Abstract
Retention of bile acids (BAs) in the liver during cholestasis plays an important role in the development of cholestatic liver injury. Several studies have reported that high concentrations of certain BAs induce cell death and inflammatory response in the liver, and BAs may promote liver tumorigenesis. Macroautophagy (hereafter referred to as autophagy) is a lysosomal degradation process that regulates organelle and protein homeostasis and serves as a cell survival mechanism under a variety of stress conditions. However, it is not known if BAs modulate autophagy in hepatocytes. In the present study, we determined autophagic flux in livers of farnesoid X receptor (FXR) knockout (KO) mice that have increased concentrations of hepatic BAs and in primary cultured mouse hepatocytes treated with BAs. The results showed that autophagic flux was impaired in livers of FXR KO mice and in BA-treated primary mouse hepatocytes. Mechanistically, BAs did not affect the activities of cathepsin or the proteasome, but impaired autophagosomal-lysosomal fusion likely due to reduction of Rab7 protein expression and targeting to autophagosomes. In conclusion, BAs suppress autophagic flux in hepatocytes by impairing autophagosomal-lysosomal fusion, which may be implicated in bile acid-induced liver tumor promotion observed in FXR KO mice.
Key Words: bile acid, autophagy, hepatocytes, farnesoid X receptor.
Bile acids (BAs) are amphipathic molecules synthesized in the liver and are important for the intestinal absorption of dietary fats and fat-soluble vitamins (Chiang, 2003). However, during cholestasis, BAs are accumulated in the liver, resulting in liver injury by inducing cell death (Schoemaker et al., 2003). Hydrophobic BAs, such as cholic acid (CA) and chenodeoxycholic acid (CDCA), are known to be toxic in cultured hepatocytes. The mechanisms for BA-induced toxicity are not fully understood, but activation of Fas death receptor, oxidative stress and endoplasmic reticulum stress may be involved (Faubion et al., 1999; Tamaki et al., 2008; Yerushalmi et al., 2001).
Autophagy is a cellular lysosomal degradation pathway that degrades cellular proteins and damaged organelles to promote cell survival in response to a variety of stresses. The autophagy process is characterized by the formation of double-membrane autophagosomes. Autophagosomes fuse with lysosomes to form autolysosomes where the autophagosome-enwrapped contents are degraded (Mizushima, 2007). The mechanisms by which autophagosomes fuse with lysosomes are not clear, but soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, the small GTP binding protein Rab7, and the homotypic fusion and vacuole protein sorting (HOPS) complex may play a role in regulating the fusion of autophagosomes with lysosomes in mammalian cells (Furuta et al., 2010; Itakura et al., 2012; Jager et al., 2004).
Autophagy is a dynamic process, and the ubiquitin-like protein, microtubule-associated protein 1 light chain 3 (LC3), is thought to be important for isolation membrane elongation and eventual closure of the autophagosomal membrane (Nakatogawa et al., 2007). LC3 is conjugated to phosphatidylethanolamine (PE) (called LC3-II) and targets the autophagosomal membrane (Kabeya et al., 2000). The LC3-II on the outer autolysosome membrane is de-conjugated and removed by the cysteine protease Atg4B and recycled, while the LC3-II on the inner membrane together with the enveloped cytosolic contents are degraded by the lysosome (Kirisako et al., 2000). Thus, the accumulation of LC3-II could be due to either the induction of autophagy or the inhibition of lysosomal functions and/or the defect of fusion of autophagosomes with lysosomes, which leads to impaired degradation of LC3-II (Klionsky et al., 2012). This dynamic process of autophagosome formation, delivery of autophagosomal cargo to lysosomes and completion of lysosomal degradation of cargo is known as autophagic flux. Therefore, autophagic flux is a more accurate indicator of autophagic process than simple measurement of the number of autophagosomes (Mizushima et al., 2010). Autophagy also selectively degrades some specific autophagy substrates such as sequestosome 1 (SQSTM1)/p62, which accumulate in autophagy-deficient cells or mouse liver (Kirkin et al., 2009; Komatsu et al., 2007; Ni et al., 2012). Thus, determination of LC3-II and p62 levels in the presence or absence of lysosomal inhibitors, such as chloroquine (CQ), has been widely used to monitor autophagic flux (Klionsky et al., 2012).
Farnesoid X receptor (FXR) is a member of the nuclear hormone receptor superfamily and highly expressed in the liver, intestine, kidney, and adrenal gland (Forman et al., 1995; Lu et al., 2001). FXR plays a critical role in maintaining cholesterol and BA homeostasis based on studies from FXR knockout (KO) mice and FXR agonists (Kim et al., 2007; Sinal et al., 2000). BAs including CA and CDCA are very potent endogenous ligands of FXR. FXR KO mice have increased hepatic apoptosis, inflammation, compensatory proliferation and hepatomegaly, and spontaneous liver tumors in aging mice (Kim et al., 2007; Yang et al., 2007). All these hepatic pathology in FXR KO mice echo those observed in liver-specific Atg7 or Atg5 KO mice, animals with impaired autophagy in the liver (Inami et al., 2011; Takamura et al., 2011). While the exact mechanisms by which FXR KO mice develop these phenotypes in the liver are under diligent investigation, FXR KO mice have increased concentrations of BAs in liver and serum, which is suggested to partially responsible for promoting liver tumor formation in mice (Kim et al., 2007; Yang et al., 2007). We thus hypothesized that FXR KO mice have impaired autophagy in the liver, which could be due to increased concentrations of BAs. Here, we report that whole body, but not liver-specific, FXR KO mice had increased accumulation of p62 and LC3-II proteins in the liver, suggesting autophagy is impaired in the livers of whole body FXR KO mice. Furthermore, we found that BAs decreased Rab7 expression and its recruitment to autophagosomes, resulting in impaired autophagosomal-lysosomal fusion and subsequent decreased autophagic flux in primary cultured mouse hepatocytes.
MATERIALS AND METHODS
Reagents and Antibodies.
CA, CDCA, and taurocholic acid (TCA) were obtained from Sigma–Aldrich. All cell culture materials were obtained from Invitrogen. Antibodies used in the study were GFP (Santa Cruz Biotechnology, Cat. # sc-9996), p62 (Abnova, Cat. # H00008878-M01), β-Actin (Sigma, Cat. # a5541), Atg5 (MBL, Cat# PM050), GAPDH (Cell Signaling, Cat. # 2118), Rab7 (Cell Signaling Cat. # 9367), and LAMP-1 and LAMP-2 (Developmental Studies Hybridoma BAsnk, Iowa City, IA, Cat. # 1D4B and Cat. # ABL-93). The rabbit polyclonal anti-LC3B antibody was described previously (Ding et al., 2009). The secondary antibodies used in this study for immunoblotting analysis were HRP-conjugated goat anti-mouse (Jackson ImmunoResearch, Cat. #115-035-062), rabbit (Jackson ImmunoResearch, Cat. #111-035-045), and rat (Jackson ImmunoResearch Cat. # 111-035-143) antibodies. The secondary antibodies used for immunostaining were Cy3-conjugated goat anti-rabbit (Jackson ImmunoResearch Cat. # 111-165-144), Cy3-conjugated goat anti-rat (Jackson ImmunoResearch Cat. #112-165-143), DyLight conjugated goat anti-mouse (Jackson ImmunoResearch Cat. #115-505-146), and AlexaFluor 488 goat anti-rabbit antibodies (Invitrogen).
Animals and Primary Mouse Hepatocyte Culture.
