Summary
Here, we show common expression of nuclear receptors (NRs) and substantial cooperative behavior of prostate cells in response to NR ligands while observing significant loss of NR expression in prostate cancer, suggesting the importance of NRs in regulating prostate homeostasis.
Abstract
The current study aimed to assess the topology of the nuclear receptor (NR) superfamily in normal prostate epithelial cells and its distortion in prostate cancer. Both in vitro and in silico approaches were utilized to profile NRs expressed in non-malignant RWPE-1 cells, which were subsequently investigated by treating cells with 132 binary NR ligand combinations. Nine significant cooperative interactions emerged including both superadditive [22(R)-hydroxycholesterol and eicosatetraenoic acid] and subadditive [1α,25(OH)2D3 and chenodeoxycholic acid] cellular responses, which could be explained in part by cooperative control of cell-cycle progression and candidate gene expression. In addition, publicly available data were employed to assess NR expression in human prostate tissue. Common and significant loss of NR superfamily expression was established in publicly available data from prostate tumors, in part predicting parallel distortion of targeting microRNA. These findings suggest that the NR superfamily in the prostate cooperatively integrates signals from dietary, hormonal and metabolic cues, and is significantly distorted in prostate cancer.
Introduction
The nuclear receptor (NR) superfamily of transcription factors relays dietary and lipid-derived hormonal signals to the genome. In turn, NRs regulate gene expression patterns involved in many important functions including cell metabolism, development, proliferation and differentiation (1–5). The 48 human NR family members can be classified broadly according to ligand affinity (6). High-affinity steroid receptors such as the androgen receptor (AR/AR) are well characterized and central to prostate homeostasis. A number of micronutrient ligands are also bound with high affinity by specific receptors, such as the active metabolites of vitamins A and D [retinoic acid and retinoid X receptors (i.e. RARA/RARα, RARB/RARβ, RARG/RARγ and RXRA/RXRα, RXRB/RXRβ, RXRG/RXRγ) and vitamin D receptor (VDR/VDR)]. A second group of receptors, often termed the adopted orphan receptors, binds with broader affinity to more abundant macronutrients such as polyunsaturated fatty acids, oxysterols, and bile acids [i.e. peroxisome proliferator-activated receptors (PPARA/PPARα, PPARD/PPARδ, PPARG/PPARγ), liver X receptors (LXR-a/LXRα, LXR-b/LXRβ) and farnesoid X receptor (FXR/FXR)]. Finally, a group of genuine orphan receptors exists for which no ligand has yet been identified, for example NGFIB/NGFIB.
Underscoring their importance in the prostate, conditional disruption of various NRs disrupts prostate tissue maintenance and can lead to hyperplasia, for example in RARγ-depleted mice (7). Recent work has similarly implicated LXRs as integral mediators of prostatic homeostasis and malignant progression, for instance with the observation of prostatic intraepithelial neoplasia in LXR− /− mice fed high-cholesterol diets (8). It is therefore not surprising that NRs are distorted by genetic and epigenetic processes in prostate cancer (PCa). For example, while distortions in AR and estrogen receptor (ESR1/ESRα, ESR2/ESRβ) signaling are recognized, so also are the antiproliferative effects of VDR, RARs and PPARs (1,9–12). Also, epidemiological and chemoprevention studies have identified that either initiation or progression of PCa may relate to reduced dietary intake of micronutrients and macronutrients associated with NR activation and signaling (13–18).
Transcriptional control through individual NRs is well understood. This process involves the well-characterized, dynamic interaction of NRs with co-activator and corepressor complexes that in turn regulate the epigenetic status of target genes (reviewed in ref. 19). By contrast, understanding their integrated functions remains enigmatic. Numerous studies have observed cross talk between NRs and their signaling pathways, with mechanisms including overlapping accumulation of NRs at genomic loci (20), cooperative regulation of gene transcription and direct transcriptional control of NR expression upon binary activation in vitro and in vivo (10,11,21). The current study was undertaken in an effort toward understanding how the NR superfamily functions cooperatively in non-malignant prostate epithelial cells to govern emergent cellular behavior.
