameters (S2) were calculated from 15N relaxation data in order to analyze motion on the ps–ns timescale. Three regions within the first 97 residues contained relatively diminished S2 values: residues 30–37, 65–70, and 83–89 (Fig. 6C). These results, in conjunction with measured ΔδCα’s that were ~50% smaller for helix-N than for helix-C, indicated that helix-N occupied a helical conformation significantly less often than helix-C (Fig. 6D).82 Variations in the magnitudes of RDC-derived local alignment tensors (Da) confirmed the enhanced backbone dynamics of this region.82 Bisaglia et al.83 measured R1 and R2 relaxation rates and heteronuclear 1H-15N NOEs for a C-terminal deletion construct of α-synuclein (residues 1–99) in the presence of SDS micelles and provided further evidence in support of a dynamic helix-N. All of the R1, R2, and NOE values for residues 24–44 indicated an increase in relative flexibility. The reduced spectral density functions, which generally report on N-H bond vector fluctuations without making assumptions regarding protein shape, were calculated at 3 different frequencies, J(0), J(ωN), and J(ωH+ωN), and verified the enhanced backbone dynamics that were observed for residues between Lys24 and Thr44.83 Thus, the effectively averaged conformations within the ensemble of NMR structures of SDS micelle-bound α-synuclein could not accurately represent the dynamic nature of helix-N.82,83 It should be noted, however, that helix-N’s larger curvature and shorter length may account for the relatively increased backbone motions with respect to helix-C.82
Figure 6. NMR-derived solution structures, general order parameters, and ΔδCα values of SDS-micelle bound α-synuclein. (A) Cluster of 20 NMR solution structures aligned to helix-C. Backbone RMSD from the average structure for residues 3–97 was 1 Å. (B) Charge distribution mapped onto the solution structure. Heat map depicted gradient from -5 (red) to +5 (blue). (C) S2 measurements plotted as a function of residue number. Dashed regions indicated residues displaying enhanced disorder relative to other helical regions. Note that the C-terminal residues were disordered, as expected. (D) ΔδCα’s upon SDS-micelle binding plotted as a function of residue number. Residues 30–37 displayed smaller ΔδCα’s, indicating diminished helical propensity. The chemical shift differences have not been corrected for the ~+0.5 ppm isotope shift induced by perdeuteration. Reprinted from reference 82 with permission. Copyright 2005 American Society for Biochemistry and Molecular Biology.
Analysis of the charge distribution throughout α-synuclein provided a structural basis for the binding of SDS micelles (Fig. 6B).82 The inner surface of each helix contained largely nonpolar residues that likely facilitated favorable interactions with lipid tails. Sideward-oriented basic side chains could thus promote binding to negatively charged headgroups, which could explain the preferential binding to acidic phospholipids (e.g., PS/PI vesicles).66,82
Vesicular Dependency of Helical Conformation
Binding to small and large unilamellar vesicles as a single 11/3-helix
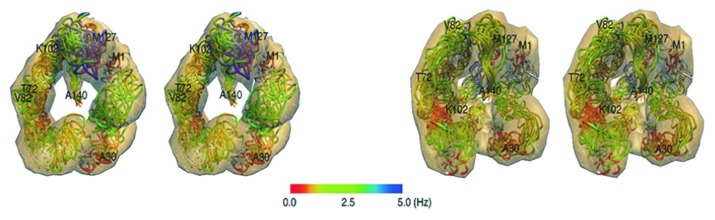
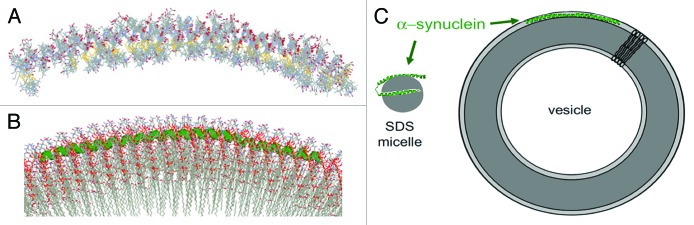
It had been suggested that vesicle size could dictate whether α-synuclein adopted 1 extended helix, or 2 broken helices.84 As noted above, SDS micelle-bound α-synuclein adopted 2 anti-parallel helices (Fig. 6A and B),82 and the authors surmised that the inter-helix linker could function as a “hinge” to mediate interactions with larger vesicles.82 Indeed, subsequent evidence confirmed that α-synuclein adopted a single, extended helix when bound to either small or large unilamellar vesicles (SUVs and LUVs, respectively).85,86 In order to avoid the size limitations of NMR, electron paramagnetic resonance (EPR) and single-molecule (sm)FRET measurements were employed as “molecular rulers.”87 Pulsed EPR data from 17 doubly spin-labeled α-synuclein variants were consistent with the formation of an extended α-helix upon binding to SUVs. By combining inter-spin label distance restraints, measurements of α-synuclein vesicular immersion, and α-helix backbone dihedral angle constraints, the group produced 9 simulated annealing molecular dynamics (SAMD) structural models (Fig. 7A), including a SUV-bound model (Fig. 7B).85 Interestingly, α-synuclein curved to the surface of the vesicle as a single, extended helix. Positioned below the phosphate head groups, central residues Lys58 and Lys60, which protruded outward and upward, established favorable contacts with acidic lipid head groups.85 Even more intriguing was the observed helical periodicity: 3.67 amino acids per turn, or an 11/3 helix, consistent with previous reports.70,72 It was thus concluded that α-synuclein altered its helical structure to accommodate the binding of larger, less-curved vesicles (Fig. 7C).85
Figure 7. EPR structures of α-synuclein (9–89) bound to SUVs. (A) Overly of 9 structures calculated from SAMD simulations. The yellow atoms indicated S-S bonds and enabled identification of spin labels. Red atoms depicted labeled side-chains. (B) Model of α-synuclein (green) interacting with the lipid surface. Note that the protein embedded itself slightly below the head groups (red) and followed the curved surface of the vesicle. (C) Model illustrating α-synuclein conformations when bound to SDS micelles vs. SUVs. The highly curved micelles cannot accommodate an extended helix, which was observed upon binding to SUVs. Adapted from reference 85 with permission. Copyright 2008 National Academy of Sciences.
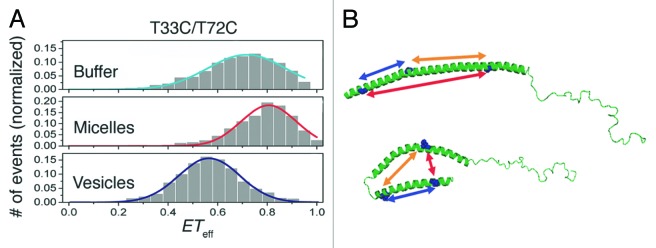
A similar study exploited FRET efficiencies (ETeff), usually dependent on the distances (r−6) between acceptor and donor fluorophores, to further characterize α-synuclein’s helical structure in the presence of both SDS micelles and LUVs.86 ETeff histograms of maleimide-labeled (S9C/T72C) α-synuclein displayed enhanced energy transfers in the presence of micelles, consistent with anti-parallel helices. However, diminished energy transfers when bound to LUVs confirmed the formation of a single, extended helix: the Förster radius of maleimide fluorophores is 54 Å, whereas the distance between spin labels on S9C and T72C in an unbroken helix is ~90 Å, a distance at which ETeff approaches zero. Upon moving the fluorophores closer together (T33C/T72C), the ETeff for micelle-bound α-synuclein was 0.81, which further supported an anti-parallel helical conformation (Fig. 8A). In the presence of LUVs, ETeff decreased to 0.57 and indicated that residues T33C and T72C had been further separated via formation of an extended helix (Fig. 8B).86
Figure 8.ETeff histograms of maleimide-labeled (T33C/T72C) α-synuclein and proposed conformations of the micelle- and membrane-bound protein. (A, I) Energy transfers in the presence of buffer; (II) in the presence of SDS micelles; and (III) in the presence of LUVs. In buffer alone, α-synuclein displayed efficient energy transfers, presumably due to freely interconverting conformations. In the presence of micelles, energy transfers in α-synuclein increased, suggesting that the distance between the fluorophores placed on T33C and T72C had diminished. Vesicles increased the distance between the 2 fluorophores and indicated formation of an extended helix. (B) Proposed helical conformations of membrane-associated α-synuclein (top: in vesicles; bottom: in micelles). Blue arrows depict energy transfers between fluorophore-labeled S9C/T33C, yellow arrows between T33C/T72C, and red arrows between S9C/T72C. Adapted with permission from reference 86. Copyright 2009 American Chemical Society.
Different modes of lipid binding, part I: Coexisting populations of broken and extended α-helices
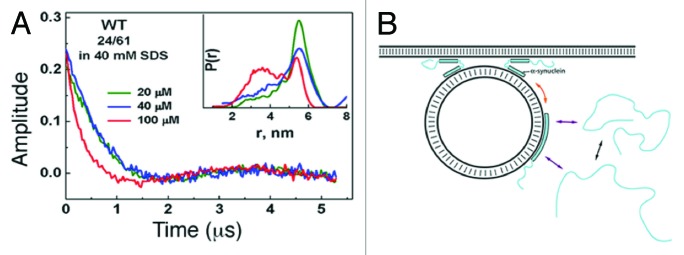
Drescher et al.88 and Georgieva et al.89 utilized EPR to probe α-synuclein’s structure in the presence of SUVs and, by suggesting the formation of broken (i.e., anti-parallel) and extend helices, respectively, the results were seemingly contradictory. However, on the basis of smFRET data, correlation analysis of FRET fluctuations (FCS-FRET), and pulsed dipolar electron spin resonance (ESR) measurements, it was subsequently proposed that, in the presence of lipids, α-synuclein interconverted between broken and extended helix conformations.90,91 Below the critical micelle concentration (CMC) of added SDS, α-synuclein labeled with fluorophores at residues G7C and G84C yielded ETeff data suggestive of 3 inter-fluorophore distances: 1 assigned to unbound α-synuclein, 1 assigned to the broken helix, and 1 assigned to the extended helix.90 At SDS concentrations above the CMC at which SDS micelles adopt cylindrical shapes, a large reduction in ETeff indicated the predominant formation of an extended helix.90 In agreement with these data, double electron-electron resonance (DEER)-based measurements of intramolecular distances from 3 doubly spin-labeled α-synuclein variants (spin-pairs incorporated at residues Q24C/E61C, Q24C/T72C, and Q24C/E83C) illustrated that the lipid-to-protein ratio, rather than the actual concentration of lipid itself, regulated the types of helices that were formed (Fig. 9A).91 At a lipid-to-protein ratio of 400, intramolecular distance distributions clearly indicated coexisting broken and extended helices.91 Increasing the ratio to 2000 shifted the distributions toward the extended helix. Varying the concentrations of SDS over a fixed amount of α-synuclein yielded similar results: high ratios (> 500) of SDS-to-protein elicited an extended helix, whereas lower ratios led to coexisting broken and extended helices. Notably, the highest SDS concentration used in these studies was 60 mM, far below that of the concentration at which SDS micelles become cylindrical (~200 mM), which suggested that α-synuclein might be able alter its lipid environment, consistent with previous reports.92,93 Similarly, recent DEER-based intramolecular distance measurements of LUV-bound α-synuclein spin-labeled at residues A27C and A56C reported the presence of both extended and broken helices.94 These results led to a proposed model in which coexisting α-synuclein helices regulated synaptic vesicle fusion at the plasma membrane (Fig. 9B).91
Figure 9. DEER-based inter-spin measurements in α-synuclein bound to LUVs and proposed model of the in vivo function of α-synuclein’s coexisting helices. (A) Spin labels were placed at residues Gln24 and Glu61. The ratio of SDS to protein was 2000 (green), 1000 (blue), or 400 (red). As the SDS to protein ratio was lowered, time domain data and distance distributions (P[r]) clearly indicated the coexistence of broken and extended α-helices. Note that the concentration of SDS (40 mM) was far below the concentration at which micelles become cylindrical (~200 mM). Adapted from reference 91. (B) Proposed model in which extended α-synuclein helices bind to synaptic vesicles and, upon nearing the plasma membrane, convert to broken helices to regulate vesicle fusion. This model has yet to be experimentally examined in vivo. Adapted with permission from reference 91. Copyright 2010, American Society for Biochemistry and Molecular Biology.