Wild type (WT) C57BL/6J, FXR KO, FXRflox/flox (Albumin Cre negative or positive) mice (FXRfl/fl, Alb Cre- or FXRfl/fl, Alb Cre+) were housed with free access to water and chow. All animals received humane care. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center. Mouse hepatocytes isolated from WT and FXR−/− mice were described previously (Ding et al., 2004), and cultured in William’s medium E with 10% fetal bovine serum, 2mM glutamine as well as routine antibiotic supplements for 2h for attachment. Cells were then cultured in the same medium without serum overnight before treatment. All cells were maintained in a 37°C incubator with 5% CO2.
Fluorescence, Confocal and Electron Microscopy and Immune Blotting Analysis.
Adenovirus expressing GFP-LC3 was used as described previously (Gao et al., 2008). To examine autophagy, primary hepatocytes were seeded in a 12 well-plate (1×105 per well) on a cover slide and infected with adenovirus-GFP-LC3 (100 viral particles per cell) overnight. Cells were treated with CA (100 µM), CDCA (100 µM), and TCA (100 µM) in the presence or absence of chloroquine (CQ, 20 µM) for 6h. For some experiments, hepatocytes were transfected with RFP-GFP-LC3 using TurboFect (Fermentas) for 24h followed by designated treatments. After treatments, cells were fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 2h at room temperature or kept at 4°C for microscopy. Fluorescence images were acquired under a Nikon Eclipse 200 fluorescence microscope with MetaMorph software. For the immunostaining assay, fixed primary hepatocytes were immunostained with anti-Lamp1 or anti-Rab7 antibodies followed by Cy3-conjugated secondary antibody and Hoechst 33342 staining as previously described (Ding et al., 2004). Cell images were then obtained using a Leica Confocal microscope (TCS SPE model). For electron microscopy (EM), hepatocytes were fixed with 2.5% glutaraldehyde in 0.1mol/l sodium cacodylate buffer (pH 7.4), followed by 1% OsO4. After dehydration, thin sections were cut and stained with uranyl acetate and lead citrate. Digital images were obtained using a JEM 1016CX electron microscope.
DQ-Bovine Serum Albumin Assay.
Primary cultured mouse hepatocytes were first loaded with DQ-BSA (10 µg/ml, Invitrogen) 1hr prior to treatment with various BAs and CQ for another 6h. DQ-BSA is a derivative of BSA that is labeled to such a high degree with BODIPY dyes, BODIPY TR-X, that the dye is strongly self-quenched. Proteolysis of the BODIPY-BSA conjugate results in de-quenching and the released protein fragment that contains isolated red fluorophore is brightly fluorescent that has excitation and emission maxima of 590 and 620nm. Following treatment, hepatocytes were washed with PBS to remove excessive DQ-BSA and lysed in 1% Triton X-100 in 50mM Tris-HCL (pH 8.8) solution. Fluorescence intensity of the lysates was quantified using a Tecan Infinite M200 Pro plate reader (excitation: 590 and emission: 620).
RNA Isolation and Real-Time qPCR.
RNA was isolated from cultured hepatocytes and livers using Qiagen RNeasy kit (Qiagen, Valencia, CA) and reverse transcribed into cDNA by RevertAid reverse transcriptase (Fermentas). Real-time PCR was performed on an Applied Biosystems Prism 7900HT real-time PCR instrument (ABI, Foster City, CA) using Maxima SYBR green/rox qPCR reagents (Fermentas). Primer sequences were as follows: β-actin forward: 5′ – TGT TAC CAA CTG GGA CGA CA – 3′; β-actin reverse: 5′ – GGG GTG TTG AAG GTC TCA AA – 3′; Map1lc3b forward: 5′ – CCGAGAAGACCTTCAAGCAG – 3′; Map1lc3b reverse: 5′ – ACACTTCGGAGATGGGAGTG – 3′; Sqstm1/p62 forward: 5′ – aga atg tgg ggg aga gtg tg– 3′; Sqstm1/p62 reverse: 5′ – tcg tct cct cct gag cag tt – 3′; Shp forward: 5′ – ctg cag gtc gtc cga cta tt – 3′; and Shp reverse: 5′ – acc tcg aag gtc aca gca tc – 3′.
Cathepsin B and Proteasome Activity Assay.
Specific fluorescence substrates were used to measure Cathepsin B and proteasome activities. Primary cultured mouse hepatocytes were treated with CA (100 µM), CDCA (100 µM), TCA (100 µM), CQ (20 µM) or E64D (10 µM) for 6h. After treatment, cells were lysed in M2 buffer (50mM Tris pH 7.5, 130mM NaCl, 10% glycerol, 0.5% NP-40, 0.5mM EDTA, and 0.5mM EGTA in dH2O) without protease inhibitors. To determine cathepsin B activity, protein lysates (15 µg) were incubated with 2 µM z-RR-AMC (Calbiochem # 219392) in assay buffer(10mM HEPES-NaOH pH 7.4, 220mM mannitol, 68mM sucrose, 2mM NaCl, 2.5mM KH2PO4, 0.5mM EGTA, 2mM MgCl2, 5mM pyruvate, 0.1mM PMSF, and 1mM DTT in dH2O) for 1hr, and fluorescence intensity was determined using a Tecan Infinite M200 Pro plate reader (excitation: 380 and emission: 460). To determine proteasome activity, protein lysates (10 µg) were incubated with 0.65 µM Suc-LLVY-AMC (Calbiochem #539142) in assay buffer (50mM Tris pH 7.5, 25mM KCl, 10mM NaCl, and1mM MgCl2) for 1hr, and the fluorescence intensity was determined using a Tecan Infinite M200 Pro plate reader (excitation: 380nm and emission: 460nm).
Immunoblot Analysis.
Cells were washed in PBS and lysed in RIPA buffer. Twenty micrograms of protein from each sample were separated by SDS–PAGE and transferred to PVDF membranes. The membranes were stained with primary antibodies followed by secondary horseradish peroxidase-conjugated antibodies. The membranes were further developed with SuperSignal West Pico chemiluminescent substrate (Pierce). Densitometry analysis was performed using Un-Scan-It software and further normalized with β-actin.
Statistical Analysis.
All experimental data were expressed as mean ± SE and subjected to a Student t-test or one-way analysis of variance with Holm–Sidak post hoc test where appropriate. *p < .05 was considered significant.