In addition, a dual aim of the current study was to characterize to what extent this important family of transcriptional regulators is altered in malignancy. The recent concerted effort through The Cancer Genome Atlas (TCGA) and other consortia to gain global genomic understanding of cancer has yielded key datasets that serve as powerful tools to query and understand how gene families are deregulated in PCa. Therefore, we utilized in silico approaches to mine publicly available prostate tumor data thereby to reveal and characterize further the targeted disruption of the NR superfamily.
Methods
Materials
All ligands (Sigma–Aldrich) were stored in ethanol or dimethyl sulfoxide at maximum concentrations recommended by manufacturer. Prior to treatment, 1000× stock solutions were prepared, diluted 1:10 in media, and treated 1:100 to final concentrations.
Cell culture
RWPE-1 (non-malignant) prostate epithelial cells (22,23) were maintained in keratinocyte-serum-free medium supplemented with epidermal growth factor, bovine pituitary extract (Invitrogen) and 1% penicillin/streptomycin. All cells were grown at 37°C in a humidified atmosphere of 5% CO2 in air.
Cell proliferation assay
Proliferation was measured by use of ViaLight® Plus Kit (Lonza Inc., Rockland, ME), which utilizes bioluminescent detection of cellular ATP as a measure of cell viability. RWPE-1 cells were previously optimized to seeding density of 4 × 103 cells per well in 96-well, white-walled plates (Corning Inc., Corning, NY) to ensure exponential growth throughout experiments. Individual wells were dosed with agents to a final volume of 100 µl. Dosing occurred at the beginning of the experiment, and cells were incubated for 96 h, with re-dosing occurring after 48 h of incubation. Luminescence was detected with SynergyTM 2 multimode microplate reader (BioTek® Instruments, Winooski, VT). Each experiment was performed in triplicate wells in triplicate experiments. The combination of individual mean treatments was summed to determine the ‘additive’ antiproliferative response. The experimentally determined mean of the combined treatments was used as the ‘observed’ antiproliferative response. These values were then compared utilizing the Student’s t-test, with superadditive effects defined as observed values significantly greater than additive values and sub-additive effects defined as observed values significantly less than additive values. If the additive antiproliferative response did not differ from that observed the effect is then defined as additive.
Cell-cycle analysis
Cells were allowed exponential growth and treated with agonists for 24 h, or 72 h with re-dosing after 48 h. Mid-exponential growth cell cultures were harvested with accutase (Invitrogen), fixed and stained with propidium iodide buffer (10 µg/ml propidium iodide, 1% (wt/vol) trisodium citrate, 0.1% (vol/vol) TritonX-100, 100 µM sodium chloride) for 45 min, on ice, in the dark. Cell-cycle distribution was determined utilizing FACSCaliburTM Flow Cytometer (Becton-Dickinson) and analyzed with ModFIT 3.1 SP3 cell-cycle analysis software.
Quantitative real-time reverse transcription–polymerase chain reaction profiling of single-gene targets
Quantitative real-time reverse transcription–polymerase chain reaction (Q-RT–PCR) was employed for detection of candidate messenger RNA (mRNA) transcripts. Total RNA was isolated via TRIzol® reagent (Invitrogen, Carlsbad, CA) and complementary DNA prepared using iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA) according to manufacturer’s instructions. Relative gene expression was subsequently quantified via ABI 7300 Real Time PCR System (Applied Biosystems). 18S VIC-labeled probe was used as an internal control, and all other targets were detected using assay-on-demand TaqMan primers and probes (ABI). All experiments were performed in biological triplicates, with at least technical duplicates. Fold changes were determined using the 2-ΔΔCt method (24), where ΔCt was calculated as the Ct of the target gene minus the Ct of18S rRNA, and ΔΔCt was calculated as the difference between treatment and respective vehicle.