Different modes of lipid binding, part II: Distinct subsets of N-terminal interactions
Somewhat contradictory to the EPR, ESR, and smFRET studies outlined above, NMR analyses led to a proposed phospholipid binding model wherein α-synuclein partitioned into distinct populations of N-terminally bound protein, encompassing either residues 1–25, residues 1–97, or a more complicated scenario at high lipid concentrations.95 Prior NMR studies had utilized SDS micelles as a lipid mimetic67,69-71,73 which, owing to the relatively small sizes of spherical SDS micelles, avoided the rapid NMR signal decay observed with slowly tumbling protein-bound SUVs. However, exploiting these “advantageous” signal losses, Bodner et al.95 implemented a clever set of NMR experiments to probe α-synuclein’s conformational dynamics in the presence of synaptic vesicle-mimicking SUVs. Addition of SUVs to [U-15N]-labeled α-synuclein did not result in the formation of any new 1H-15N HSQC resonances, nor were any of the resonances of unbound protein perturbed. Rather, the signal intensities of a subset of NMR cross-peaks exhibited non-uniform attenuations.95 Thus, for any given residue, such decreases in signal intensities quantitatively described the fraction of molecules that had bound SUVs and had subsequently became “NMR invisible.” In the attenuation profile at low lipid concentrations, a significant plateau between residues Gln25 and Ala30, within otherwise continuous stretches of similar signals, suggested that α-synuclein bound to lipids in two distinct modes: SL1, in which only the N-terminal residues 1–25 were lipid-bound (i.e. NMR invisible), and SL2, with residues 1–97 NMR invisible.95 SL1 populations of lipid-bound α-synuclein, with only residues 1–25 immobilized, likely exposed the amyloidogenic NAC region, which could initiate pre-fibril formation.95 These data, which were supported by isothermal titration calorimetry- (ITC),96 tryptophan fluorescence-,97 and EPR-based98,99 analyses of α-synuclein’s N-terminal lipid binding affinities, substantiated the notion that low phospholipid concentrations enhanced fibrilization via distinct N-terminus–lipid interactions.100 Intriguingly, high SUV concentrations produced data that were indicative of more than two distinct SUV-bound states (SLn), with additional signal attenuation transitions observed at, in addition to the previously observed SL1/SL2 junction, Asn65, Ala85, and Pro120—implicating possible roles of C-terminal residues in lipid binding (Box 1).95 Dynamic light scattering (DLS), transmission electron microscopy (TEM), and cryo-EM experiments further revealed that SUVs underwent extensive rearrangements to form large aggregates and tubular structures in the presence of α-synuclein at concentrations used for NMR analyses.95 However, possible artifacts from TEM and cryo-EM sample preparation (i.e., dehydration, flash freezing, etc.) raised inherent uncertainty regarding their effects on SUV morphologies, and thus, NMR pulsed field gradient (PFG) techniques were used to measure the translational diffusion rates of α-synuclein-bound SUVs. At low SUV concentrations, the corresponding radius of hydration, 37 ± 2 Å, and molecular weights of the α-synuclein-SUV particles, ~150 kDa, pointed to the formation of compact oligomers, with binding modes that were incompatible with previously outlined EPR/ESR/FRET models (see above). As such, it was suggested that a bundle of α-synuclein helices surrounding a phospholipid core mediated SUV binding, in agreement with the observed slow exchange rates (1–10 s−1) between free and SUV-bound protein95
Box 1.
Rather surprisingly, the observation that C-terminal residues participated in SUV binding, as shown by 2D 1H-15N HSQC signal attenuation at high SUV concentrations, begs the question of whether or not cis-trans isomerization of the 5 C-terminal Xaa-Pro bonds plays a role in modulating lipid binding.95 Breaks in the attenuation profile coincide with the positions of proline residues 108, 117, 120, 128, and 138, and it has been shown that nonpolar environments indeed promote the formation of cis Xaa-Pro bonds.95,101 Proline is a “disorder-promoting” amino acid with a cis/trans energy barrier approaching 20 kcal/mol.102,103 Depending on the protein’s secondary structure and local environment, one isomer may be favored over the other—with cis and trans bonds promoting turn-like structures and extended conformations, respectively.103 During the process of protein folding, however, the inherently low rate of cis-trans isomerization (~10−3 s−1) frequently manifests as the rate-limiting step.104 Enzymes belonging to the class of peptidyl-prolyl cis-trans isomerases (PPIases) enhance this rate of cis-trans interconversion, culminating in a thermodynamically accelerated folding process.105 It is thus tantalizing to propose that, either at high concentrations of vesicles or in the presence of PPIases, the formation of cis Xaa-Pro bonds within α-synuclein’s C-terminal region could lead to enhanced aggregation and fibrilization. This speculation is supported by the presence of the PPIases Pin1 and FKBP12 in Lewy bodies and by elegant studies that have illustrated an enhanced rate of α-synuclein fibrilization in the presence of FKB12 in vitro.106-110
Pfefferkorn et al.111 utilized neutron reflectometry (NR), MD simulations, and tryptophan fluorescence (F4W α-synuclein) to monitor the binding depths of full-length α-synuclein and N-terminal peptides into lipid vesicles and a surface-stabilized sparsely tethered bilayer lipid membrane (stBLM). Brominated lipid-induced tryptophan fluorescence quenching data of F4W α-synuclein suggested that the full-length protein penetrated 6–11 Å into the bilayer. Neutron reflection data of the stBLM incubated with α-synuclein were in agreement with tryptophan fluorescence results, and indicated that a significant portion of α-synuclein remained solvent exposed (63 Å) while a subset of protein was membrane-bound (13 Å penetration depth).111 However, mean estimated distances between C-terminal α-synuclein residues 94–136, which remain disordered upon lipid binding, encompassed a range of 33–43 Å in solution.75 The observed 63 Å of solvent-exposed α-synuclein supported NMR data of Bodner et al.95 and demonstrated that, upon lipid binding, residues upstream of the disordered C-terminus remained accessible to solvent.111 A set of F4W α-synuclein peptides, designated P4 (residues 1–4), P6 (1–6), P10 (1–10), and P15 (1–15), were constructed to further evaluate membrane binding interactions localized at the N-terminus. Lipid addition changed tryptophan fluorescence most significantly for α-synuclein peptides P10 and P15 and the associated membrane partition constants were reminiscent of full-length α-synuclein.111 Upon lipid addition, changes in 1H NMR spectra of isolated α-synuclein peptides confirmed that lipid binding affinity was dependent on peptide length (P15 > P10 > P6 > P4), and CD spectra of P15 showed lipid-induced coil–to–helix conformational transitions. Finally, the group performed MD simulations of P15 and observed bilayer penetration depths of 5.5–11 Å—similar to those of full-length protein—and dynamic binding interactions between α-synuclein residues 1–5 and the lipid bilayer,111 which were intriguing given that physiological α-synuclein is acetylated at Met1.60
Disorder in the Court: Order?
Tetrameric α-synuclein
Despite overwhelming evidence supporting α-synuclein’s intrinsically disordered and monomeric nature, Bartels et al.60 suggested that native α-synuclein instead existed as a stable and helix-rich tetramer. This study was based on a non-denaturing purification protocol, whereas many of the prior studies used heat precipitation and detergent-based purifications. Native gel electrophoresis of lysates from α-synuclein-expressing M17D, HEK, HeLa, and COS cell lines, along with mouse frontal cortex and human red blood cells (RBCs), all displayed the presence of 45–50 kDa species. Western blot analysis revealed that these species were all α-synuclein-immunoreactive, and clear-native PAGE (CN-PAGE) migration patterns suggested a molecular weight of 55–60 kDa, which was indicative of a tetrameric α-synuclein conformation.60 Isoelectric focusing determined that the apparent tetramers had the same pI as monomeric α-synuclein, which led the group to conclude that the oligomers were indeed homo-tetramers.60 However, while these analyses hinted at oligomeric species, the applied techniques relied strongly on a protein’s intrinsic charge and conformation, factors that and have since been shown to serve as poor indicators of an IDP’s conformational state and true molecular weight.112
Nevertheless, the presence of α-synuclein tetramers was confirmed by 2 analytical techniques that typically evaluate protein mass and oligomeric status independent of protein conformation: scanning transmission electron microscopy (STEM) and sedimentation analytical ultracentrifugation (SE-AUC). STEM generated an unbiased molecular weight histogram from 1,000 automatically selected RBC-derived α-synuclein particles (purified under non-denaturing conditions) and yielded a distribution peak ~55 kDa.60 SE-AUC analyses led to an average native RBC α-synuclein mass of 57.8 kDa, which was consistent with a tetramer. Additionally, mass spectrometry indicated that subunits within the RBC-derived α-synuclein tetramer had undergone Nα-acetylation (14,505 Da) whereas recombinantly produced samples had not (14,462 Da).60
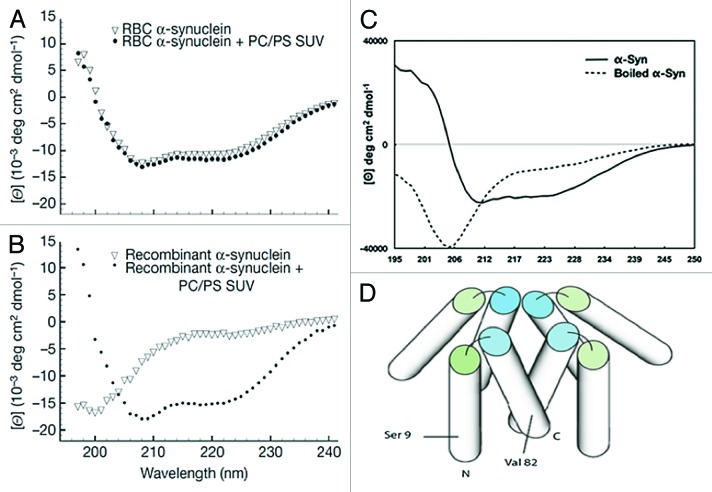
Of great surprise, the CD spectra of non-denatured α-synuclein homo-tetramers revealed two minima at 222 and 208 nm that were unaffected by the addition of SUVs (Fig. 10A). This highly helical structure was in stark contrast to previously reported data, in which purified α-synuclein was largely disordered and adopted helical structures only when added to lipid vesicles (Fig. 10B; see above). Because of their non-denaturing purification protocol, the group hypothesized that lipids may still be bound to α-synuclein and likely elicited the observed helical conformations that formed independent of exogenously added lipids. Incubation with a reagent (Lipidex 1000) that removed protein-bound lipids did not appreciably alter the CD spectra, and prompted the conclusion that lipids were not required to stabilize helical α-synuclein tetramers. More significantly, and perhaps indicating a physiological role, the tetramer bound negatively charged vesicles with an apparent Kd of 56 +/− 61 nM—2 orders of magnitude lower than that of the monomer.60,112
Figure 10. CD spectra of both native and recombinant α-synuclein in the presence and absence of acidic phospholipids, and the proposed dynamic tetramer model. (A) CD spectra of the putative tetramer from human red blood cells in the presence and absence of lipids. The non-denaturing protocol used in Bartels et al.60 reportedly preserved helical structure in native α-synuclein tetramers, independent of lipid addition. (B) CD spectra of recombinant free and lipid-bound α-synuclein monomer purified under typical denaturing conditions. The protein is denatured, as expected, and transitioned to α-helical conformations only in the presence of negatively charged vesicles. Reprinted with permission from Macmillan Publishers Ltd: Nature, reference 60, 2011. (C) CD spectra of a GST fusion construct of α-synuclein, which contained an additional 10 residues at its N-terminus, were similar to Bartels et al.60 in (A) Boiling the samples resulted in disordered protein. (D) Putative dynamic α-synuclein tetramer model with dynamic helices. NOE-data established transient helical conformations. Measured PREs indicated that each subunit formed anti-parallel helices as in Figure 6A. Subunits were aligned in parallel (i.e., C- to C-termini interactions). Ser9 and Val82 represented the sites of Cys substitution used for PRE analyses. Reprinted from reference 61 with permission.
In support of these findings, Wang et al.61 reported that a GST fusion construct of α-synuclein, which contained 10 extra N-terminal residues after removal of the tag, formed a dynamic tetramer under non-denaturing conditions. CD spectra of the free tetramer revealed 65% helix, 17% turn, and 8% disorder, and boiling the sample yielded disordered α-synuclein, which could not reform the tetramer (Fig. 10C). TALOS+113 software-mediated assignment of the chemical shifts of [U-13C, 15N]-labeled tetrameric α-synuclein suggested that transient helices were formed. 15N-edited NOESY spectra confirmed these results, and weak Hα, HN i, i+3 NOEs established that the helices were fractionally occupied.61 To determine the relative orientations of each of the subunits within the tetramer, the group incorporated a MTSL spin label at S9C and mixed spin-labeled, non-isotope-labeled α-synuclein with equimolar amounts of [U-15N]-labeled α-synuclein and monitored intermolecular PREs upon tetramer formation. Each subunit was found to align in parallel with their N-terminal regions forming the exterior of the tetramer (Fig. 10D) and maximum PREs were observed near Val37.61 Additionally, and somewhat suspiciously, to record inter-subunit PREs the tetramer was reconstituted from distinctly labeled populations of disordered, monomeric α-synuclein (i.e., tetrameric subunits). Yet, Wang et al.61 had previously shown that dissociating the tetramer in vitro irreversibly produced disordered monomers.61 Moreover, these PRE data did not agree with the anti-parallel N-to-C-terminal interchain contacts that had been derived from an earlier PRE study of MTSL-labeled A19C α-synuclein,114 nor with the anti-parallel alignment of oligomers found in 2 in vivo fluorescence studies.115,116 Given the fact that N- and C-terminal portions of isolated α-synuclein formed transient interactions in solution,73 the construct used in the study by Wang et al.,61 which contained 10 additional residues at its N-terminus, likely modulated these contacts.