RESULTS
Increased Accumulation of Hepatic p62 and LC3-II Proteins in FXR KO Mice
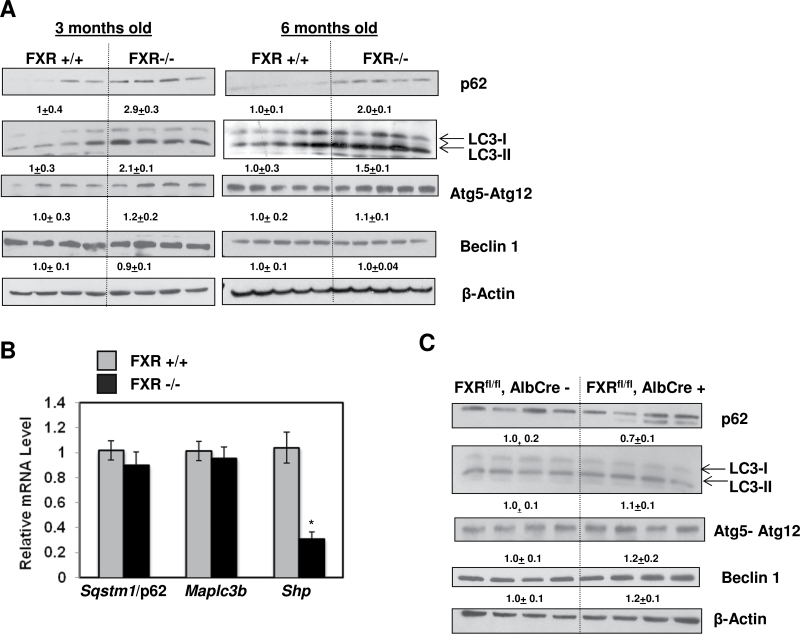
Since mice with autophagy-deficiency in the liver and FXR KO mice all develop spontaneous liver tumors, we hypothesized that FXR KO mice may have impaired autophagy in the liver. Indeed, similar to liver-specific Atg7 or Atg5 KO mice, both 3- and 6-month-old FXR KO mice had increased accumulation of p62 proteins in the liver (Fig. 1A). Interestingly, hepatic LC3-II protein levels were also increased in 3- and 6-month-old FXR KO mice compared to their age-matched WT controls. In contrast, other autophagy proteins that regulate the upstream events for the formation of autophagosome such as Atg5-Atg12 and Beclin 1 seemed to be no difference between WT and FXR KO mice. Moreover, no difference in hepatic mRNA levels of Sqstm1/p62 or Map1lc3b (Microtubule-associated protein 1 light chain 3 beta) between WT and FXR KO mice, whereas the level of Shp, a FXR target gene, was dramatically diminished in FXR KO mice (Fig. 1B). It is known that FXR KO mice have increased hepatic and serum BAs (Kim et al., 2007; Sinal et al., 2000), we next determined whether the increased levels of p62 and LC3-II was due to the increased BAs or the lack of FXR. We thus determined the levels of p62 and LC3-II in liver-specific FXR KO mice, which we previously showed that these mice had normal hepatic BAs (Borude et al., 2012). No significant differences were found for p62, LC3-II, Atg5-Atg12, and Beclin 1 protein expression between liver-specific FXR KO mice and their WT littermates (Fig. 1C). Altogether, these data indicate that FXR KO mice had increased hepatic levels of p62 and LC3-II, which is less likely due to the direct effects of lack of FXR.
FIG. 1.
Increased p62 and LC3-II protein expression in FXR KO mouse livers. A, Total lysates from 3- and 6-month-old FXR+/+ and FXR−/− mouse livers were subjected to immunoblot analysis. Digital data are densitometry analysis and presented as a ratio of wild type mice (n = 4–5). B, mRNA was isolated from around 3-month-old FXR+/+ and FXR−/− KO mouse livers and qRT-PCR was performed. The gene expression levels were normalized to β-actin and shown as fold increase over wild type mice (n = 6–7). C, Total liver lysates from 3-month-old FXRfl/fl:Alb-Cre- and FXRfl/fl:Alb Cre+ mouse livers were subjected to immunoblot and densitometry analysis.
BAs Increased p62 and LC3-II Proteins in Primary Cultured Mouse Hepatocytes
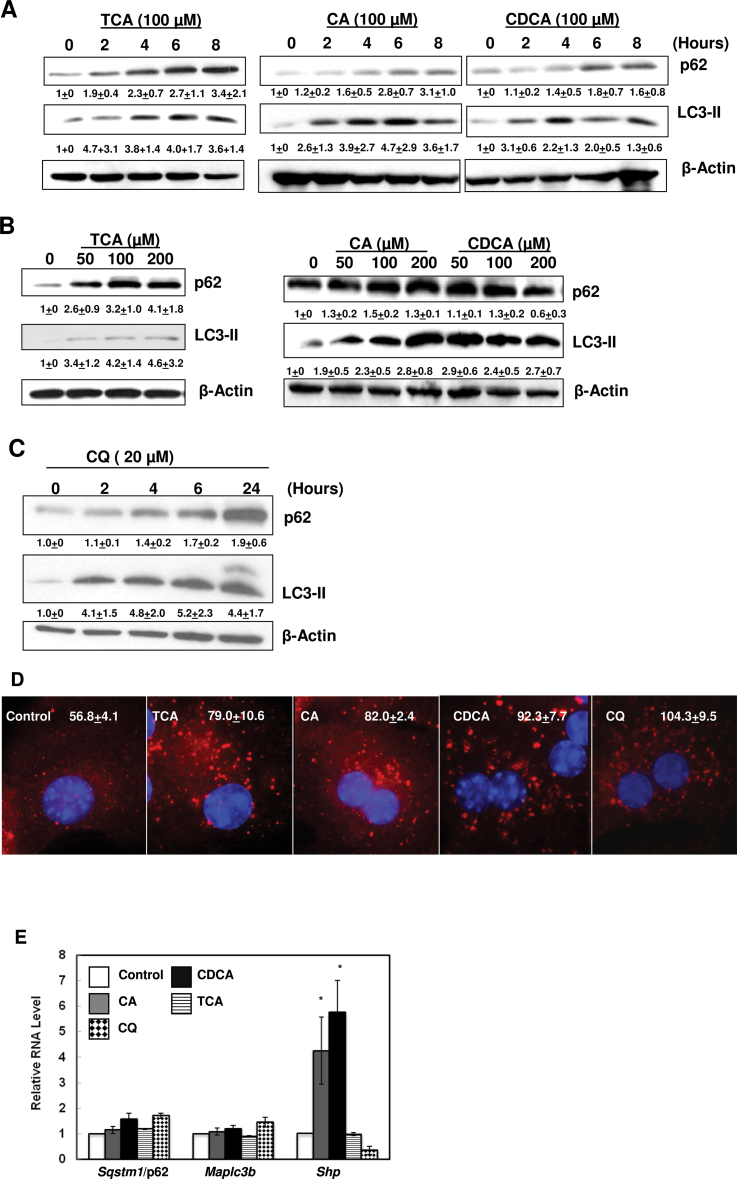
Since hepatic BA levels were elevated in FXR KO mice (Kim et al., 2007; Sinal et al., 2000), we determined whether BAs would directly modulate p62 and LC3-II levels. Three major primary BAs were chosen for the in vitro studies, TCA, CA, and CDCA. TCA was chosen because serum and hepatic levels of TCA increased to around 1mM in BDL mice (Zhang et al., 2012). CA was chosen because serum levels of CA increased to 200–250 µM in mice subjected to BDL for 7 days (Marschall et al., 2006), and CA has been widely used in feeding experiments on mice that show significant toxicity in mouse liver (Kim et al., 2007; Sinal et al., 2000). We also chose CDCA because serum levels of CDCA increase in BDL mice, and CDCA increases expression of inflammatory genes in cultured mouse hepatocytes (Allen et al., 2011). Exposure of primary hepatocytes to TCA, CA and CDCA increased p62 and LC3-II protein levels in a time- and dose-dependent manner (Figs. 2A and 2B). We have previously found that unlike the liver tissue, it was difficult to detect the LC3-I form in the primary cultured mouse hepatocytes. This either could be due to the high basal level of autophagy in primary cultured or the antibody that we used preferentially binding to LC3-II form (Ding et al., 2009, 2010). Exposure of primary hepatocytes to CQ, which suppresses autophagy by increasing lysosomal pH, also increased p62 and LC3-II protein levels in a time-dependent manner (Fig. 2C). Consistent with the immunoblotting analysis, BA- or CQ-treated hepatocytes had increased number and size of p62 dots (Fig. 2D). BA treatment did not increase mRNA levels of Map1lc3b or Sqstm1/p62, but both CA and CDCA increased the mRNA level of Shp, a well-known FXR target gene that is induced by BAs (Fig. 2E). CQ treatment slightly increased the expression of Map1lc3b and Sqstm1/p62 but decreased the expression of Shp (Fig. 2E). These results indicate that BA-induced p62 and LC3-II protein accumulations are due to post-translational regulation but not due to regulation of gene transcription in primary mouse hepatocytes.