Q-RT–PCRM profiling of multiple NR
Multitarget microfluidic Q-RT–PCR(Q-RT–PCRM) measurement of multiple gene transcripts was undertaken on custom-designed TaqManTM Low Density Array (ABI 7900HT Fast Real-Time PCR System), as described previously (3). Briefly, the array included probes and primers for 18S and NRs (i.e. high and broad affinity such as VDR and PPARs); NR choice was guided by serial analysis of gene expression data from normal prostate tissue (25). mRNA from cell-cycle sorted cells was quantified in triplicate samples measured in duplicate as described previously (3). Basal expression of NRs was determined relative to the housekeeping gene 18S rRNA and normalized to the average relative expression of all NRs and fold change calculated by the 2^-ΔΔCt.
Microarray analysis of NR expression in RWPE-1
Basal gene expression of non-malignant prostate epithelial (RWPE-1) cells was carried out by using microarray. Total RNA was isolated with TRIzol® reagent and, after quality assurance, was hybridized to Illumina HT-12 V4.0 BeadChip for determination of gene expression. Normalized, log2 transformed intensities for each NR gene were compared with the mean normalized, log2 intensity across the entire array.
Analysis of NR expression from Memorial Sloan-Kettering Cancer Center, TCGA and the human protein Atlas datasets
The Memorial Sloan-Kettering Cancer Center (MSKCC) cBio Cancer Genomics Portal (26) (http://cbioportal.org) was used to interrogate a PCa dataset (27). In our analysis, only primary tumors from white, non-Hispanic patients containing mRNA (n = 98) and microRNA (miRNA, n = 78) expression data were considered. The significance threshold for NR expression alteration was set to Z = ±2. Bootstrapping analysis (n = 10 000) was performed on the clinical samples to determine significance of the percentage of altered expression in the NR superfamily (n = 48 genes). Specifically, the percentage of altered expression was measured in 10 000 sets of 48 randomly selected gene sets and an empirical P value was then determined for the expression of the gene set comprising the NR superfamily. To help verify these findings in another patient cohort, NR expression was queried and examined in PCa tissue samples and matched normal tissue from data made available from TCGA, and visualized using the Cancer Genomics Browser (UC Santa Cruz). NR protein expression was interrogated from The Human Protein Atlas Project, a comprehensive database characterizing protein expression across normal and malignant human tissues utilizing antibody-based methods (28). Thirty-one NRs have thus far been characterized across tissue types, including normal prostate and PCa tissues, and were utilized for this study. All tissues have been annotated by certified pathologists, and overall staining intensity of a given protein was determined as negative, weak, moderate, or strong. Normal prostate staining is an overall representation of three available non-malignant prostate tissue samples, whereas PCa tissues (n ≥ 12) are individually quantified and stratified by staining intensity level to give a representation of the distribution protein level associated with tumor tissues. The staining intensity level most represented among PCa tissues was determined to calculate overall protein detection in tumor tissue.
MicroRNA prediction analysis from miRWalk database
All the miRNAs present in latest version of miRbase were analyzed to find miRNAs that are predicted to target NRs. Target prediction analysis was performed using ‘miRWalk’, which provides miRNA prediction results from 10 different prediction algorithms (DIANA-mT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4, PICTAR5, PITA, RNA22, TargetScan). To be conservative in our selection, we considered only those predictions between a miRNA and a given NR positive if 5/10 algorithms recognized the interaction. Once the miRNA prediction list was formulated, it was consolidated to assess which predicted miRNAs targeted the most deregulated NRs. Only those miRNAs predicted to target at least four deregulated NRs (altered in >18% of patients) were considered for expression analysis. Expression data of miRNAs were downloaded from the MSKCC cBio Cancer Genomics Portal dataset for the same tumor cohort used for gene expression analysis and the significance analysis was executed similarly as discussed above for NRs.