Both Bartels et al.60 and Wang et al.61 reported that the α-synuclein tetramers exhibited reduced aggregation tendencies. Furthermore, tetramer-formed fibrils were non-toxic when added to cultured neuronal cells.60,61 In conjunction with the discovery that tetrameric α-synuclein bound lipids with enhanced affinity, the authors proposed that, under physiological conditions, α-synuclein existed as a stable tetramer. Dissociation of the tetramer into respective monomeric subunits would promote toxic aggregation and the onset of neurodegenerative diseases.60,61 Yet, contrary to these reports, stable oligomers of a GFP-α-construct reportedly enhanced cytotoxicity.115
In-cell NMR: α-synuclein is a disordered monomer inside bacteria
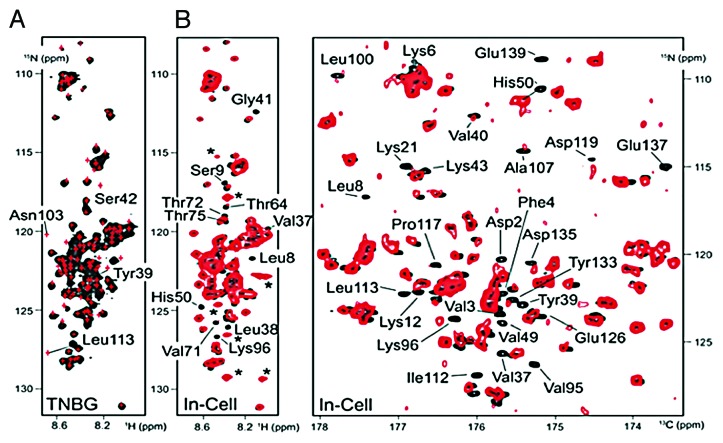
Using the BMRB-deposited 1H-15N HSQC chemical shift data of tetrameric α-synuclein, Binolfi et al.117 generated an artificial NMR spectrum and compared it to the 2D 1H-15N HSQC spectrum of monomeric α-synuclein purified under denaturing conditions, but resuspended into the same buffer that was used by Wang et al.61 in their NMR study (Fig. 11A).117 Conspicuously, the 2 spectra were strikingly identical; differences were only observed for Tyr39 and Leu113 (Ser42, Asn103, and all Gly residues experienced line broadening, which was attributed to known unfavorable chemical exchange effects at 25°C and pH 7.4,118 and optimizing buffer conditions reversed these effects). Upon closer inspection of the NMR spectrum found in Supplemental Figure 3 of Wang et al.,61 it became clear that the deposited 1H-15N chemical shifts for Tyr39 and Leu113 did not agree with the published data. According to the BMRB deposition, the cross peak for Leu113 should be found at (1H, 15N ppm) (8.37, 127.1), yet the labeled peak in Supplemental Figure 3 placed it near (8.17, 126.5). In addition, the deposited position for Tyr39 was (7.86, 121.7), whereas in the spectrum Tyr39 was located at (8.00, 122.0). The chemical shifts for Tyr39 and Leu113 in Supplemental Figure 3, which were different from deposited data in the BMRB, were consistent with those in Binolfi et al.117 Thus, the authors concluded that NMR spectra of “tetrameric α-synuclein with dynamic helices” more accurately resembled those of the intrinsically disordered monomer.117
Figure 11. In vitro and in-cell NMR spectra of α-synuclein. (A) 1H-15N SOFAST-HMQC spectra of α-synuclein purified under denaturing conditions (black) and placed in the same buffer conditions and temperature setting as in Wang et al.61 Overlaid in red are the positions of the BMRB-deposited 1H-15N HSQC peaks of the putative α-synuclein tetramer from Wang et al.61 Note the chemical shift differences for Tyr39 and Leu113 in the 2 spectra (see text). (B) Left panel: overlay of 1H-15N SOFAST-HMQC spectra of monomeric α-synuclein in vitro (black) and in vivo (red). Right panel: overlay of (1H-flip) 13CO-15N spectra, as colored in the left panel. Asterisks denote E. coli background signals. Reproduced with permission from reference 117. Copyright 2012, The Biochemical Society.
Aiming to examine the protein’s structural features inside intact bacteria (i.e., without applying any purification protocol), the group collected 2D in-cell NMR spectra of [U-13C, 15N]-labeled α-synuclein overexpressed in Escherichia coli. The close correspondence of 2D in-cell 1H-15N SOFAST-HMQC and 2D (1H-flip) 13CO-15N spectra to previously collected in vitro NMR spectra confirmed that α-synuclein displayed the same intrinsically disordered monomeric protein characteristics inside cells, as after purification with denaturing or non-denaturing protocols (Fig. 11B).117 Binolfi et al.117 duly noted that, in the tetramer hypothesis, the helical N-terminus (i.e., the first 100 residues) would not yield the observed high quality in-cell NMR spectra. In the crowded intracellular environment (~400 g of macromolecules/L), folded globular proteins experience the increased viscosity of the bacterial cytoplasm, which greatly affects rotational diffusion and, consequentially, the spectral characteristics of a protein’s NMR signals. This was recently demonstrated in an elegant in-cell NMR study of a [U-15N]-labeled ubiquitin-α-synuclein fusion construct. In-cell 2D 1H-15N HSQC spectra only revealed cross-peaks corresponding to the disordered α-synuclein portion of the construct, whereas the smaller, folded ubiquitin portion remained NMR invisible (Fig. 12A).119 When these in-cell NMR samples were lysed, and HSQC spectra directly recorded on the resulting extract slurries, NMR signals of the folded ubiquitin moieties were additionally detected (Fig. 12B).119 Similarly, folded and/or dynamic intracellular tetramers of α-synuclein would be expected to yield low-quality in-cell NMR spectra with line-broadened peak characteristics, if detectable at all, and chemical shifts that were perturbed from the resonances of monomeric α-synuclein in vitro. However, the high-qualities of the in-cell NMR spectra presented by Binolfi et al.117 (Fig. 11B) unambiguously confirmed that α-synuclein was a disordered monomer inside intact E. coli cells, which was consistent with previous studies.59,120-123
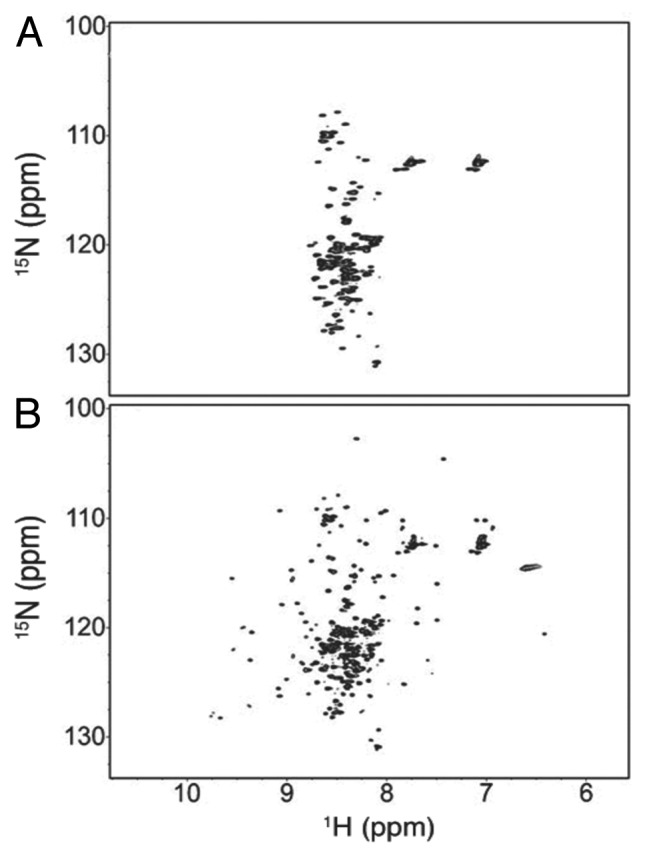
Figure 12. 2D 1H-15N HSQC spectra of the ubiquitin-α-synuclein fusion protein. (A) In-cell NMR spectrum displaying low 1H peak dispersion that was indicative of disordered α-synuclein. Macromolecular crowding and excluded volume in vivo rendered the ubiquitin portion of the construct NMR invisible. (B) NMR spectrum recorded 1.5 h after cells were lysed. Additional signals with enhanced dispersion are attributed to the globular ubiquitin upon dilution of macromolecules. Copyright 2011 Wiley. Used with permission from reference 119.
Reevaluating the tetramer hypothesis
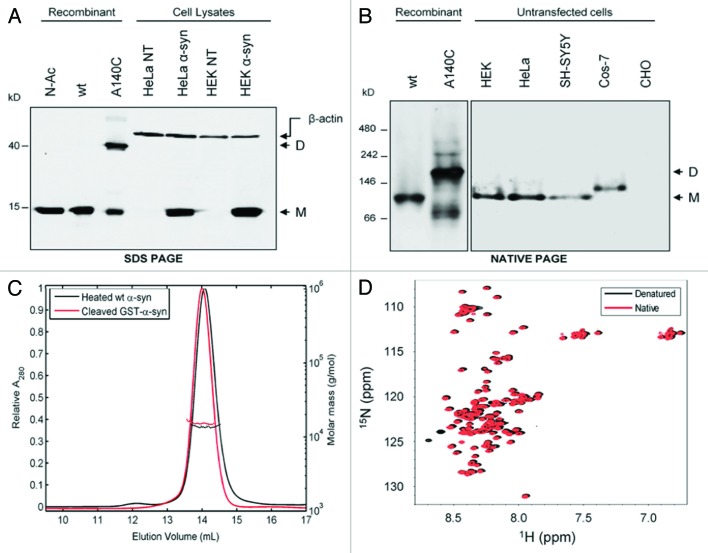
In further support of intrinsically disordered, monomeric α-synuclein, 7 independent laboratories collectively published an extensive study analyzing the protein under various purification procedures and from various biological sources.124 Combinations of electrospray ionization mass spectrometry (ESI-MS), 2D NMR, SDS-PAGE, CN-PAGE, CD spectroscopy, DLS, ELISA, in vivo cross-linking, western blotting, and analytical gel filtration were unable to detect structural, or functional differences between RBC-, mammalian-, and E. coli-derived α-synuclein—regardless of whether it had been Nα-acetylated, fused to GST tags, or purified under denaturing vs. non-denaturing conditions (Fig. 13A–D). These data clearly illustrated that α-synuclein existed as an intrinsically disordered monomer. However, the authors did not exclude the possibility that α-synuclein could exist in a functionally oligomeric state under defined physiological conditions (i.e., in the presence of specific protein-protein or lipid-based interactions).124
Figure 13. Collection of figures from 7 laboratories that all confirmed α-synuclein was intrinsically disordered and monomeric, regardless of purification protocols or biological sources. (A) SDS-PAGE of cell lysates obtained from HEK293T and HeLa cells were either transfected with plasmids encoding WT human α-synuclein or not transfected at all (NT). Notice that both recombinant Nα-acetylated and WT α-synuclein (Lanes 1 and 2) co-migrated with α-synuclein from HeLa and HEK293T cells (Lanes 4 and 6). A140C α-synuclein (Lane 3) was crosslinked with disuccinimidyl suberate (DSS) and formed dimers (D) All other lanes exclusively consisted of monomeric protein (M). (B) Same conditions as (A), but proteins were instead resolved via CN-PAGE. WT, HEK, HeLa, and SH-SY5Y α-synuclein samples all migrated to a single band of ~66 kDa. Cos-7 α-synuclein migrated slightly slower. Yet, native PAGE cannot accurately resolve the molecular weight of IDPs (see text). (C) Analytical gel filtration/light scattering analysis of purified and cleaved GST-α-synuclein. Note that both boiled α-synuclein and GST-tag-cleaved α-synuclein yielded the same elution profiles. (D) 1H-15N HSQC spectra of denatured [U-15N]-labeled α-synuclein (black) overlaid with a spectrum of non-denatured [U-15N]-labeled α-synuclein (red). Resonances from both denatured and native α-synuclein superimposed extremely well, which suggested that native and denatured protein adopted similar conformations in solution. Reprinted with permission from reference 124. Copyright 2012 American Society for Biochemistry and Molecular Biology.