FIG. 2.
BAs increased p62 and LC3-II protein levels in hepatocytes in a time- and dose-dependent manner. Primary hepatocytes were treated with CA, CDCA, or TCA for 0, 2, 4, 6, and 8h (A) or (B) with different concentrations of BAs for 6h. Whole cell lysates were subjected to immunoblot analysis. Densitometry analysis data are presented as a ratio of control (n = 3–4). C, Primary hepatocytes were treated with CQ for various time points and total cell lysates were subjected to immunoblot analysis, and densitometry analysis was performed as described in (A) (n = 3). D, Hepatocytes were treated with 100 µM of TCA, CA, CDCA or CQ (20 µM) for 6h and immunostained for p62 followed by fluorescence microscopy. Representative images are shown. The number of p62 puncta per cell was quantified (>20 cells were counted in each experiment from at least three independent experiments). E, Primary hepatocytes were treated as in (D) and qRT-PCR was performed. The gene expression levels were normalized to β-actin and shown as fold increase over control (n = 3).
No Increased Autophagic Flux in BA-Treated Primary Mouse Hepatocytes
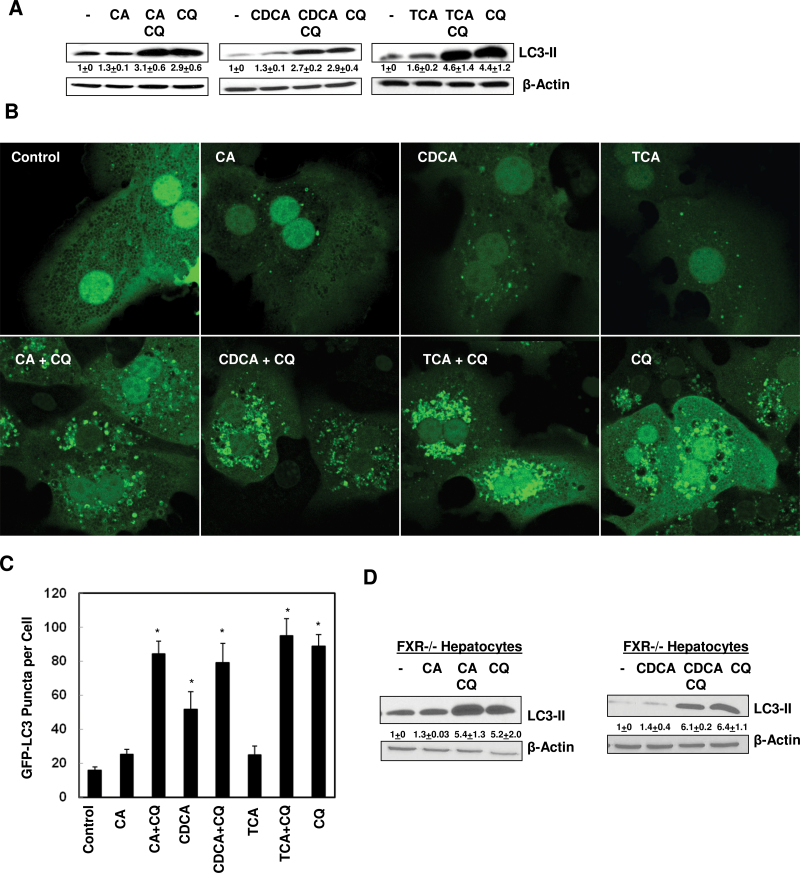
We next performed autophagic flux assays in BA-treated hepatocytes. Primary hepatocytes were exposed to BAs in the presence or absence of CQ for 6h. As can be seen, LC3-II levels were increased in BA-treated cells and were further increased in the presence of CQ, but the co-treatment of BAs with CQ had almost the same LC3-II levels as treatment with CQ alone (Fig. 3A). Furthermore, the number of GFP-LC3 puncta increased in BA-treated cells, but the number of GFP-LC3 puncta was not further increased in cells co-treated with BAs and CQ compared to cells that were treated with CQ alone (Figs. 3B and 3C). To further determine whether BAs-induced impaired autophagic flux would require FXR, we treated primary cultured FXR−/− hepatocytes with BAs in the presence or absence of CQ for 6h. Similar to the results that we observed in WT hepacoytes, both CA and CDCA did not further increase the LC3-II and p62 levels in the presence of CQ compared to the CQ alone treatment (Fig. 3D). These results indicate that BAs did not increase autophagic flux in both WT and FXR−/− hepatocytes. The lack of increased autophagic flux, together with increased p62 levels by BAs, suggests that BAs may impair autophagic degradation in hepatocytes independent of FXR.
FIG. 3.
BAs did not increase autophagic flux in hepatocytes. A, Primary hepatocytes were treated with 100 µM of CA, CDCA, or TCA with or without CQ (20 µM) for 6h. Total cell lysates were subjected to immunoblot analysis followed by densitometry analysis. Data were presented as a ratio of control (n = 4–8). B, Primary hepatocytes were infected with adenoviral GFP-LC3 (100 viral particles per cell) overnight and treated with as in (A) followed by fluorescence microscopy. Representative GFP-LC3 images are shown. C, The number of GFP-LC3 dots per cell was determined (>20 cells were counted in each experiment from at least three independent experiments). D, Primary FXR−/− hepatocytes were treated with 100 µM of CA or CDCA, with or without CQ (20 µM) for 6h. Total cell lysates were subjected to immunoblot analysis followed by densitometry analysis. Data were presented as a ratio of control (n = 3).