Results
Evidence of NR expression in prostate epithelial cells
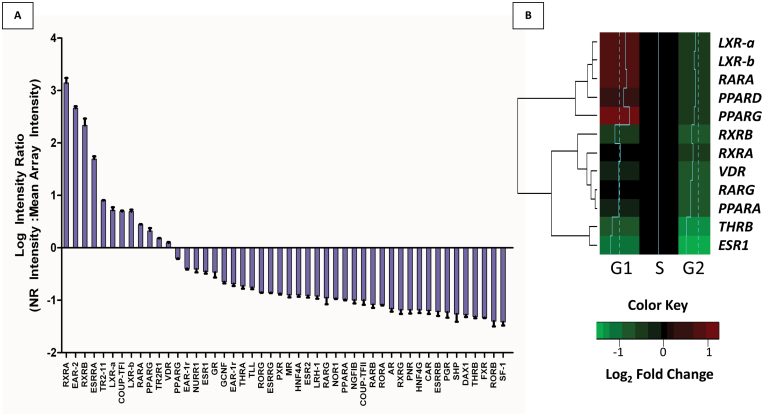
To determine the NR expression profile in non-malignant prostate epithelia, we undertook a microarray approach in RWPE-1 cells (Figure 1A). A table providing the nomenclature for all members of the NR superfamily is provided in Supplementary Table S1, available at Carcinogenesis Online. Our results indicate that a broad range of NRs were expressed. Specifically, we found strong expression (transcripts detectable at levels greater than the average of all genes) of 12 NRs including RARs, RXRs, VDR, LXRs, PPARs, and the orphan receptor EAR-2, among others. These findings were largely confirmed from analysis of publicly available datasets from RWPE-1 cells (ArrayExpress Archive; Supplementary Figure S1A, available at Carcinogenesis Online). Subsequently, we applied Q-RT–PCRM to assess expression levels of NRs, specifically across the phases of the cell cycle (Figure 1B). This method supported the NR microarray expression patterns, with detectable expression of LXRs, PPARs, VDR, RXRs and RARs, and undetectable levels of several NRs that also had low expression as detected by microarray, including PXR, CAR and FXR (Supplementary Figure S1B, available at Carcinogenesis Online). Interestingly, although AR levels are low or absent in these cells, they are rapidly inducible upon treatment with AR ligands (23). Furthermore, elevated NR expression occurred prominently in G0/G1 with significant dampening and reduction of expression levels in G2/M compared with S phase (P < 0.0001, two-way analysis of variance; Figure 1B). For example, LXRs and PPARs maintained highest expression in G0/G1, whereas several NRs maintained highest expression in S phase (downregulated in G0/G1) including ESR1 and THRB. To expand the impact of these findings beyond cell models, an in silico query for NRs was undertaken in normal human prostate epithelia through the use of serial analysis of gene expression (SAGE) data (25) (Supplementary Figure S2, available at Carcinogenesis Online). Again, robust expression of RARs, RXRs, LXRs, EAR-2, AR and FXR re-enforces the concept that multiple NRs are expressed in normal prostate epithelial cells in vivo as well as in vitro.
Fig. 1.
Expression of the NR network in non-malignant prostate epithelial cells (RWPE-1). (A) Basal expression of the 48-member human NR superfamily from microarray analysis of untreated RWPE-1 cells. Expression is represented as the log2 intensity ratio, with each individual NR normalized intensity set relative to the mean normalized intensity across arrays. (B) RWPE-1 basal expression of NRs across the phases of the cell cycle. Expression of NR was measured in FACS separated, basal RWPE-1 cells by Q-RT–PCRM and normalized to 18S expression in each phase of the cell cycle. Heatmap represents Log2 fold changes relative to S phase expression. Histograms (blue): dotted line represents 0 Log2 FC, and histograms represent relative change of given gene (right=up, left=down). Results were determined in triplicate experiments.
A summary of NRs detected by different methods and across normal and malignant tissue is listed in Table I. In all, 35 NRs were detected by at least one of the three methods (microarray, Q-PCR and SAGE) utilized to determine expression in normal prostate cells. Examination of NR protein levels in The Human Protein Atlas suggested that NR mRNA levels were indicative of protein levels. This database includes immunohistochemical analysis across multiple tissue types, including prostate, using carefully characterized antibodies (Supplementary Figure S3, available at Carcinogenesis Online). Most NRs detectable by SAGE were detectable at the protein level, although this was incomplete due to a lack of a complete set of validated antibodies against NRs. Out of 48 human NRs, in total 31 were available in the database and most NRs detectable by SAGE were detectable at the protein level in normal prostate tissue. Protein level in normal prostate tissue ranged from weak staining (i.e. RARβ, RXRγ) to intermediate staining (LXRα, FXR, RXRβ) to strong staining (i.e. RARα, RARγ; Table I). A table summarizing publicly available datasets utilized for NR expression analysis is available in Supplementary Table S2, available at Carcinogenesis Online. In total, these initial observations suggested the presence of a range of NRs in normal prostate epithelial cells and raised the possibility of these receptors working cooperatively to sense and respond to lipophilic compounds.