Burre et al.125 reported similar findings in their analyses of native brain α-synuclein. Size exclusion chromatography (SEC) profiles of both native and recombinant α-synuclein, which were purified in the absence of detergents, or heat precipitation, were identical and led to an apparent molecular mass of 63 kDa. Similarly, CN-PAGE migration patterns for each α-synuclein sample revealed a single migrating species of ~65 kDa.125 Boiling the samples did not alter their migration properties. Further analysis of native brain α-synuclein with SEC–multi-angle laser-light scattering (SEC-MALS) indicated the presence of a monomer, but also of small amounts (less than 5%) of higher molecular weight forms of α-synuclein (~58 kDa), which could indicate the presence of a tetrameric species at relatively low abundance.125 Molecular weights of Nα-acetylated α-synuclein tetramer subunits (14.5 kDa), as reported by Bartels et al.,60 remained inconsistent with the observed mass of the monomeric form of native brain α-synuclein, which was determined to be 16.4 kDa.125 What exactly caused this ~2 kDa difference in molecular weights was not pursued further in this study, although it raises the question of whether Nα-acetylation or any other PTM affected oligomerization of the protein. Indeed, CD spectra of native brain α-synuclein contained 21–24% α-helical content, which was much higher than the 2% observed for any form of the recombinant protein.66,124 The authors concluded that native brain α-synuclein “primarily consists of an unstructured monomer,” which is supported by the SEC-MALS and immunostaining results, but did not speculate on the possible causes for the large helical contents in the α-synuclein CD spectra.125
In vivo cross-linking: An unknown bioorganic molecule stabilizes the tetramer in vivo?
Aiming to further analyze the proposed oligomeric state(s) of α-synuclein in vivo and without experiments that require protein overexpression, Dettmer et al.126 utilized the human erythroleukemia (HEL) cell line that endogenously expresses high levels of the protein. Cross-linking of intact HEL cells via the amine-reactive disuccinimidyl glutarate (DSG) revealed that 60 kDa α-synuclein species constituted the “predominant form” of the protein, followed by lesser amounts of 80 and 100 kDa α-synuclein. However, significant amounts of monomeric α-synuclein were present throughout the study, and even cross-linked dimeric and trimeric forms of the protein appeared.126 Repeating the DSG cross-linking protocol in vitro after cell lysis failed to produce tetramers, although, conspicuously, other homo-oligomeric control proteins, e.g., DJ-1, VDAC, GAPDH, DRP-1, and HSP70, were efficiently cross-linked in vitro at the DSG concentrations used for prior in vivo cross-linking analyses. Challenging the supposedly “predominant” nature of tetrameric α-synuclein, increased in vitro DSG concentrations (1–5 mM) captured higher-order α-synuclein oligomers (> 80 kDa) and indicated that the protein was capable of cross-linking, albeit no oligomers below 80 kDa were observed.126 Following the hypothesis that the inability to recover 60 kDa α-synuclein species in vitro may have resulted from reduced levels of macromolecular crowding in diluted cell lysates, the group repeated in vitro cross-linking experiments with lysates of higher total protein concentrations. A protein concentration-dependent recovery of the 60 kDa α-synuclein tetramer was observed, which led the authors to propose that either the high degree of intracellular macromolecular crowding127,128 was required to stabilize the tetramer conformation, or other cellular components, e.g., small lipids, were necessary for forming tetramers and higher oligomers. Upon cell lysis and dilution of any of these biological activities, α-synuclein oligomers may have dissociated into disordered monomers.126
Rather intriguingly, α-synuclein oligomers cross-linked in vitro fell apart following exposure to sonication-induced heat, or shearing, yet oligomers cross-linked in vivo were able to withstand extremely harsh environmental conditions that included treatment with 5 M urea and 10 min of boiling.126 When Wang et al.61 boiled their tetramer samples, they observed pronounced structural rearrangements that culminated in irreversible dissociation into disordered monomers. The observation that tetramers isolated from HEL cells depended on high α-synuclein concentrations and/or the presence of lipids contradicted previous data regarding putative α-synuclein tetramers. Bartels et al.60 and Wang et al.61 analyzed RBC- and E. coli-derived α-synuclein, respectively, and reported helical tetrameric conformations at relatively dilute protein concentrations in vitro and in the absence of exogenously added or co-purified lipids. Additionally, Coelho-Cerqueira et al.129 presented a glutaraldehyde (GHD)-mediated α-synuclein cross-linking study in E. coli cells and reported that the protein predominantly existed as a disordered monomer in dynamic equilibrium with a small population (3–5%) of dimers. Cross-linked tetramers were observed only at extremely high concentrations of GHD (25 mM), whereas dimers formed at GHD concentrations as low as 1 mM. Purification protocols were based on heat or acid extractions, and osmotic shock or sonication cell lysis, and all procedures yielded disordered, monomeric α-synuclein.129 These results supported those of Fauvet et al.124 and argued strongly against the purification-dependent α-synuclein tetramer formation in E. coli presented by Wang et al.61
Computational analyses provide insight into the oligomeric populations of α-synuclein
A recent computational study constructed a large ensemble of monomeric α-synuclein structures.130 The structural calculations utilized replica exchange molecular dynamics (REMD) and a Bayesian weighting algorithm in which the weight (i.e., relative stability) of each conformer was related to its respective agreement with experimentally derived NMR chemical shift, RDC, and SAXS data. The average Rg of the structural ensemble was 41 ± 1 Å, in excellent agreement with the previously reported value.65,130 Remarkably, however, the distribution of radii of gyration within the ensemble encompassed values ranging from approximately 20 Å all the way to 60 Å—indicating that α-synuclein adopted conformations accessible to both a random coil (Rg = 51 Å) and a globular protein (Rg = 15.1 Å) of 140 amino acids in length.130 Additionally, the calculated α-helical and β-sheet content for the ensemble average was 2% and 11%, respectively, with certain structures approaching values as high as 28% sheet and 20% helix. Within this monomeric α-synuclein subpopulation of relatively higher helical propensities, it was suggested that intermolecular association of amphipathic helices via hydrophobic interactions could enable the formation of helical tetramers.130
A follow-up computational study reported on the dynamic nature of α-synuclein multimers.131 Two types of models were generated: one by constructing trimers and tetramers via interhelical contacts within selected species of the monomeric structural ensemble presented above, and another by incorporating the available NMR data from Wang et al.,61 which were derived from a 10 residue N-terminally extended construct of α-synuclein. These additional residues were not included in the modeling routines.131 Because the lack of sufficient, experimentally obtained NOE data hindered the calculations of high-resolution tetramer structures in the first place, low-resolution tetramer models were employed as initial seed structures. From these, 533 REMD structures were calculated, of which 64.1 ± 6.4% were monomeric, 7.7 ± 3.6% were trimeric, and 28.2 ± 6% were tetrameric.131 Within the tetrameric population, the structures adopted either predominantly helical, or predominantly sheet conformations, with β-sheet tetramers preferred (only 5.1 ± 2.9% of the total structures were helical tetramers). Surprisingly, most β-sheet tetramers exposed the amyloidogenic NAC region in extended conformations, which Gurry et al.131 speculated could point to pathologically relevant on-pathway intermediates to amyloid structures. Additionally, these β-sheet-rich tetramers, being the major tetrameric species, contradicted the extensive helical content found in the tetramers from Bartels et al.60 and Wang et al.61 It was noted, however, that the minority helical tetramers did minimize the solvent exposure of the amyloidogenic NAC region, and thereby permitted a mechanism for nontoxic α-synuclein storage.131
Nα-Acetylation: Modulator of α-Synuclein Structure and Function?
Introduction to protein Nα-acetylation
In eukaryotes, Nα-acetyltransferases (NATs) utilize the cofactor acetyl-coenzyme A to cotranslationally acetylate the N-terminal residues of many proteins, with recent estimates suggesting that 85% of all human proteins undergo Nα-acetylation.132,133 The biological implications of Nα-acetylation remain unclear, although recent studies suggest roles in modulating subcellular localization, rates of protein synthesis, and protein-protein interactions.134-136 Substrate specificity of particular NATs depends on the presence, or absence of an initiator methionine and the sequence identities of the following residues. The NatB complex for example, exclusively acetylates N-terminal initiator Met residues that are followed by either Asp, Asn, Gln, or Glu residues,137 which is the case for α-synuclein. Despite the widespread Nα-acetylation of human proteins, most in vitro studies on recombinant α-synuclein had utilized E. coli to express the protein. However, owing to inherent differences in the acetylation pathways,138 E. coli do not acetylate α-synuclein, or any other recombinantly expressed protein for that matter. Nα-acetylation of α-synuclein was recognized as an important mammalian PTM following the observation by Bartels et al.60 that the alleged α-synuclein tetramer subunits from human RBCs contained Nα-acetyl groups (14,505 Da), whereas the recombinantly produced protein did not (14,462 Da). Aggregated α-synuclein isolated from PD and DLB deposits were found to be Nα-acetylated, as well.139 To investigate whether Nα-acetylation of α-synuclein (Ac-α-synuclein) exerted modulatory effects on its structure and function, multiple groups have implemented a recently developed NatB bacterial co-expression system to produce large quantities of the modified protein for biophysical analyses.140
Nα-acetylation leads to the formation of α-synuclein oligomers?
Trexler and Rhoades112 employed CD spectroscopy, SEC, and SE-AUC to comparatively study Ac-α-synuclein and α-synuclein purified by either ammonium sulfate precipitation (“harsh”), or by glycerol and octyl β-D-glucopyranoside (BOG) detergent extraction (“mild”). The combination of Nα-acetylation and a mild purification protocol was required to promote the formation of structured α-synuclein oligomers of ~90 kDa, as determined by SE-AUC. However, both the native gel migration patterns and the SEC elution volumes of the monomer and oligomer were nearly identical. These findings, as the authors noted, demonstrated the inability of SEC and PAGE to accurately determine the molecular weights of IDPs.112 Mildly purified Ac-α-synuclein oligomers exhibited ~20% helical content by CD spectroscopy, which prompted Trexler and Rhoades112 to speculate that BOG molecules, omnipresent throughout the extraction, may have remained bound to Ac-α-synuclein and elicited the observed increase in helicity and enhanced aggregation behavior.
Monomeric Nα-acetylated free α-synuclein forms a transient N-terminal helix
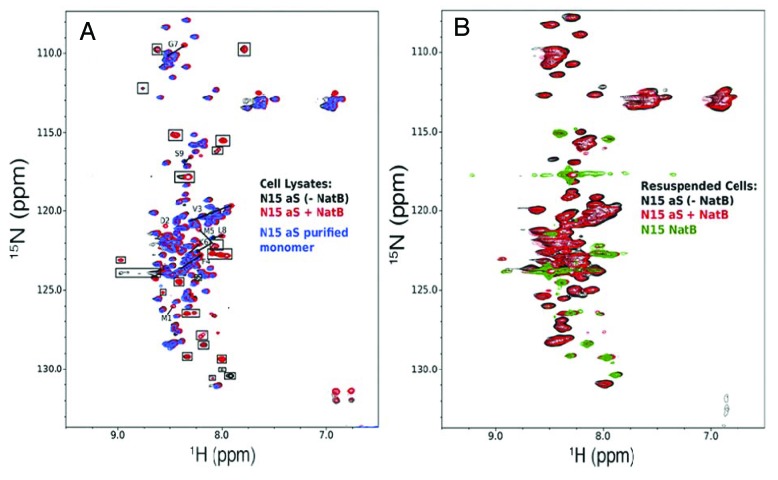
Three other studies found no evidence for a stable helical structure of Ac-α-synuclein and concluded that the modified protein remained largely disordered.141-143 Maltsev et al.141 compared 1H-15N HSQC spectra of Ac-α-synuclein and α-synuclein and observed chemical shift differences for the first 12 residues that indicated a 6% relative increase in helical content upon acetylation. However, levels of detectable transient helicity rapidly diminished with increasing distance from the acetyl group.141 Measured differences in three-bond scalar coupling constants (3JHN-Hα) and ΔδCα’s of residues 1–5 of Ac-α-synuclein reported at 17% increase in α-helical content of the modified protein. No other differences were observed in the NMR spectra of differentially acetylated α-synucleins.141 In agreement with these results, Kang et al.142 also detected a similar increase in helicity for residues 1–12, as well as small reductions in β-sheet propensities near residues 28–31, 43–46, and 50–66. To address whether Nα-acetylation impacted the oligomerization behavior of the protein, the group performed ion mobility and ESI-MS (ESI-IMS-MS) measurements and determined that 90–95% of recombinantly expressed and purified Ac-α-synuclein existed in monomeric conformations, while the other 5–10% formed dimers.142 These results were no different from unmodified α-synuclein. In fact, the 2 samples yielded identical native PAGE migration patterns and SEC profiles.141 Similarly, Fauvet et al.143 examined the oligomerization status of semisynthetic Ac-α-synuclein that had been produced by fusing a synthetic Nα-acetylated α-synuclein peptide (residues 1–10) to residues 11–140 of recombinant α-synuclein. SEC, CN-PAGE, and CD analyses uniformly confirmed that both native (NatB co-expression) and semisynthetic (fusion) Ac-α-synuclein were unfolded monomers in solution.143 In addition, they compared 2D 1H-15N HSQC NMR spectra of purified recombinant α-synuclein with those of cell lysates of E. coli grown on 15N-labeled medium and transfected with plasmids carrying α-synuclein alone, or α-synuclein and NatB. Besides acetylation-induced chemical shift perturbations of N-terminal α-synuclein residues, which corroborated the observed increase in helicity, all other protein NMR signals superimposed well (Fig. 14A).143 In-cell NMR spectra of unmodified and modified α-synuclein showed line broadening of residues 1–10, which implied that, irrespective of the acetylation status, the N-terminus of α-synuclein experienced conformational and/or chemical exchange, and/or interacted with intracellular bacterial components (Fig. 14B).142 These results, together with previously published in-cell NMR data on α-synuclein inside bacteria,117-123 substantiated the notion that Ac-α-synuclein adopted a primarily disordered conformation inside and outside cells, similar to what had been determined for the unmodified protein.124
Figure 14.Nα-acetylation did not significantly alter the structural properties of α-synuclein. (A) 2D 1H-15N HSQC spectra of lysates from E. coli cells transfected with α-synuclein alone (black) or α-synuclein with Nα-acetyltransferase (NatB) (red) (i.e., to produce Ac-α-synuclein) were overlaid upon the spectrum of purified recombinant α-synuclein (blue). Resonances attributed to NatB, which did not pertain to the analysis, were boxed. Aside from resonances of N-terminal residues (labeled M1, D2, V3, F4, M5, K6, G7, L8, and S9) that showed chemical shift differences upon acetylation consistent with the formation of a transient helix, all other resonances colocalized, which indicated that the protein adopted a monomeric and disordered structure with or without acetylation. (B) Same spectra as (A) but instead recorded in living E. coli cells. Green resonances were attributed to NatB and did not pertain to the analysis. Red resonances corresponded to Ac-α-synuclein and were superimposed with black resonances of α-synuclein, which suggested that acetylation did not appreciably alter α-synuclein’s structure. Reprinted from reference 143. Copyright 2012 American Society for Biochemistry and Molecular Biology.