BAs Did Not Affect Lysosome or Proteasome Function in Hepatocytes
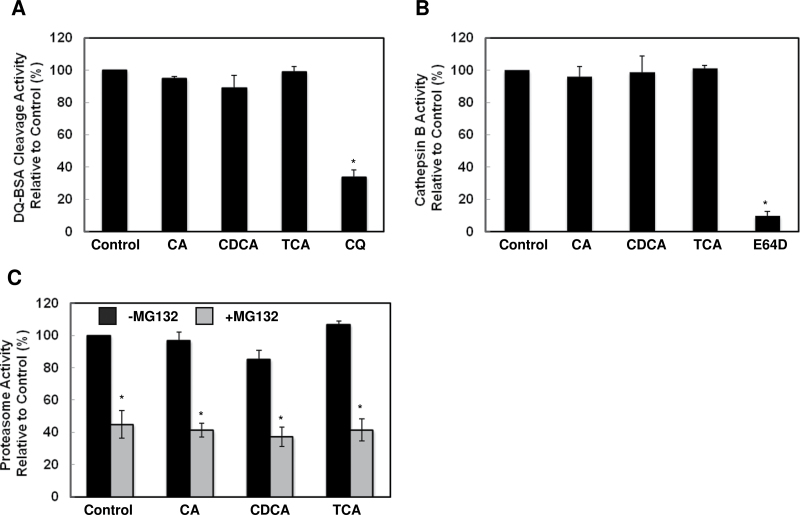
Since BAs increased LC3-II and p62 levels in hepatocytes, a phenotype typically induced by the lysosomal inhibitor CQ although the effects induced by BAs were less potent compared to CQ. We next determined the effects of BAs on lysosomal function. DQ-BSA is BSA that is heavily labeled with a BODIPY dye that it self-quenches. Once it enters lysosomal compartments, DQ-BSA is cleaved by lysosomal proteases, which causes release of a single, dye-labeled fluorescent peptide. Thus, the fluorescence intensity of DQ-BSA can be used to reflect lysosomal function (Klionsky et al., 2012). We found BAs did not alter the fluorescence intensity released from DQ-BSA compared to control cells, but CQ treatment significantly decreased the fluorescence intensity (Fig. 4A). Furthermore, cathepsin B activity was similar among cellular lysates prepared from BA-treated hepatocytes compared to lysates from control cells. As a positive control, lysates from E64D (a cysteine protease inhibitor)-treated cells had significantly decreased cathepsin B activity (Fig. 4B). Because p62 protein levels may also be regulated by the proteasome, we next determined proteasome activity in BA-treated hepatocytes. Treatment with BAs did not alter the proteasome activity in hepatocytes but the addition of MG132 (a proteasome inhibitor) in the lysates decreased proteasome activity (Fig. 4C). Collectively, these results suggest that unlike CQ, BAs do not directly disrupt lysosomal integrity or function. The accumulated p62 and LC3-II in BA-treated hepatocytes could be due to an impaired autophagic process upstream of lysosomal degradation, but it is not due to impaired proteasome function.
FIG. 4.
BAs did not directly affect lysosomal or proteasome functions. A, Primary hepatocytes were pre-loaded with DQ-BSA (5 µg/ml) for 1hr and then treated with BAs for 6h. Fluorescence intensity from cleaved DQ-BSA was measured using total cell lysates. Hepatocytes were treated with BAs for 6h, and total cell lysates were either incubated with z-RR-AMC for cathepsin B activity (B) or Suc-LLVY-AMC with or without MG132 (2 µm) for proteasome activity measurement (C) (n = 3).
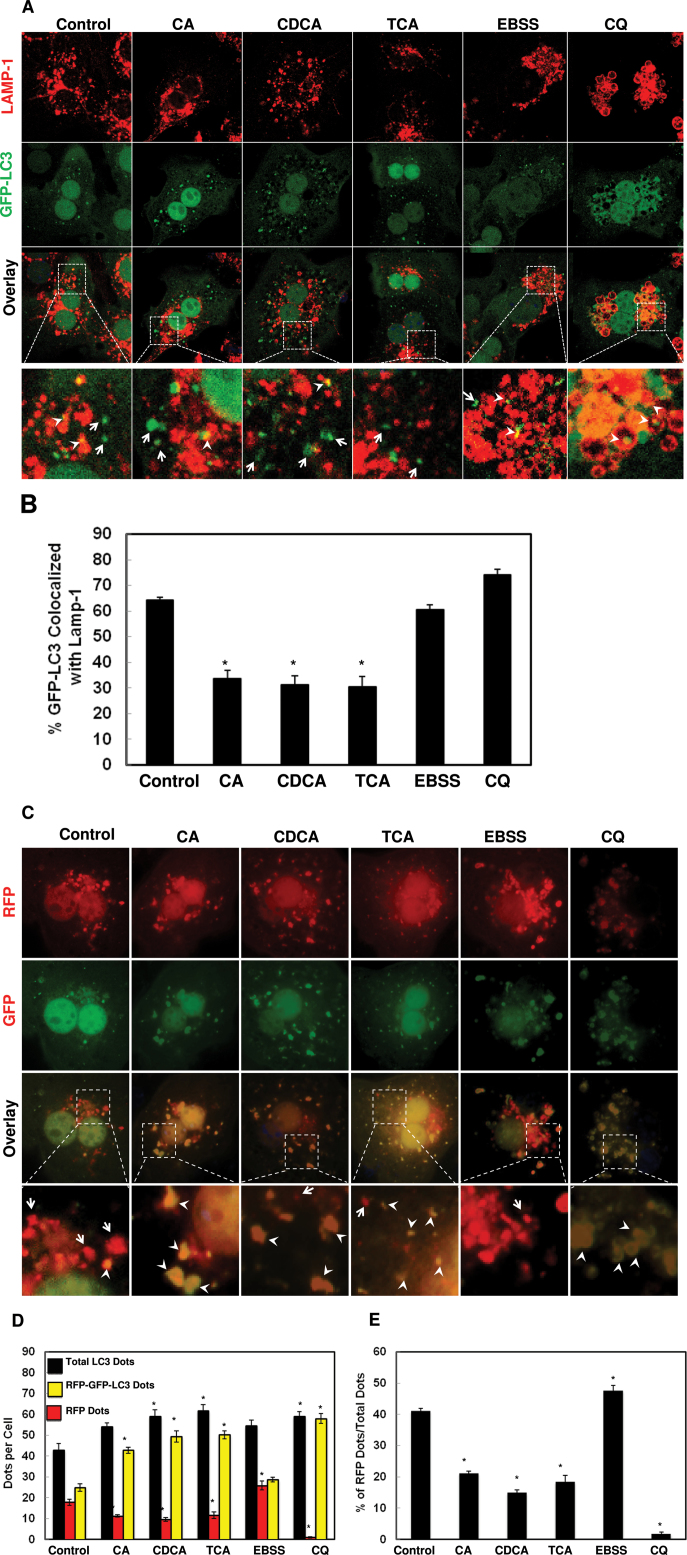
BAs Inhibited Autophagosomal-Lysosomal Fusion
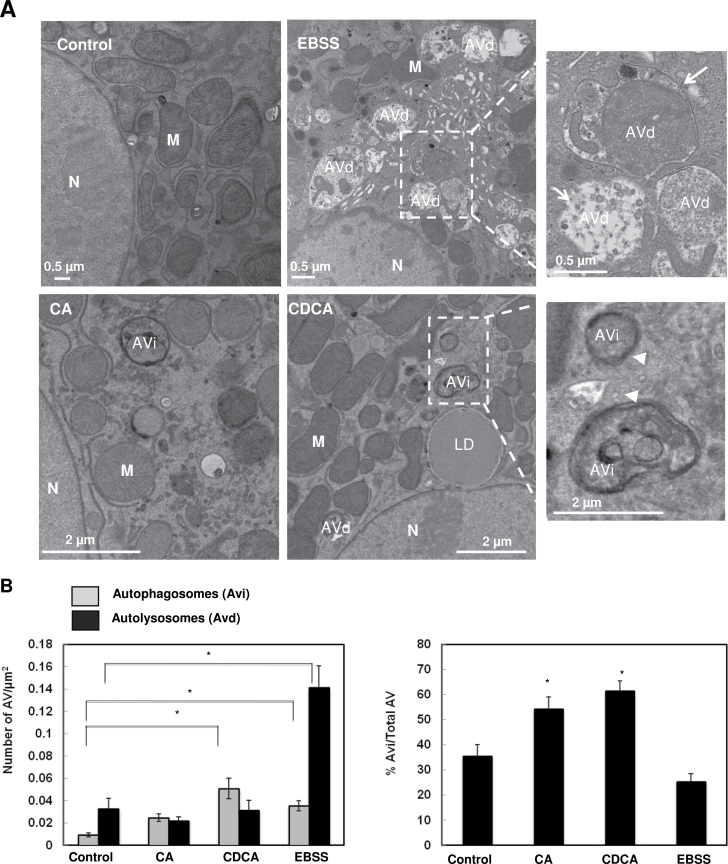
To determine whether BAs would affect the fusion of autophagosomes with lysosomes, we performed immunostaining for the lysosomal associated membrane protein 1 (LAMP-1), a lysosomal outer membrane protein, and quantified the colocalization of LAMP-1 with GFP-LC3 puncta after BA treatment. We found that cells treated with BAs decreased the colocalization of GFP-LC3 puncta with LAMP-1 compared to control cells or cells cultured in Earle’s Balanced Salt Solution (EBSS), which represent basal autophagy or starvation-induced autophagy (Figs. 5A and 5B), respectively. We also found that CQ treatment slightly increased the colocalization of GFP-LC3 puncta with LAMP-1 compared to untreated cells, suggesting that CQ did not affect the fusion of autophagosomes with lysosomes. To further confirm that BAs might impair the maturation of autophagosomes, we transfected hepatocytes with tandem RFP-GFP-LC3. In this assay, RFP fluorescence is more stable in acidic compartments, and GFP fluorescence is rapidly quenched, so mature autolysosomes will have red puncta. Blocking the fusion of an autophagosome with a lysosome or suppressing lysosomal degradation (ie, increase lysosomal pH by CQ) increases the number of yellow puncta (Kimura et al., 2007; Ni et al., 2011). Consistent with the GFP-LC3 and LAMP-1 colocalization results, the number of RFP only positive dots was significantly higher in control cells or amino acid-starved cells than in cells treated with BAs or CQ (Figs. 5C–E). EM analysis also revealed that amino acid-starved cells had more single membrane autolysosomes (Avd) with degraded contents (Figs. 6A, arrows, B and C), whereas cells treated with BAs had more double membrane early autophagosomes (Avi) enclosing undegraded contents (Fig. 6A, arrow heads, and B and C). Taken together, these data collectively suggest that BAs may impair the fusion of autophagosomes with lysosomes in hepatocytes.