Table I.
List of NR ligands utilized in proliferation assays, their determined growth inhibitory ED25 concentrations and detection of NR expression by different methods
| NR | Ligands examined | ED25 (nM) | Cell line | Prostate tissue | ||||
|---|---|---|---|---|---|---|---|---|
| RWPE-1 | Normal | Cancer | ||||||
| microarray | Q-RT–PCRM | SAGE | Protein Atlas | SAGE | Protein Atlas | |||
| RXRA | 9cRA | 1 | det | det | det | Strong | det | Moderate |
| RXRB | 9cRA | 1 | det | det | det | Moderate | det | Strong |
| RXRG | 9cRA | 1 | ND | — | det | Weak | ND | Negative |
| RARA | 9cRA, ATRA | 1, 10 | det | det | ND | — | ND | — |
| RARB | 9cRA, ATRA | 1, 10 | ND | ND | det | Weak | ND | Weak |
| RARG | 9cRA, ATRA | 1, 10 | det | det | ND | Strong | det | Moderate |
| LXR-a | 22-HC | 1000 | det | det | det | Moderate | ND | Moderate |
| LXR-b | 22-HC | 1000 | det | det | det | — | det | — |
| PPARA | EPA, ETYA, Bez | 10, 100, 0.1 | ND | det | det | — | ND | — |
| PPARD | EPA, ETYA, Bez, GW501 | 10, 100, 0.1, 5 | det | det | det | Negative | det | Negative |
| PPARG | EPA, ETYA, Bez | 10, 100, 0.1 | det | det | ND | Negative | ND | Weak |
| ESR1 | E2 | 10000 | det | det | det | Negative | ND | Negative |
| ESR2 | E2 | 10000 | ND | det | det | Strong | ND | Moderate |
| FXR | LCA, CDA | 100, 1000 | ND | ND | det | Moderate | det | Moderate |
| AR | DHT | 1000 | ND | ND | det | — | ND | Moderate |
| VDR | 1α,25(OH)2D3, LCA | 100, 100 | det | det | ND | — | ND | — |
| EAR-2 | — | — | det | — | det | — | det | — |
| ESRRA | — | — | det | — | det | — | det | — |
| ESRRB | — | — | ND | — | ND | — | det | — |
| ESRRG | — | — | ND | — | ND | Weak | ND | Negative |
| COUP-TFI | — | — | ND | — | det | Weak | ND | Negative |
| COUP-TFII | — | — | det | — | det | — | det | — |
| EAR-1 | — | — | det | — | ND | Strong | ND | Moderate |
| EAR-1R | — | — | det | — | det | Weak | det | Moderate |
| GR | — | — | det | — | det | Strong | det | Moderate |
| TR2R1 | — | — | det | — | det | Moderate | det | Moderate |
| THRA | — | — | ND | — | det | Weak | det | Negative |
| THRB | — | — | det | det | ND | Weak | ND | Weak |
| LRH-1 | — | — | ND | — | det | Strong | det | Strong |
| NGFIB | — | — | ND | — | det | — | det | — |