The function of α-synuclein Nα-acetylation remains unclear
These studies have also produced conflicting results with regard to the lipid binding affinities and rates of fibril elongation of Ac-α-synuclein. In the hands of Maltsev et al.,141 Ac-α-synuclein was found to bind SUVs with an affinity twice as large as that of the unmodified protein, which resulted in a greatly enhanced helical content at relatively lower SUV/protein ratios. Thus, formation of the acetytlation-dependent transient helix within the N-terminus of α-synuclein has been proposed to function as the triggering event in lipid binding.141 On the other hand, Fauvet et al.143 reported no differences in lipid-binding affinities between Ac-α-synuclein and α-synuclein using LUVs or murine synaptosomes. In one study141 the apparent rates of in vitro fibril elongation were unaffected by acetylation, whereas in another,142 a significantly diminished rate of elongation was reported for Ac-α-synuclein—perhaps due to transient stabilization of the N-terminal helix, or altered long-range electrostatic interactions between N- and C-terminal α-synuclein regions.
Conclusions
Nearly 20 y after its original discovery,1 the first high-resolution structure of SDS micelle-bound α-synuclein provided enormous insight into its structural dynamics.82 Earlier studies had utilized lower resolution SAXS,63 CD,65 and FTIR spectroscopy65 to hypothesize that the protein was intrinsically disordered in solution, yet adopted a highly α-helical structure upon binding to negatively charged vesicles.66 The presence of 7 imperfect 11-residue repeats encoding amphipathic helices led to predictions of 2 non-canonical α11/3-helices,70,72 1 unbroken α-helix,67 and 5 lipid-binding helices.66 Subsequent EPR85,89,94 and FRET measurements86,90 indicated that α-synuclein could indeed bind to small and large unilamellar vesicles as a single, unbroken helix, although heterogeneous in nature through the coexistence of broken helices.90,91,94 Ironically, 2 anti-parallel α-helices were observed in the original SDS micelle-bound structure,82 yet α11/3-helices were found in the unbroken helix model of SUV-bound α-synuclein.85 Contrary to the widely accepted and intrinsically disordered nature of α-synuclein, 2 publications in 2011 described the presence of structured tetramers and concluded that these tetramers represented the physiologically relevant species.60,61 However, by comparing 2D NMR spectra collected from in vitro monomeric α-synuclein and from E. coli overexpressing α-synuclein, it was concluded that the majority of intracellular α-synuclein existed as disordered monomers.118,143 Furthermore, substantial evidence came from the combined efforts of 7 laboratories, whose extensive characterizations of RBC-, mammalian-, and E. coli-derived α-synuclein, with and without denaturing protocols, proved that the protein was intrinsically disordered.124 Contesting the above results, in vivo cross-linking in HEL cells displayed the formation of a “primarily” tetrameric α-synuclein species126 with substantial amounts of monomer still present. The group proposed that the presence of a bioorganic molecule of yet unknown identity stabilizes α-synuclein oligomers in vivo.126 A subsequent cross-linking investigation in E. coli cells reported primarily disorderd monomeric α-synuclein, with very small populations of dimers.128 Many prior biophysical investigations had utilized α-synuclein produced by bacterial overexpression,63,65,70,72,85,90 an expression system that does not yield Nα-acetylated proteins.100 Strikingly, α-synuclein isolated from RBCs was uniformly Nα-acetylated60 and prompted analyses on the structural and functional impact of Nα-acetylation.112,141-143 Three independent groups reported that the presence, or absence of the Nα-acetyl group did not affect oligomeric status of the protein.141-143 However, the impact of this modification on α-synuclein lipid binding and its fibrilization rates remains unclear.124,141,143 Despite all of the biophysical investigations on α-synuclein, the primary physiological function of this protein is still poorly defined. Because of its direct involvement in PD, i.e., undergoing a change from a disordered monomer, or structured tetramer to toxic, β-sheet-rich fibrils, or soluble oligomers, α-synuclein represents a medically relevant problem in modern structural biology. Future applications of high-resolution techniques in biophysics and structural biology are well poised to unravel the molecular mechanisms that underlie α-synuclein’s role in PD.
Glossary
Abbreviations:
- AD
Alzheimer disease
- NAC
non-Aβ component
- WT
wild-type
- FISH
fluorescence in situ hybridization
- PD
Parkinson disease
- NACP
NAC precursor
- LBs
Lewy bodies
- PTM
post translational modification
- LBDs
Lewy body diseases
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- VAMP2
vesicle-associated membrane protein 2
- IDP
intrinsically disordered protein
- CD
circular dichroism
- SAXS
small-angle X-ray scattering
- Rg
radius of gyration
- FTIR
Fourier transform infrared
- NMR
nuclear magnetic resonance
- 2D
two-dimensional
- HSQC
heteronuclear single quantum coherence
- [U-]
uniformly-
- 3D
three-dimensional
- ΔδCα
secondary 13Cα chemical shift
- ppm
parts per million
- R1
rate of longitudinal magnetic relaxation
- R2
rate of transverse magnetic relaxation
- SDS
sodium dodecyl sulfate
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- TOCSY
total correlation spectroscopy
- PRE
paramagnetic resonance enhancement
- -OH-TEMPO
-hydroxy-2,2,6,6-tetramethylpiperidin-1-oxyl
- MTSL
(1-oxy-2,2,5,5-tetramethyl-D-pyrroline-3-methyl)-methanethiosulfonate
- RDC
residual dipolar coupling
- C8E5
n-octyl-penta(ethylene glycol)/octanol
- FET
fluorescence energy-transfer
- ET
electron transfer
- DA
donor-acceptor
- MD
molecular dynamics
- EDTA
ethylenediaminetetraacetic acid
- S2
general order parameter
- Da
local alignment tensor
- J(ωH+ωN)
reduced spectral density function at 1H and 15N frequencies
- PS
phosphatidylserine
- PI
phosphatidylinositol
- SUV
small unilamellar vesicle
- LUV
large unilamellar vesicle
- EPR
electron paramagnetic resonance
- FRET
Förster resonance energy transfer
- smFRET
single-molecule FRET
- SAMD
simulated annealing molecular dynamics
- ETeff
FRET efficiency
- FCS-FRET
correlation analysis of FRET fluctuations
- ESR
electron spin resonance
- CMC
critical micelle concentration
- DEER
double electron-electron resonance
- DLS
dynamic light scattering
- TEM
transmission electron microscopy
- cryo-EM
cryo-electron microscopy
- PFG
pulsed field gradient
- PPIase
peptidyl-prolyl isomerase
- ITC
isothermal titration calorimetry
- NR
neutron reflectometry
- stBLM
sparsely tethered bilayer lipid membrane
- RBC
red blood cell
- CN-PAGE
clear native-polyacrylamide gel electrophoresis
- STEM
scanning TEM
- SE-AUC
sedimentation analytical ultracentrifugation
- PC
phosphatidylcholine
- GST
glutathione S-transferase
- GFP
green fluorescent protein
- BMRB
Biological Magnetic Resonance Database
- SOFAST-HMQC
band-selective optimized-flip-angle short-transient heteronuclear multiple quantum coherence
- ESI-MS
electospray ionization mass spectrometry
- ELISA
enzyme-linked immunosorbent assay
- SEC
size exclusion chromatography
- SEC-MALS
SEC with multi-angle light scattering
- PTM
post-transitional modification
- HEL
human erythroleukemia
- DSG
disuccinimidyl glutarate
- GHD
glutaraldehyde
- REMD
replica exchange molecular dynamics
- NAT
Nα-acetyl transferase
- BOG
octyl β-D-glucopyranoside
- 3J
three-bond coupling constant
- ESI-IMS-MS
ESI-MS with ion mobility spectrometry
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Jin Hae Kim, William Ford Freyberg, and members of the Philipp Selenko research group for critically reading the manuscript and offering insightful comments. This work was supported by National Institutes of Health grant U01 GM094622
References
- 1.Maroteaux L, Campanelli JT, Scheller RH. . Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci 1988; 8:2804 - 15; PMID: 3411354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uéda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero DA, Kondo J, Ihara Y, Saitoh T. . Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A 1993; 90:11282 - 6; http://dx.doi.org/ 10.1073/pnas.90.23.11282; PMID: 8248242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwai A, Yoshimoto M, Masliah E, Saitoh T. . Non-A beta component of Alzheimer’s disease amyloid (NAC) is amyloidogenic. Biochemistry 1995; 34:10139 - 45; http://dx.doi.org/ 10.1021/bi00032a006; PMID: 7640267 [DOI] [PubMed] [Google Scholar]
- 4.Bodles AM, Guthrie DJ, Greer B, Irvine GB. . Identification of the region of non-Abeta component (NAC) of Alzheimer’s disease amyloid responsible for its aggregation and toxicity. J Neurochem 2001; 78:384 - 95; http://dx.doi.org/ 10.1046/j.1471-4159.2001.00408.x; PMID: 11461974 [DOI] [PubMed] [Google Scholar]
- 5.Iwai A, Masliah E, Yoshimoto M, Ge N, Flanagan L, de Silva HA, Kittel A, Saitoh T. . The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron 1995; 14:467 - 75; http://dx.doi.org/ 10.1016/0896-6273(95)90302-X; PMID: 7857654 [DOI] [PubMed] [Google Scholar]
- 6.Withers GS, George JM, Banker GA, Clayton DF. . Delayed localization of synelfin (synuclein, NACP) to presynaptic terminals in cultured rat hippocampal neurons. Brain Res Dev Brain Res 1997; 99:87 - 94; http://dx.doi.org/ 10.1016/S0165-3806(96)00210-6; PMID: 9088569 [DOI] [PubMed] [Google Scholar]
- 7.Culvenor JG, McLean CA, Cutt S, Campbell BC, Maher F, Jäkälä P, Hartmann T, Beyreuther K, Masters CL, Li QX. . Non-Abeta component of Alzheimer’s disease amyloid (NAC) revisited. NAC and alpha-synuclein are not associated with Abeta amyloid. Am J Pathol 1999; 155:1173 - 81; http://dx.doi.org/ 10.1016/S0002-9440(10)65220-0; PMID: 10514400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayer TA, Jäkälä P, Hartmann T, Havas L, McLean C, Culvenor JG, Li QX, Masters CL, Falkai P, Beyreuther K. . Alpha-synuclein accumulates in Lewy bodies in Parkinson’s disease and dementia with Lewy bodies but not in Alzheimer’s disease beta-amyloid plaque cores. Neurosci Lett 1999; 266:213 - 6; http://dx.doi.org/ 10.1016/S0304-3940(99)00311-0; PMID: 10465711 [DOI] [PubMed] [Google Scholar]
- 9.Chen X, de Silva HA, Pettenati MJ, Rao PN, St George-Hyslop P, Roses AD, Xia Y, Horsburgh K, Uéda K, Saitoh T. . The human NACP/alpha-synuclein gene: chromosome assignment to 4q21.3-q22 and TaqI RFLP analysis. Genomics 1995; 26:425 - 7; http://dx.doi.org/ 10.1016/0888-7543(95)80237-G; PMID: 7601479 [DOI] [PubMed] [Google Scholar]
- 10.Shibasaki Y, Baillie DA, St Clair D, Brookes AJ. . High-resolution mapping of SNCA encoding alpha-synuclein, the non-A beta component of Alzheimer’s disease amyloid precursor, to human chromosome 4q21.3-->q22 by fluorescence in situ hybridization. Cytogenet Cell Genet 1995; 71:54 - 5; http://dx.doi.org/ 10.1159/000134061; PMID: 7606927 [DOI] [PubMed] [Google Scholar]
- 11.Golbe LI, Di Iorio G, Sanges G, Lazzarini AM, La Sala S, Bonavita V, Duvoisin RC. . Clinical genetic analysis of Parkinson’s disease in the Contursi kindred. Ann Neurol 1996; 40:767 - 75; http://dx.doi.org/ 10.1002/ana.410400513; PMID: 8957018 [DOI] [PubMed] [Google Scholar]