FIG. 5.
BAs impaired autophagosomal-lysosomal fusion. A, Primary hepatocytes were infected with adenoviral GFP-LC3 (100 viral particles per cell) overnight and then treated with 100 µM of BAs, or 20 µM of CQ or EBSS for 6h. Cells were immunostained for LAMP-1 followed by confocal microscopy. Representative images are shown, and the lower panels are enlarged images from the boxed areas. Arrow heads: yellow dots (colocalization of GFP-LC3 puncta with LAMP-1) and arrows: GFP-LC3 puncta only. B, Percentage of GFP-LC3 puncta colocalized with LAMP-1 (>20 cells were counted in each experiment from at least three independent experiments). C, Primary hepatocytes were transfected with RFP-GFP-LC3 plasmid for 24h and then treated as in (A). Representative images are shown, and the lower panels are enlarged images from the boxed areas. Arrow heads: yellow dots (RFP-GFP-LC3 puncta) and arrows: RFP-LC3 puncta only. D, Total RFP-LC3 and RFP-GFP-LC3 puncta were quantified and percentage of RFP-LC3 only puncta was calculated (E) (>20 cells were counted in each experiment from at least three independent experiments).
FIG. 6.
BAs induced accumulation of early autophagosomes. A, Hepatocytes were treated with 100 µM of CA or CDCA or EBSS for 6h. Representative EM images are shown. Right panels are enlarged images from the boxed areas. Arrow heads: early autophagosome (Avi) and arrows: autolysosome (Avd). M, mitochondria; N, nuclei. B, Early autophagosomes (AVi) and late autophagosomes (AVd) were quantified and AVi/AVd ratio is presented (>20 different cell sections).
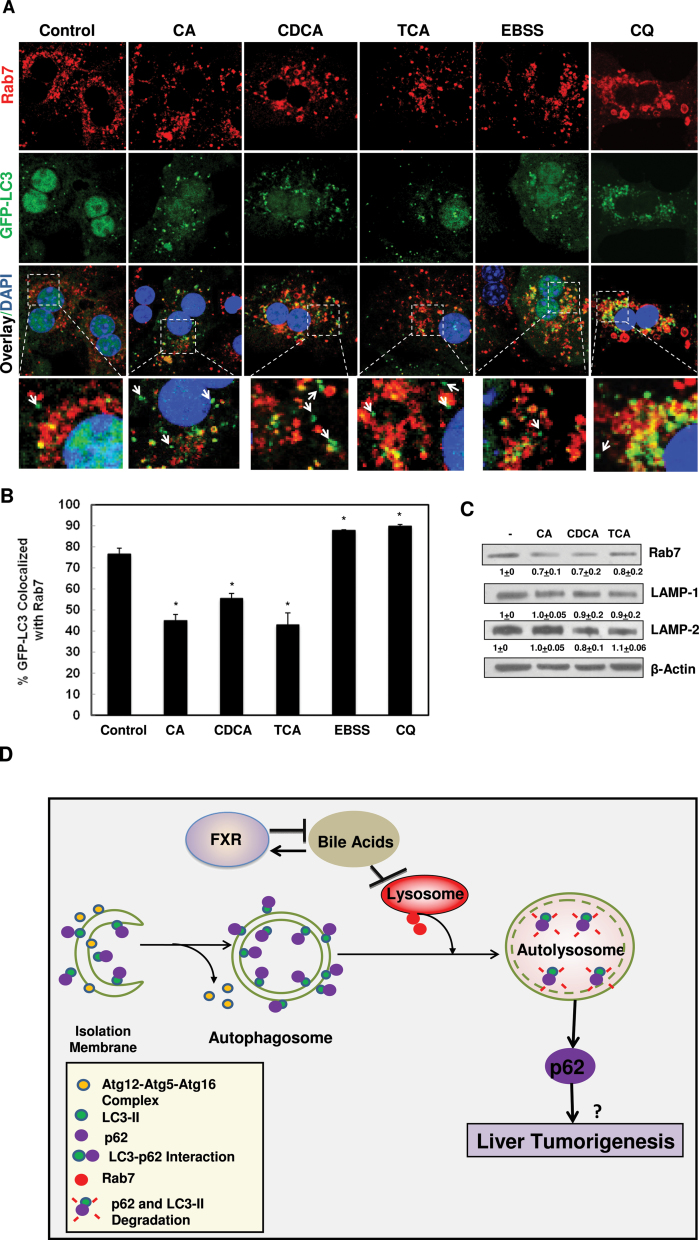
BAs Decreased Levels of Rab7 and Targeting to Autophagosomes in Hepatocytes
Rab7, a small GTPase protein, is important for regulating the fusion of autophagosomes with lysosomes (Chua et al., 2011; Jager et al., 2004). Therefore, we determined if BAs affected Rab7-mediated fusion of autophagosomes with lysosomes. Indeed, the Rab7 staining displayed a peri-nucler pattern in non-treated hepatocytes that is quite similar to the LAMP1 staining, which is consistent with the notion that most Rab7 were localized on the late endosomal/lysosomal compartments. More importantly, we found that around 78% of GFP-LC3 puncta were colocalized with Rab7 positive compartments, which was decreased to approximately 40%–55% in hepatocytes treated with BAs (Figs. 7A and 7B). In contrast, almost 90% of GFP-LC3 puncta were colocalized with Rab7 positive compartments in amino acid-starved or CQ-treated hepatocytes. Furthermore, treatment with BAs also decreased the protein levels of Rab7, but not LAMP1 or LAMP2 (Fig. 7C). These results suggest that BAs impair autophagosomal-lysosomal fusion likely due to decreased Rab7-mediated fusion events. The possible cellular events induced by BAs that might cause the inhibition of autophagic flux in hepatocytes were proposed in Figure 7D.