| SF1 | — | — | ND | — | det | — | det | — |
| DAX1 | — | — | ND | ND | det | — | ND | — |
| HNF4A | — | — | ND | — | det | Negative | ND | Negative |
| HNF4G | — | — | ND | — | ND | Moderate | ND | Strong |
| RORA | — | — | ND | — | det | Moderate | ND | Moderate |
| RORB | — | — | ND | — | det | Negative | ND | Negative |
| RORG | — | — | ND | — | ND | — | ND | — |
| NURR1 | — | — | det | — | ND | Moderate | det | Moderate |
| TR2-11 | — | — | det | — | ND | — | det | — |
| GCNF | — | — | det | — | ND | — | ND | — |
| PNR | — | — | ND | — | ND | — | ND | — |
| PGR | — | — | ND | — | ND | Negative | ND | Negative |
| MR | — | — | ND | — | ND | Strong | ND | Moderate |
| PXR | — | — | ND | ND | ND | ND | — | |
| CAR | — | — | ND | ND | ND | Moderate | ND | Negative |
| NOR1 | — | — | ND | — | ND | Moderate | ND | Moderate |
| TLX | — | — | ND | — | ND | Negative | ND | Negative |
| SHP | — | — | ND | — | ND | — | ND | — |
Approximate ED25 concentrations were experimentally determined by sigmoidal growth curves. A given NR was determined to be expressed if (i) SAGE captured at least 1 tag per 200 000 (Supplementary Figure S2, available at Carcinogenesis Online), (ii) Q-RT–PCRM threshold detection of Ct < 35 (Supplementary Figure S1B, available at Carcinogenesis Online), (iii) microarray intensity > −1 LIR from mean array intensity (Figure 1A). NRs were detectable (det) or were not detectable (ND) by such criteria. Protein expression is indicated by relative staining intensity (negative, weak, moderate, strong) as determined by the Human Protein Atlas (Supplementary Figure S3, available at Carcinogenesis Online ).
RWPE-1 prostate epithelial cells respond to wide panel of nuclear receptor ligands
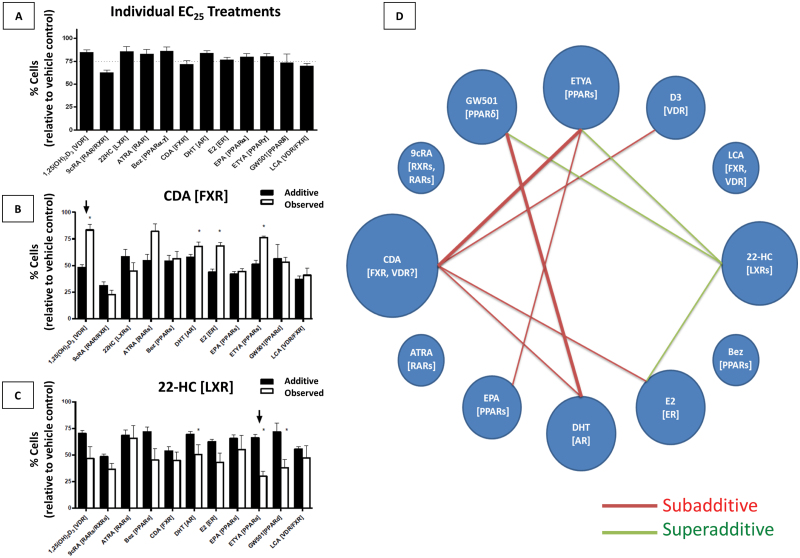
To establish to what extent this pattern of NR expression was biologically relevant, we undertook a comprehensive series of proliferation assays in RWPE-1. We screened the ligand response toward the 12 most abundantly expressed NRs including AR (given its inducible nature). Specifically, we examined the actions of well-established natural and synthetic ligands and generated non-sigmoidal dose-response curves from which we interpolated growth inhibitory ED25 values (Figure 2A, Table I, and data not shown).
Fig. 2.