- 12.Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, Di Iorio G, Sanges G, Stenroos ES, Pho LT, Schaffer AA, et al. . Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science 1996; 274:1197 - 9; http://dx.doi.org/ 10.1126/science.274.5290.1197; PMID: 8895469 [DOI] [PubMed] [Google Scholar]
- 13.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. . Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997; 276:2045 - 7; http://dx.doi.org/ 10.1126/science.276.5321.2045; PMID: 9197268 [DOI] [PubMed] [Google Scholar]
- 14.Krishnan S, Chi EY, Wood SJ, Kendrick BS, Li C, Garzon-Rodriguez W, Wypych J, Randolph TW, Narhi LO, Biere AL, et al. . Oxidative dimer formation is the critical rate-limiting step for Parkinson’s disease alpha-synuclein fibrillogenesis. Biochemistry 2003; 42:829 - 37; http://dx.doi.org/ 10.1021/bi026528t; PMID: 12534296 [DOI] [PubMed] [Google Scholar]
- 15.Krüger R, Kuhn W, Müller T, Woitalla D, Graeber M, Kösel S, Przuntek H, Epplen JT, Schöls L, Riess O. . Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet 1998; 18:106 - 8; http://dx.doi.org/ 10.1038/ng0298-106; PMID: 9462735 [DOI] [PubMed] [Google Scholar]
- 16.Zarranz JJ, Alegre J, Gómez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atarés B, et al. . The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 2004; 55:164 - 73; http://dx.doi.org/ 10.1002/ana.10795; PMID: 14755719 [DOI] [PubMed] [Google Scholar]
- 17.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. . Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004; 364:1167 - 9; http://dx.doi.org/ 10.1016/S0140-6736(04)17103-1; PMID: 15451224 [DOI] [PubMed] [Google Scholar]
- 18.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. . alpha-Synuclein locus triplication causes Parkinson’s disease. Science 2003; 302:841; http://dx.doi.org/ 10.1126/science.1090278; PMID: 14593171 [DOI] [PubMed] [Google Scholar]
- 19.Hoffman-Zacharska D, Koziorowski D, Ross OA, Milewski M, Poznański J, Jurek M, Wszolek ZK, Soto-Ortolaza A, Sławek J, Janik P, et al. . Novel A18T and pA29S substitutions in α-synuclein may be associated with sporadic Parkinson’s disease. Parkinsonism Relat Disord 2013; http://dx.doi.org/ 10.1016/j.parkreldis.2013.07.011; PMID: 23916651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proukakis C, Dudzik CG, Brier T, MacKay DS, Cooper JM, Millhauser GL, Houlden H, Schapira AH. . A novel α-synuclein missense mutation in Parkinson disease. Neurology 2013; 80:1062 - 4; http://dx.doi.org/ 10.1212/WNL.0b013e31828727ba; PMID: 23427326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesage S, Anheim M, Letournel F, Bousset L, Honoré A, Rozas N, Pieri L, Madiona K, Dürr A, Melki R, et al. , for the French Parkinson’s Disease Genetics (PDG) Study Group. . G51D α-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann Neurol 2013; http://dx.doi.org/ 10.1002/ana.23894; PMID: 23526723 [DOI] [PubMed] [Google Scholar]
- 22.Kara E, Lewis PA, Ling H, Proukakis C, Houlden H, Hardy J. . α-Synuclein mutations cluster around a putative protein loop. Neurosci Lett 2013; 546:67 - 70; http://dx.doi.org/ 10.1016/j.neulet.2013.04.058; PMID: 23669636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrer MJ. . Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet 2006; 7:306 - 18; http://dx.doi.org/ 10.1038/nrg1831; PMID: 16543934 [DOI] [PubMed] [Google Scholar]
- 24.Gazewood JD, Richards DR, Clebak K. . Parkinson disease: an update. Am Fam Physician 2013; 87:267 - 73; PMID: 23418798 [PubMed] [Google Scholar]
- 25.Moriarty GM, Janowska MK, Kang L, Baum J. . Exploring the accessible conformations of N-terminal acetylated α-synuclein. FEBS Lett 2013; 587:1128 - 38; http://dx.doi.org/ 10.1016/j.febslet.2013.02.049; PMID: 23499431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickson DW. Parkinson's disease and parkinsonism: neuropathology. Cold Spring Harb Perspect Med 2012; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN. . Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci (Landmark Ed) 2009; 14:5188 - 238; http://dx.doi.org/ 10.2741/3594; PMID: 19482612 [DOI] [PubMed] [Google Scholar]
- 28.Breydo L, Wu JW, Uversky VN. . Α-synuclein misfolding and Parkinson’s disease. Biochim Biophys Acta 2012; 1822:261 - 85; http://dx.doi.org/ 10.1016/j.bbadis.2011.10.002; PMID: 22024360 [DOI] [PubMed] [Google Scholar]
- 29.Uversky VN. . A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn 2003; 21:211 - 34; http://dx.doi.org/ 10.1080/07391102.2003.10506918; PMID: 12956606 [DOI] [PubMed] [Google Scholar]
- 30.Silva BA, Breydo L, Uversky VN. . Targeting the chameleon: a focused look at α-synuclein and its roles in neurodegeneration. Mol Neurobiol 2013; 47:446 - 59; http://dx.doi.org/ 10.1007/s12035-012-8334-1; PMID: 22940885 [DOI] [PubMed] [Google Scholar]
- 31.Drescher M, Huber M, Subramaniam V. . Hunting the chameleon: structural conformations of the intrinsically disordered protein alpha-synuclein. Chembiochem 2012; 13:761 - 8; http://dx.doi.org/ 10.1002/cbic.201200059; PMID: 22438319 [DOI] [PubMed] [Google Scholar]
- 32.Cabin DE, Shimazu K, Murphy D, Cole NB, Gottschalk W, McIlwain KL, Orrison B, Chen A, Ellis CE, Paylor R, et al. . Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha-synuclein. J Neurosci 2002; 22:8797 - 807; PMID: 12388586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouberai MM, Wang J, Swann MJ, Galvagnion C, Guilliams T, Dobson CM, Welland ME. . α-Synuclein Senses Lipid Packing Defects and Induces Lateral Expansion of Lipids Leading to Membrane Remodeling. J Biol Chem 2013; 288:20883 - 95; http://dx.doi.org/ 10.1074/jbc.M113.478297; PMID: 23740253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. . Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med 2002; 8:600 - 6; http://dx.doi.org/ 10.1038/nm0602-600; PMID: 12042811 [DOI] [PubMed] [Google Scholar]
- 35.Saha AR, Ninkina NN, Hanger DP, Anderton BH, Davies AM, Buchman VL. . Induction of neuronal death by alpha-synuclein. Eur J Neurosci 2000; 12:3073 - 7; http://dx.doi.org/ 10.1046/j.1460-9568.2000.00210.x; PMID: 10971650 [DOI] [PubMed] [Google Scholar]
- 36.Lotharius J, Brundin P. . Impaired dopamine storage resulting from alpha-synuclein mutations may contribute to the pathogenesis of Parkinson’s disease. Hum Mol Genet 2002; 11:2395 - 407; http://dx.doi.org/ 10.1093/hmg/11.20.2395; PMID: 12351575 [DOI] [PubMed] [Google Scholar]
- 37.Ellis CE, Murphy EJ, Mitchell DC, Golovko MY, Scaglia F, Barceló-Coblijn GC, Nussbaum RL. . Mitochondrial lipid abnormality and electron transport chain impairment in mice lacking alpha-synuclein. Mol Cell Biol 2005; 25:10190 - 201; http://dx.doi.org/ 10.1128/MCB.25.22.10190-10201.2005; PMID: 16260631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, et al. . Inhibition of mitochondrial fusion by α-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J 2010; 29:3571 - 89; http://dx.doi.org/ 10.1038/emboj.2010.223; PMID: 20842103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeWitt DC, Rhoades E. . α-Synuclein can inhibit SNARE-mediated vesicle fusion through direct interactions with lipid bilayers. Biochemistry 2013; 52:2385 - 7; http://dx.doi.org/ 10.1021/bi4002369; PMID: 23528131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi BK, Choi MG, Kim JY, Yang Y, Lai Y, Kweon DH, Lee NK, Shin YK. . Large α-synuclein oligomers inhibit neuronal SNARE-mediated vesicle docking. Proc Natl Acad Sci U S A 2013; 110:4087 - 92; http://dx.doi.org/ 10.1073/pnas.1218424110; PMID: 23431141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loeb V, Yakunin E, Saada A, Sharon R. . The transgenic overexpression of alpha-synuclein and not its related pathology associates with complex I inhibition. J Biol Chem 2010; 285:7334 - 43; http://dx.doi.org/ 10.1074/jbc.M109.061051; PMID: 20053987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. . Mitochondrial α-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett 2010; 486:235 - 9; http://dx.doi.org/ 10.1016/j.neulet.2010.09.061; PMID: 20887775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu M, Qin ZJ, Hu D, Munishkina LA, Fink AL. . Alpha-synuclein can function as an antioxidant preventing oxidation of unsaturated lipid in vesicles. Biochemistry 2006; 45:8135 - 42; http://dx.doi.org/ 10.1021/bi052584t; PMID: 16800638 [DOI] [PubMed] [Google Scholar]
- 44.Zakharov SD, Hulleman JD, Dutseva EA, Antonenko YN, Rochet JC, Cramer WA. . Helical alpha-synuclein forms highly conductive ion channels. Biochemistry 2007; 46:14369 - 79; http://dx.doi.org/ 10.1021/bi701275p; PMID: 18031063 [DOI] [PubMed] [Google Scholar]
- 45.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. . Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A 2005; 102:10427 - 32; http://dx.doi.org/ 10.1073/pnas.0502066102; PMID: 16020533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Souza JM, Giasson BI, Lee VM, Ischiropoulos H. . Chaperone-like activity of synucleins. FEBS Lett 2000; 474:116 - 9; http://dx.doi.org/ 10.1016/S0014-5793(00)01563-5; PMID: 10828462 [DOI] [PubMed] [Google Scholar]
- 47.George JM, Jin H, Woods WS, Clayton DF. . Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron 1995; 15:361 - 72; http://dx.doi.org/ 10.1016/0896-6273(95)90040-3; PMID: 7646890 [DOI] [PubMed] [Google Scholar]
- 48.Chandra S, Fornai F, Kwon HB, Yazdani U, Atasoy D, Liu X, Hammer RE, Battaglia G, German DC, Castillo PE, et al. . Double-knockout mice for alpha- and beta-synucleins: effect on synaptic functions. Proc Natl Acad Sci U S A 2004; 101:14966 - 71; http://dx.doi.org/ 10.1073/pnas.0406283101; PMID: 15465911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dev KK, Hofele K, Barbieri S, Buchman VL, van der Putten H. . Part II: alpha-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacology 2003; 45:14 - 44; http://dx.doi.org/ 10.1016/S0028-3908(03)00140-0; PMID: 12814657 [DOI] [PubMed] [Google Scholar]
- 50.Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC. . Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 2010; 329:1663 - 7; http://dx.doi.org/ 10.1126/science.1195227; PMID: 20798282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burré J, Sharma M, Südhof TC. . Systematic mutagenesis of α-synuclein reveals distinct sequence requirements for physiological and pathological activities. J Neurosci 2012; 32:15227 - 42; http://dx.doi.org/ 10.1523/JNEUROSCI.3545-12.2012; PMID: 23100443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diao J, Burré J, Vivona S, Cipriano DJ, Sharma M, Kyoung M, Südhof TC, Brunger AT. . Native α-synuclein induces clustering of synaptic-vesicle mimics via binding to phospholipids and synaptobrevin-2/VAMP2. Elife 2013; 2:e00592; http://dx.doi.org/ 10.7554/eLife.00592; PMID: 23638301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bekei B, Rose HM, Herzig M, Dose A, Schwarzer D, Selenko P. . In-cell NMR in mammalian cells: part 1. Methods Mol Biol 2012; 895:43 - 54; http://dx.doi.org/ 10.1007/978-1-61779-927-3_4; PMID: 22760311 [DOI] [PubMed] [Google Scholar]
- 54.Skora L, Cho MK, Kim HY, Becker S, Fernandez CO, Blackledge M, Zweckstetter M. . Charge-induced molecular alignment of intrinsically disordered proteins. Angew Chem Int Ed Engl 2006; 45:7012 - 5; http://dx.doi.org/ 10.1002/anie.200602317; PMID: 17009286 [DOI] [PubMed] [Google Scholar]
- 55.Salmon L, Nodet G, Ozenne V, Yin G, Jensen MR, Zweckstetter M, Blackledge M. . NMR characterization of long-range order in intrinsically disordered proteins. J Am Chem Soc 2010; 132:8407 - 18; http://dx.doi.org/ 10.1021/ja101645g; PMID: 20499903 [DOI] [PubMed] [Google Scholar]