FIG. 7.
BAs decreased the colocalization of GFP-LC3 with Rab7. A, Primary hepatocytes were infected with adenovirus GFP-LC3 (100 viral particles per cell) overnight and then treated with BAs, CQ or EBSS for 6h. Cells were then immunostained with Rab7 followed by confocal microscopy. Representative images are shown, and the lower panels are enlarged images from the boxed areas. Arrows: GFP-LC3 puncta only. B, Percentage of GFP-LC3 colocalized with Rab7 was quantified (>20 cells were counted in each experiment from at least three independent experiments). C, Hepatocytes were treated with BAs for 6h and total cell lysates were subjected to immunoblot analysis followed by densitometry analysis as described in Figure 1A (n = 3). D, A proposed model that BAs inhibit autophagic flux by suppressing Rab7-mediated autophagsomal-lysosomal fusion in hepatocytes. During autophagy induction, small pieces of isolation membranes grow to form double-membrane autophagosomes, a process that is regulated by Atg5-Atg12-Atg16 complex and LC3-PE conjugation (LC3-II). LC3 also directly interacts with autophagy substrate protein p62 and recruit p62 into autophagosomes. Autophagosomes then fuse with lysosomes to form autolysosomes, where LC3-II and p62 are degraded. This process is mediated by Rab7 and other fusion proteins on either autophagosomes or lysosomes. BAs activate FXR whereas activated FXR negatively regulates synthesis of hepatic BAs. BAs inhibit the fusion of autophagosomes with lysosomes in hepatocytes likely due to decreased expression of Rab7 and its targeting to autophagosomes, resulting in impaired autophagic flux. Impaired autophagic flux leads to the accumulation of LC3-II and p62. Accumulated p62 may contribute to liver tumorigenesis.
DISCUSSION
BAs are known to exert hepatotoxicity and induce apoptosis by activating either the death receptor or extrinsic apoptosis pathways (Jaeschke et al., 2002). However, the role of BAs in hepatic autophagy is not clear. A previous study showed that deoxycholic acid (DCA) treatment increased the number of GFP-LC3 puncta in primary rat hepatocytes (Zhang et al., 2008). While the authors concluded that DCA induced autophagy in primary rat hepatocytes, their conclusion was questionable because no autophagic flux assay was conducted in their study. We also found that BAs increased LC3-II protein levels as well as the number of GFP-LC3 puncta, which are in agreement with this previous report. However, results from further autophagic flux assays clearly indicated that BAs inhibit autophagic flux in hepatocytes.
How do BAs inhibit autophagic flux? Several mechanisms may lead to decreased autophagic flux, which include: (1) inhibition of the upstream induction of autophagy and autophagosome biogenesis; (2) direct inhibition of lysosomal function; or (3) inhibition of autophagosomal-lysosomal fusion. We found that BA-treatment alone caused increased LC3-II levels and GFP-LC3 puncta; therefore, it is less likely that BAs inhibit upstream autophagosome biogenesis. Furthermore, our results also demonstrated that BAs did not affect cathepsin B or lysosomal proteolytic activity, suggesting that BAs may not directly impair lysosomal function. This is in agreement with a previous study reporting that TCA did not inhibit cathepsin activity or alter lysosomal pH in rat liver or in isolated rat hepatocytes (Larocca et al., 1999). Therefore it is likely that BAs might decrease autophagic flux by impairing autophagosomal-lysosomal fusion. Indeed, results from multiple different approaches including colocalization of GFP-LC3 with LAMP-1, tandem RFP-GFP-LC3 assay and EM studies strongly support that BAs inhibit autophagosomal-lysosomal fusion. We also found that BA treatment not only decreased Rab7 protein levels, but also decreased its targeting to GFP-LC3 positive compartments, which could eventually lead to decreased autophagosomal-lysosomal fusion. Increased intracellular levels of Ca2+ by thapsigargin has been shown to block the recruitment of Rab7 to autophagosomes resulting in defects for autophagosomal-lysosomal fusion (Ganley et al., 2011). Interestingly, BAs have also been shown to induce endoplasmic reticulum stress and elevated intracellular Ca2+ in rat hepatocytes (Tsuchiya et al., 2006). It would be interesting to determine the possible role of intracellular Ca2+ in BA-mediated inhibition of autophagosomal-lysosomal fusion.
What is the significance of the inhibition of hepatic autophagy by BAs? A large body of evidence suggests that FXR may act as a tumor suppressor against liver tumorigenesis. FXR may act on multiple levels to suppress liver tumorigenesis via control of BA homeostasis, prevention of hepatocyte apoptosis, reduction of reactive oxygen species production, inhibition of hepatic inflammation, as well as activation of the expression of Shp (Wang et al., 2013). In contrast, an aberrant high level of BAs has been shown to act as a liver tumor promoter. Firstly, clinical studies reveal a close association between cholestatic liver diseases and liver cancer (Jansen, 2007). Secondly, a CA-enriched diet strongly promotes diethylnitrosamine initiated liver tumorigenesis in mice (Yang et al., 2007). Thirdly, FXR KO mice, which have elevated levels of hepatic BAs, develop spontaneous liver tumors that are significantly reduced by feeding the mice with cholestyramine, a BA-sequestering resin (Yang et al., 2007). However, it is not clear how BAs promote liver tumorigenesis, although it has been suggested that high levels of BAs might induce DNA damage to inactivate tumor suppressor genes, cell death, and the inflammatory response in the liver (Wang et al., 2013). Autophagy has been known to serve as a tumor suppressor. Beclin1 heterozygous mice that have decreased expression of Beclin 1, a gene involved in the autophagy process, have increased spontaneous tumors in multiple tissues including liver (Qu et al., 2003). Furthermore, liver-specific Atg5 or Atg7 KO mice also have increased liver tumors but the tumor progression was significantly blunted in the Atg7/p62 double KO mice (Inami et al., 2011; Takamura et al., 2011), supporting the notion that autophagy is a bona fide tumor suppressor and increased p62 levels could be one of the important factors responsible for the tumorigenesis associated with impaired-autophagy. We previously found that FXR can directly bind to the Sqstm1/p62 gene as determined by chromatin immunoprecipitation (Williams et al., 2012). However, when mice were treated with a synthetic FXR agonist, we found that the mRNA and protein expression levels of Sqstm1/p62 only increased in mouse ileum but not in liver. How FXR differentially regulate Sqstm1/p62 expression in different mouse tissues is still not clear. However, we found that p62 protein accumulated in FXR KO mouse liver, suggesting that other transcriptional factor(s) would be important in the regulating the expression of Sqstm1/p62. Nevertheless, the findings that BAs inhibited autophagic flux in primary cultured mouse hepatocytes and in mouse liver in the present study suggest that decreased autophagy may be a novel mechanism that accounts for the liver tumorigenesis induced by BAs, and for the tumorigenesis observed in FXR ko mice.
In conclusion, we demonstrated that BAs inhibit autophagic degradation in vitro and may also play a role in the impaired hepatic autophagy in FXR KO mice in vivo. We further demonstrated that BAs decreased the expression of Rab7 and targeting to the autophagosome resulting in decreased autophagosomal-lysosomal fusion. Autophagy deficiency has been shown to cause liver injury and promote liver tumorigenesis, and increased concentrations of BAs have been linked to spontaneous liver tumor development in FXR KO mice. Therefore, our results suggest a possible link between BAs and impaired autophagy in BA-induced hepatotoxicity and liver tumorigenesis. Modulating autophagy function could be a promising therapeutic approach for preventing cholestasis and related liver tumorigenesis.
FUNDING
National Institute on Alcohol Abuse and Alcoholism (R01 AA020518-01); National Center for Research Resources (5P20RR021940-07); National Institute of General Medical Sciences (8 P20 GM103549-07), as well as DK031343 and DK090036 (G.L.G).
ACKNOWLEDGMENTS
Sharon Manley is a recipient of the Biomedical Research Training Program Fellowship from University of Kansas Medical Center.
REFERENCES
- Allen K., Jaeschke H., Copple B. L. (2011). Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am. J. Pathol. 178, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borude P., Edwards G., Walesky C., Li F., Ma X., Kong B., et al. (2012). Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology 56(6), 2344–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. Y. (2003). Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 284, G349–G356 [DOI] [PubMed] [Google Scholar]
- Chua C. E., Gan B. Q., Tang B. L. (2011). Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell. Mol. Life Sci. 68, 3349–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W. X., Li M., Chen X., Ni H. M., Lin C. W., Gao W., et al. (2010). Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 139(5), 1740–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W. X., Ni H. M., DiFrancesca D., Stolz D. B., Yin X. M. (2004). Bid-dependent generation of oxygen radicals promotes death receptor activation-induced apoptosis in murine hepatocytes. Hepatology 40, 403–413 [DOI] [PubMed] [Google Scholar]
- Ding W. X, Ni H. M., Gao W., Chen X., Kang J. H., Stolz D. B., et al. (2009). Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol. Cancer Ther. 8(7), 2036–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubion W. A., Guicciardi M. E., Miyoshi H., Bronk S. F., Roberts P. J., Svingen P. A., et al. (1999). Toxic bile salts induce rodent hepatocyte apoptosis via direct activation of Fas. J. Clin. Invest. 103(1), 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., et al. (1995). Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81(5), 687–693 [DOI] [PubMed] [Google Scholar]
- Furuta N., Fujita N., Noda T., Yoshimori T., Amano A. (2010). Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol. Biol. Cell 21, 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I. G., Wong P. M., Gammoh N., Jiang X. (2011). Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell 42, 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Ding W. X., Stolz D. B., Yin X. M. (2008). Induction of macroautophagy by exogenously introduced calcium. Autophagy 4, 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y., Waguri S., Sakamoto A., Kouno T., Nakada K., Hino O, et al. (2011). Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell. Biol. 193(2), 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Mizushima N. (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 [DOI] [PubMed] [Google Scholar]
- Jaeschke H., Gores G. J., Cederbaum A. I., Hinson J. A., Pessayre D., Lemasters J. J. (2002). Mechanisms of hepatotoxicity. Toxicol. Sci. 65, 166–176 [DOI] [PubMed] [Google Scholar]
- Jager S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., et al. (2004). Role for Rab7 in maturation of late autophagic vacuoles. J. Cell. Sci. 117(Pt 20), 4837–4848 [DOI] [PubMed] [Google Scholar]
- Jansen P. L. (2007). Endogenous bile acids as carcinogens. J. Hepatol. 47, 434–435 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19(21), 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Morimura K., Shah Y., Yang Q., Ward J. M., Gonzalez F. J. (2007). Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 28, 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., et al. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell. Biol. 151(2), 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y. S., Bjorkoy G., Nunn J. L., Bruun J. A., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33(4), 505–516 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo-Arozena A., Adeli K., et al. (2012). Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8(4), 445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., et al. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131(6), 1149–1163 [DOI] [PubMed] [Google Scholar]
- Larocca M. C., Pellegrino J. M., Rodriguez Garay E. A., Marinelli R. A. (1999). Taurocholate-induced inhibition of hepatic lysosomal degradation of horseradish peroxidase. Biochim. Biophys. Acta 1428, 341–347 [DOI] [PubMed] [Google Scholar]
- Lu T. T., Repa J. J., Mangelsdorf D. J. (2001). Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J. Biol. Chem. 276, 37735–37738 [DOI] [PubMed] [Google Scholar]
- Marschall H. U., Wagner M., Bodin K., Zollner G., Fickert P., Gumhold J., et al. (2006). Fxr(-/-) mice adapt to biliary obstruction by enhanced phase I detoxification and renal elimination of bile acids. J. Lipid Res. 47(3), 582–592 [DOI] [PubMed] [Google Scholar]
- Mizushima N. (2007). Autophagy: process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. (2010). Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- Ni H. M., Bockus A., Wozniak A. L., Jones K., Weinman S., Yin X. M., et al. (2011). Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 7(2), 188–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H. M., Boggess N., McGill M. R., Lebofsky M., Borude P., Apte U., et al. (2012). Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol. Sci. 127(2), 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., et al. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112(12), 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker M. H., Gommans W. M., Conde de la Rosa L., Homan M., Klok P., Trautwein C., et al. (2003). Resistance of rat hepatocytes against bile acid-induced apoptosis in cholestatic liver injury is due to nuclear factor-kappa B activation. J. Hepatol. 39(2), 153–161 [DOI] [PubMed] [Google Scholar]
- Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. (2000). Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 [DOI] [PubMed] [Google Scholar]
- Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., et al. (2011). Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25(8), 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki N., Hatano E., Taura K., Tada M., Kodama Y., Nitta T., et al. (2008). CHOP deficiency attenuates cholestasis-induced liver fibrosis by reduction of hepatocyte injury. Am. J. Physiol. Gastrointest. Liver Physiol. 294(2), G498–G505 [DOI] [PubMed] [Google Scholar]
- Tsuchiya S., Tsuji M., Morio Y., Oguchi K. (2006). Involvement of endoplasmic reticulum in glycochenodeoxycholic acid-induced apoptosis in rat hepatocytes. Toxicol. Lett. 166, 140–149 [DOI] [PubMed] [Google Scholar]
- Wang X., Fu X., Van Ness C., Meng Z., Ma X., Huang W. (2013). Bile acid receptors and liver cancer. Curr. Pathobiol. Rep. 1, 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. A., Thomas A. M., Li G., Kong B., Zhan L., Inaba Y., et al. (2012). Tissue specific induction of p62/Sqstm1 by farnesoid X receptor. PLoS ONE 7(8), e43961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang X., Yi T., Yen Y., Moore D. D., Huang W. (2007). Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67, 863–867 [DOI] [PubMed] [Google Scholar]
- Yerushalmi B., Dahl R., Devereaux M. W., Gumpricht E., Sokol R. J. (2001). Bile acid-induced rat hepatocyte apoptosis is inhibited by antioxidants and blockers of the mitochondrial permeability transition. Hepatology 33, 616–626 [DOI] [PubMed] [Google Scholar]
- Zhang G., Park M. A., Mitchell C., Walker T., Hamed H., Studer E., et al. (2008). Multiple cyclin kinase inhibitors promote bile acid-induced apoptosis and autophagy in primary hepatocytes via p53-CD95-dependent signaling. J. Biol. Chem. 283(36), 24343–24358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hong J. Y., Rockwell C. E., Copple B. L., Jaeschke H., Klaassen C. D. (2012). Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int. 32, 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]