- 56.Roche J, Ying J, Maltsev AS, Bax A. . Impact of Hydrostatic Pressure on an Intrinsically Disordered Protein: A High-Pressure NMR Study of α-Synuclein. Chembiochem 2013; http://dx.doi.org/ 10.1002/cbic.201300244; PMID: 23813793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cino EA, Karttunen M, Choy WY. . Effects of molecular crowding on the dynamics of intrinsically disordered proteins. PLoS One 2012; 7:e49876; http://dx.doi.org/ 10.1371/journal.pone.0049876; PMID: 23189168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu ST, Bertoncini CW, Dobson CM. . Use of protonless NMR spectroscopy to alleviate the loss of information resulting from exchange-broadening. J Am Chem Soc 2009; 131:7222 - 3; http://dx.doi.org/ 10.1021/ja902307q; PMID: 19432443 [DOI] [PubMed] [Google Scholar]
- 59.Bertini I, Felli IC, Gonnelli L, Kumar M V V, Pierattelli R. . 13C direct-detection biomolecular NMR spectroscopy in living cells. Angew Chem Int Ed Engl 2011; 50:2339 - 41; PMID: 21351349 [DOI] [PubMed] [Google Scholar]
- 60.Bartels T, Choi JG, Selkoe DJ. . α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 2011; 477:107 - 10; http://dx.doi.org/ 10.1038/nature10324; PMID: 21841800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, Perovic I, Chittuluru J, Kaganovich A, Nguyen LT, Liao J, Auclair JR, Johnson D, Landeru A, Simorellis AK, et al. . A soluble α-synuclein construct forms a dynamic tetramer. Proc Natl Acad Sci U S A 2011; 108:17797 - 802; http://dx.doi.org/ 10.1073/pnas.1113260108; PMID: 22006323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uversky VN, Eliezer D. . Biophysics of Parkinson’s disease: structure and aggregation of alpha-synuclein. Curr Protein Pept Sci 2009; 10:483 - 99; http://dx.doi.org/ 10.2174/138920309789351921; PMID: 19538146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT Jr.. . NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 1996; 35:13709 - 15; http://dx.doi.org/ 10.1021/bi961799n; PMID: 8901511 [DOI] [PubMed] [Google Scholar]
- 64.Gast K, Damaschun H, Eckert K, Schulze-Forster K, Maurer HR, Müller-Frohne M, Zirwer D, Czarnecki J, Damaschun G. . Prothymosin alpha: a biologically active protein with random coil conformation. Biochemistry 1995; 34:13211 - 8; http://dx.doi.org/ 10.1021/bi00040a037; PMID: 7548085 [DOI] [PubMed] [Google Scholar]
- 65.Uversky VN, Li J, Fink AL. . Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem 2001; 276:10737 - 44; http://dx.doi.org/ 10.1074/jbc.M010907200; PMID: 11152691 [DOI] [PubMed] [Google Scholar]
- 66.Davidson WS, Jonas A, Clayton DF, George JM. . Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 1998; 273:9443 - 9; http://dx.doi.org/ 10.1074/jbc.273.16.9443; PMID: 9545270 [DOI] [PubMed] [Google Scholar]
- 67.Eliezer D, Kutluay E, Bussell R Jr., Browne G. . Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol 2001; 307:1061 - 73; http://dx.doi.org/ 10.1006/jmbi.2001.4538; PMID: 11286556 [DOI] [PubMed] [Google Scholar]
- 68.Wishart DS, Sykes BD. . Chemical shifts as a tool for structure determination. Methods Enzymol 1994; 239:363 - 92; http://dx.doi.org/ 10.1016/S0076-6879(94)39014-2; PMID: 7830591 [DOI] [PubMed] [Google Scholar]
- 69.Bussell R Jr., Eliezer D. . Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol Chem 2001; 276:45996 - 6003; http://dx.doi.org/ 10.1074/jbc.M106777200; PMID: 11590151 [DOI] [PubMed] [Google Scholar]
- 70.Bussell R Jr., Eliezer D. . A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol 2003; 329:763 - 78; http://dx.doi.org/ 10.1016/S0022-2836(03)00520-5; PMID: 12787676 [DOI] [PubMed] [Google Scholar]
- 71.Chandra S, Chen X, Rizo J, Jahn R, Südhof TC. . A broken alpha -helix in folded alpha -Synuclein. J Biol Chem 2003; 278:15313 - 8; http://dx.doi.org/ 10.1074/jbc.M213128200; PMID: 12586824 [DOI] [PubMed] [Google Scholar]
- 72.Bussell R Jr., Ramlall TF, Eliezer D. . Helix periodicity, topology, and dynamics of membrane-associated alpha-synuclein. Protein Sci 2005; 14:862 - 72; http://dx.doi.org/ 10.1110/ps.041255905; PMID: 15741347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. . Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A 2005; 102:1430 - 5; http://dx.doi.org/ 10.1073/pnas.0407146102; PMID: 15671169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernadó P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M. . Defining long-range order and local disorder in native alpha-synuclein using residual dipolar couplings. J Am Chem Soc 2005; 127:17968 - 9; http://dx.doi.org/ 10.1021/ja055538p; PMID: 16366524 [DOI] [PubMed] [Google Scholar]
- 75.Lee JC, Langen R, Hummel PA, Gray HB, Winkler JR. . Alpha-synuclein structures from fluorescence energy-transfer kinetics: implications for the role of the protein in Parkinson’s disease. Proc Natl Acad Sci U S A 2004; 101:16466 - 71; http://dx.doi.org/ 10.1073/pnas.0407307101; PMID: 15536128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JC, Gray HB, Winkler JR. . Tertiary contact formation in alpha-synuclein probed by electron transfer. J Am Chem Soc 2005; 127:16388 - 9; http://dx.doi.org/ 10.1021/ja0561901; PMID: 16305213 [DOI] [PubMed] [Google Scholar]
- 77.Lee JC, Lai BT, Kozak JJ, Gray HB, Winkler JR. . Alpha-synuclein tertiary contact dynamics. J Phys Chem B 2007; 111:2107 - 12; http://dx.doi.org/ 10.1021/jp068604y; PMID: 17279794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. . Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc 2005; 127:476 - 7; http://dx.doi.org/ 10.1021/ja044834j; PMID: 15643843 [DOI] [PubMed] [Google Scholar]
- 79.Allison JR, Varnai P, Dobson CM, Vendruscolo M. . Determination of the free energy landscape of alpha-synuclein using spin label nuclear magnetic resonance measurements. J Am Chem Soc 2009; 131:18314 - 26; http://dx.doi.org/ 10.1021/ja904716h; PMID: 20028147 [DOI] [PubMed] [Google Scholar]
- 80.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. . The Xplor-NIH NMR molecular structure determination package. J Magn Reson 2003; 160:65 - 73; http://dx.doi.org/ 10.1016/S1090-7807(02)00014-9; PMID: 12565051 [DOI] [PubMed] [Google Scholar]
- 81.Crowther RA, Jakes R, Spillantini MG, Goedert M. . Synthetic filaments assembled from C-terminally truncated alpha-synuclein. FEBS Lett 1998; 436:309 - 12; http://dx.doi.org/ 10.1016/S0014-5793(98)01146-6; PMID: 9801138 [DOI] [PubMed] [Google Scholar]
- 82.Ulmer TS, Bax A, Cole NB, Nussbaum RL. . Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 2005; 280:9595 - 603; http://dx.doi.org/ 10.1074/jbc.M411805200; PMID: 15615727 [DOI] [PubMed] [Google Scholar]
- 83.Bisaglia M, Tessari I, Pinato L, Bellanda M, Giraudo S, Fasano M, Bergantino E, Bubacco L, Mammi S. . A topological model of the interaction between alpha-synuclein and sodium dodecyl sulfate micelles. Biochemistry 2005; 44:329 - 39; http://dx.doi.org/ 10.1021/bi048448q; PMID: 15628875 [DOI] [PubMed] [Google Scholar]
- 84.Jao CC, Der-Sarkissian A, Chen J, Langen R. . Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc Natl Acad Sci U S A 2004; 101:8331 - 6; http://dx.doi.org/ 10.1073/pnas.0400553101; PMID: 15155902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. . Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci U S A 2008; 105:19666 - 71; http://dx.doi.org/ 10.1073/pnas.0807826105; PMID: 19066219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trexler AJ, Rhoades E. . Alpha-synuclein binds large unilamellar vesicles as an extended helix. Biochemistry 2009; 48:2304 - 6; http://dx.doi.org/ 10.1021/bi900114z; PMID: 19220042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rabenstein MD, Shin YK. . Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci U S A 1995; 92:8239 - 43; http://dx.doi.org/ 10.1073/pnas.92.18.8239; PMID: 7667275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Drescher M, Godschalk F, Veldhuis G, van Rooijen BD, Subramaniam V, Huber M. . Spin-label EPR on alpha-synuclein reveals differences in the membrane binding affinity of the two antiparallel helices. Chembiochem 2008; 9:2411 - 6; http://dx.doi.org/ 10.1002/cbic.200800238; PMID: 18821550 [DOI] [PubMed] [Google Scholar]
- 89.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. . Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc 2008; 130:12856 - 7; http://dx.doi.org/ 10.1021/ja804517m; PMID: 18774805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. . Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A 2009; 106:5645 - 50; http://dx.doi.org/ 10.1073/pnas.0809232106; PMID: 19293380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. . The lipid-binding domain of wild type and mutant alpha-synuclein: compactness and interconversion between the broken and extended helix forms. J Biol Chem 2010; 285:28261 - 74; http://dx.doi.org/ 10.1074/jbc.M110.157214; PMID: 20592036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pandey AP, Haque F, Rochet JC, Hovis JS. . Clustering of alpha-synuclein on supported lipid bilayers: role of anionic lipid, protein, and divalent ion concentration. Biophys J 2009; 96:540 - 51; http://dx.doi.org/ 10.1016/j.bpj.2008.10.011; PMID: 19167303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Madine J, Hughes E, Doig AJ, Middleton DA. . The effects of alpha-synuclein on phospholipid vesicle integrity: a study using 31P NMR and electron microscopy. Mol Membr Biol 2008; 25:518 - 27; http://dx.doi.org/ 10.1080/09687680802467977; PMID: 18949627 [DOI] [PubMed] [Google Scholar]
- 94.Robotta M, Braun P, van Rooijen B, Subramaniam V, Huber M, Drescher M. . Direct evidence of coexisting horseshoe and extended helix conformations of membrane-bound alpha-synuclein. Chemphyschem 2011; 12:267 - 9; http://dx.doi.org/ 10.1002/cphc.201000815; PMID: 21275016 [DOI] [PubMed] [Google Scholar]
- 95.Bodner CR, Dobson CM, Bax A. . Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. J Mol Biol 2009; 390:775 - 90; http://dx.doi.org/ 10.1016/j.jmb.2009.05.066; PMID: 19481095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bartels T, Ahlstrom LS, Leftin A, Kamp F, Haass C, Brown MF, Beyer K. . The N-terminus of the intrinsically disordered protein α-synuclein triggers membrane binding and helix folding. Biophys J 2010; 99:2116 - 24; http://dx.doi.org/ 10.1016/j.bpj.2010.06.035; PMID: 20923645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pfefferkorn CM, Lee JC. . Tryptophan probes at the alpha-synuclein and membrane interface. J Phys Chem B 2010; 114:4615 - 22; http://dx.doi.org/ 10.1021/jp908092e; PMID: 20229987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robotta M, Hintze C, Schildknecht S, Zijlstra N, Jüngst C, Karreman C, Huber M, Leist M, Subramaniam V, Drescher M. . Locally resolved membrane binding affinity of the N-terminus of α-synuclein. Biochemistry 2012; 51:3960 - 2; http://dx.doi.org/ 10.1021/bi300357a; PMID: 22494024 [DOI] [PubMed] [Google Scholar]
- 99.Drescher M, Veldhuis G, van Rooijen BD, Milikisyants S, Subramaniam V, Huber M. . Antiparallel arrangement of the helices of vesicle-bound alpha-synuclein. J Am Chem Soc 2008; 130:7796 - 7; http://dx.doi.org/ 10.1021/ja801594s; PMID: 18512917 [DOI] [PubMed] [Google Scholar]
- 100.Necula M, Chirita CN, Kuret J. . Rapid anionic micelle-mediated alpha-synuclein fibrillization in vitro. J Biol Chem 2003; 278:46674 - 80; http://dx.doi.org/ 10.1074/jbc.M308231200; PMID: 14506232 [DOI] [PubMed] [Google Scholar]
- 101.Torchia DA. . Evidence for cis peptide bonds in copolypeptides of glycine and proline. Biochemistry 1972; 11:1462 - 8; http://dx.doi.org/ 10.1021/bi00758a021; PMID: 5021599 [DOI] [PubMed] [Google Scholar]
- 102.Dugave C, Demange L. . Cis-trans isomerization of organic molecules and biomolecules: implications and applications. Chem Rev 2003; 103:2475 - 532; http://dx.doi.org/ 10.1021/cr0104375; PMID: 12848578 [DOI] [PubMed] [Google Scholar]
- 103.Theillet F-X, Kalmar L, Tompa P, Han K-H, Selenko P, Dunker AK, Daughdrill GW, Uversky VN. . Act like a Pro: On the abundance and roles of proline residues in intrinsically disorderd proteins. Intrinsically Disordered Proteins 2013; 1 Forthcoming [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wedemeyer WJ, Welker E, Scheraga HA. . Proline cis-trans isomerization and protein folding. Biochemistry 2002; 41:14637 - 44; http://dx.doi.org/ 10.1021/bi020574b; PMID: 12475212 [DOI] [PubMed] [Google Scholar]
- 105.Lu KP, Finn G, Lee TH, Nicholson LK. . Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 2007; 3:619 - 29; http://dx.doi.org/ 10.1038/nchembio.2007.35; PMID: 17876319 [DOI] [PubMed] [Google Scholar]
- 106.Ryo A, Togo T, Nakai T, Hirai A, Nishi M, Yamaguchi A, Suzuki K, Hirayasu Y, Kobayashi H, Perrem K, et al. . Prolyl-isomerase Pin1 accumulates in lewy bodies of parkinson disease and facilitates formation of alpha-synuclein inclusions. J Biol Chem 2006; 281:4117 - 25; http://dx.doi.org/ 10.1074/jbc.M507026200; PMID: 16365047 [DOI] [PubMed] [Google Scholar]
- 107.Meuvis J, Gerard M, Desender L, Baekelandt V, Engelborghs Y. . The conformation and the aggregation kinetics of α-synuclein depend on the proline residues in its C-terminal region. Biochemistry 2010; 49:9345 - 52; http://dx.doi.org/ 10.1021/bi1010927; PMID: 20828147 [DOI] [PubMed] [Google Scholar]
- 108.Gerard M, Debyser Z, Desender L, Kahle PJ, Baert J, Baekelandt V, Engelborghs Y. . The aggregation of alpha-synuclein is stimulated by FK506 binding proteins as shown by fluorescence correlation spectroscopy. FASEB J 2006; 20:524 - 6; PMID: 16410343 [DOI] [PubMed] [Google Scholar]
- 109.Gerard M, Debyser Z, Desender L, Baert J, Brandt I, Baekelandt V, Engelborghs Y. . FK506 binding protein 12 differentially accelerates fibril formation of wild type alpha-synuclein and its clinical mutants A30P or A53T. J Neurochem 2008; 106:121 - 33; http://dx.doi.org/ 10.1111/j.1471-4159.2008.05342.x; PMID: 18346205 [DOI] [PubMed] [Google Scholar]
- 110.Deleersnijder A, Gerard M, Debyser Z, Baekelandt V. . The remarkable conformational plasticity of alpha-synuclein: blessing or curse?. Trends Mol Med 2013; 19:368 - 77; http://dx.doi.org/ 10.1016/j.molmed.2013.04.002; PMID: 23648364 [DOI] [PubMed] [Google Scholar]
- 111.Pfefferkorn CM, Heinrich F, Sodt AJ, Maltsev AS, Pastor RW, Lee JC. . Depth of α-synuclein in a bilayer determined by fluorescence, neutron reflectometry, and computation. Biophys J 2012; 102:613 - 21; http://dx.doi.org/ 10.1016/j.bpj.2011.12.051; PMID: 22325285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trexler AJ, Rhoades E. . N-Terminal acetylation is critical for forming α-helical oligomer of α-synuclein. Protein Sci 2012; 21:601 - 5; http://dx.doi.org/ 10.1002/pro.2056; PMID: 22407793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen Y, Delaglio F, Cornilescu G, Bax A. . TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 2009; 44:213 - 23; http://dx.doi.org/ 10.1007/s10858-009-9333-z; PMID: 19548092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu KP, Baum J. . Detection of transient interchain interactions in the intrinsically disordered protein alpha-synuclein by NMR paramagnetic relaxation enhancement. J Am Chem Soc 2010; 132:5546 - 7; http://dx.doi.org/ 10.1021/ja9105495; PMID: 20359221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Outeiro TF, Putcha P, Tetzlaff JE, Spoelgen R, Koker M, Carvalho F, Hyman BT, McLean PJ. . Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS One 2008; 3:e1867; http://dx.doi.org/ 10.1371/journal.pone.0001867; PMID: 18382657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klucken J, Outeiro TF, Nguyen P, McLean PJ, Hyman BT. . Detection of novel intracellular alpha-synuclein oligomeric species by fluorescence lifetime imaging. FASEB J 2006; 20:2050 - 7; http://dx.doi.org/ 10.1096/fj.05-5422com; PMID: 17012257 [DOI] [PubMed] [Google Scholar]
- 117.Binolfi A, Theillet FX, Selenko P. . Bacterial in-cell NMR of human α-synuclein: a disordered monomer by nature?. Biochem Soc Trans 2012; 40:950 - 4; http://dx.doi.org/ 10.1042/BST20120096; PMID: 22988846 [DOI] [PubMed] [Google Scholar]
- 118.Croke RL, Sallum CO, Watson E, Watt ED, Alexandrescu AT. . Hydrogen exchange of monomeric alpha-synuclein shows unfolded structure persists at physiological temperature and is independent of molecular crowding in Escherichia coli. Protein Sci 2008; 17:1434 - 45; http://dx.doi.org/ 10.1110/ps.033803.107; PMID: 18493022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barnes CO, Monteith WB, Pielak GJ. . Internal and global protein motion assessed with a fusion construct and in-cell NMR spectroscopy. Chembiochem 2011; 12:390 - 1; http://dx.doi.org/ 10.1002/cbic.201000610; PMID: 21290539 [DOI] [PubMed] [Google Scholar]
- 120.Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. . Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy. J Am Chem Soc 2008; 130:6310 - 1; http://dx.doi.org/ 10.1021/ja801020z; PMID: 18419123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barnes CO, Pielak GJ. . In-cell protein NMR and protein leakage. Proteins 2011; 79:347 - 51; http://dx.doi.org/ 10.1002/prot.22906; PMID: 21120863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barnes CO, Pielak GJ. . In-cell protein NMR and protein leakage. Proteins 2011; 79:347 - 51; http://dx.doi.org/ 10.1002/prot.22906; PMID: 21120863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McNulty BC, Young GB, Pielak GJ. . Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J Mol Biol 2006; 355:893 - 7; http://dx.doi.org/ 10.1016/j.jmb.2005.11.033; PMID: 16343531 [DOI] [PubMed] [Google Scholar]
- 124.Fauvet B, Mbefo MK, Fares MB, Desobry C, Michael S, Ardah MT, Tsika E, Coune P, Prudent M, Lion N, et al. . α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J Biol Chem 2012; 287:15345 - 64; http://dx.doi.org/ 10.1074/jbc.M111.318949; PMID: 22315227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Burre J, Vivona S, Diao J, Sharma M, Brunger AT, Sudhof TC. . Properties of native brain alpha-synuclein. Nature 2013; 498:E4 - 6 E - 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dettmer U, Newman AJ, Luth ES, Bartels T, Selkoe D. . In vivo cross-linking reveals principally oligomeric forms of α-synuclein and β-synuclein in neurons and non-neural cells. J Biol Chem 2013; 288:6371 - 85; http://dx.doi.org/ 10.1074/jbc.M112.403311; PMID: 23319586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ellis RJ, Minton AP. . Cell biology: join the crowd. Nature 2003; 425:27 - 8; http://dx.doi.org/ 10.1038/425027a; PMID: 12955122 [DOI] [PubMed] [Google Scholar]
- 128.Ellis RJ, Minton AP. . Protein aggregation in crowded environments. Biol Chem 2006; 387:485 - 97; http://dx.doi.org/ 10.1515/BC.2006.064; PMID: 16740119 [DOI] [PubMed] [Google Scholar]
- 129.Coelho-Cerqueira E, do Carmo-Gonçalves P, Pinheiro AS, Cortines J, Follmer C. . α-Synuclein as an intrinsically disordered monomer: fact or artifact?. FEBS J 2013; http://dx.doi.org/ 10.1111/febs.12471; PMID: 23927048 [DOI] [PubMed] [Google Scholar]
- 130.Ullman O, Fisher CK, Stultz CM. . Explaining the structural plasticity of α-synuclein. J Am Chem Soc 2011; 133:19536 - 46; http://dx.doi.org/ 10.1021/ja208657z; PMID: 22029383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gurry T, Ullman O, Fisher CK, Perovic I, Pochapsky T, Stultz CM. . The dynamic structure of α-synuclein multimers. J Am Chem Soc 2013; 135:3865 - 72; http://dx.doi.org/ 10.1021/ja310518p; PMID: 23398399 [DOI] [PubMed] [Google Scholar]
- 132.Hollebeke J, Van Damme P, Gevaert K. . N-terminal acetylation and other functions of Nα-acetyltransferases. Biol Chem 2012; 393:291 - 8; http://dx.doi.org/ 10.1515/hsz-2011-0228; PMID: 22718636 [DOI] [PubMed] [Google Scholar]
- 133.Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, et al. . Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A 2009; 106:8157 - 62; http://dx.doi.org/ 10.1073/pnas.0901931106; PMID: 19420222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Forte GM, Pool MR, Stirling CJ. . N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol 2011; 9:e1001073; http://dx.doi.org/ 10.1371/journal.pbio.1001073; PMID: 21655302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kamita M, Kimura Y, Ino Y, Kamp RM, Polevoda B, Sherman F, Hirano HN. . N(α)-Acetylation of yeast ribosomal proteins and its effect on protein synthesis. J Proteomics 2011; 74:431 - 41; http://dx.doi.org/ 10.1016/j.jprot.2010.12.007; PMID: 21184851 [DOI] [PubMed] [Google Scholar]
- 136.Yi CH, Pan H, Seebacher J, Jang IH, Hyberts SG, Heffron GJ, Vander Heiden MG, Yang R, Li F, Locasale JW, et al. . Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell 2011; 146:607 - 20; http://dx.doi.org/ 10.1016/j.cell.2011.06.050; PMID: 21854985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Starheim KK, Gevaert K, Arnesen T. . Protein N-terminal acetyltransferases: when the start matters. Trends Biochem Sci 2012; 37:152 - 61; http://dx.doi.org/ 10.1016/j.tibs.2012.02.003; PMID: 22405572 [DOI] [PubMed] [Google Scholar]
- 138.Jones JD, O’Connor CD. . Protein acetylation in prokaryotes. Proteomics 2011; 11:3012 - 22; http://dx.doi.org/ 10.1002/pmic.201000812; PMID: 21674803 [DOI] [PubMed] [Google Scholar]
- 139.Ohrfelt A, Zetterberg H, Andersson K, Persson R, Secic D, Brinkmalm G, Wallin A, Mulugeta E, Francis PT, Vanmechelen E, et al. . Identification of novel α-synuclein isoforms in human brain tissue by using an online nanoLC-ESI-FTICR-MS method. Neurochem Res 2011; 36:2029 - 42; http://dx.doi.org/ 10.1007/s11064-011-0527-x; PMID: 21674238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johnson M, Coulton AT, Geeves MA, Mulvihill DP. . Targeted amino-terminal acetylation of recombinant proteins in E. coli. PLoS One 2010; 5:e15801; http://dx.doi.org/ 10.1371/journal.pone.0015801; PMID: 21203426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maltsev AS, Ying J, Bax A. . Impact of N-terminal acetylation of α-synuclein on its random coil and lipid binding properties. Biochemistry 2012; 51:5004 - 13; http://dx.doi.org/ 10.1021/bi300642h; PMID: 22694188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kang L, Moriarty GM, Woods LA, Ashcroft AE, Radford SE, Baum J. . N-terminal acetylation of α-synuclein induces increased transient helical propensity and decreased aggregation rates in the intrinsically disordered monomer. Protein Sci 2012; 21:911 - 7; http://dx.doi.org/ 10.1002/pro.2088; PMID: 22573613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fauvet B, Fares MB, Samuel F, Dikiy I, Tandon A, Eliezer D, Lashuel HA. . Characterization of semisynthetic and naturally Nα-acetylated α-synuclein in vitro and in intact cells: implications for aggregation and cellular properties of α-synuclein. J Biol Chem 2012; 287:28243 - 62; http://dx.doi.org/ 10.1074/jbc.M112.383711; PMID: 22718772 [DOI] [PMC free article] [PubMed] [Google Scholar]