Proliferative inhibition via NR activation in prostate epithelia individually and in dual combination. (A) Proliferative inhibition of RWPE-1 cells by individual treatment of agonists known to activate particular NRs (denoted in brackets). Ligands were dosed at approximate ED25 concentrations (Table I). (B) Combination treatments involving CDA. The ED25 concentration of CDA dosed in combination with the ED25 concentration of each agonist from (A). ‘Additive’ value is determined by addition of growth inhibitory effects of individual agonist treatments in (A). ‘Observed’ value is determined experimentally. The arrow denotes the subadditive response of CDA in combination with 1α,25(OH)2D3 (D3), determined as an observed growth inhibitory response significantly less than the predicted response (P < 0.01). (C) Combination treatments involving 22(R)-hydroxycholesterol (22-HC). The arrow denotes the superadditive response of 22-HC with ETYA, determined as an observed growth inhibitory response significantly greater than the predicted response (P < 0.01). (D) Ligand interactions within the proposed prostate NR network. The network diagram is a representation of all significant, non-purely additive interactions observed from the ligand combination studies; the thickness of the line represents the degree of significance, either P < 0.05 (thin lines) or P < 0.01 (thick lines) (Supplementary Figure S3, available at Carcinogenesis Online). Each node represents an individual ligand treatment, with their characteristic NRs listed in brackets. Red lines between nodes denote an observed subadditive growth inhibitory response upon dual treatment with given ligands, inhibiting growth of RWPE-1 cells significantly less in combination than predicted from individual experiments. Green lines denote an observed superadditive growth inhibitory response upon dual treatment with given ligands, inhibiting growth of RWPE-1 cells significantly more in combination than predicted from individual experiments. The relative size of each node is proportional to its absolute number of interactions within the NR network. Normal lines represent significance (P < 0.01), whereas bold lines represent greater significance (P < 0.001) as determined by two-tailed t-test with Welch’s correction.
Several micronutrient ligands showed potent inhibition of proliferation of RWPE-1 cells at nanomolar concentrations, including retinoids [all-trans retinoic acid, (ATRA), ED25 = 10 nM), 9-cis retinoic acid (9cRA, ED25 = 1 nM)] and 1α,25(OH)2D3, (ED25 = 1 nM). There was also an acute response to a panel of ligands for broad affinity receptors, notably PPARs [eicosopentanoic acid (EPA), eicosotetraenoic acid (ETYA) and bezafibrate (Bez)], FXR [chenodeoxycholic acid (CDA) and lithocholic acid (LCA)], and LXRs [22-hydroxycholesterol (22-HC)]. The hormones estradiol (E2, ED25 = 10 µM) and dihydrotestosterone (DHT, ED25 = 1 µM) also inhibited growth albeit at concentrations above physiological levels. At physiological levels (0.1–1 nM), DHT stimulated RWPE-1 growth.
Binary treatment assays indicate NR ligand interactions within the prostate
To reveal the degree of integrated control of cellular proliferation by NR activation, we examined all possible binary ligand combinations by using each ligand at the determined ED25 ligand concentrations (Figure 2, Supplementary Figure S4, available at Carcinogenesis Online). The additive proliferative inhibitory response was determined from summing the inhibitory response of a given pair of individual ligand treatments and comparing this to the experimentally observed values. Combinations where the observed response was not significantly different from the additive response were classified as additive and no interaction could be verified. Ligand combinations that resulted in significantly dampened growth inhibitory response compared with the additive growth inhibitory response were deemed subadditive interactions (Figure 2B, arrow). By contrast, those where observed inhibitory response was significantly elevated compared with the additive response were deemed superadditive (Figure 2C, arrow). Of the 132 combinations examined, 9 significant interactions were identified (3 superadditive, 6 subadditive). Figure 2D provides a summary of identified interactions and shows the number of ligand interactions that are significantly additive or subadditive (the targeted receptor is indicated in parentheses).
The most interactive nodes identified centered on ligands for the broad affinity receptors for bile acid (CDA), lipid (ETYA) and cholesterol (22-HC) ligands, and also involved classical steroid hormones and secosteroid hormones [DHT, E2, 1α,25(OH)2D3]. More specifically, the presence of CDA significantly dampened antiproliferative signaling of several compounds, namely DHT, 1α,25(OH)2D3, E2 and ETYA. In contrast, the presence of 22-HC significantly amplified the antiproliferative signaling of ETYA, E2 and the synthetic PPARδ ligand GW501.
Combined regulation of cell-cycle status and target gene expression
Two classes of interacting nodes were examined further for their impact upon cell-cycle progression and gene regulation; the subadditive inhibitory combination of CDA with 1α,25(OH)2D3, and the superadditive inhibitory combination of 22-HC with ETYA (Figures 3 and 4).
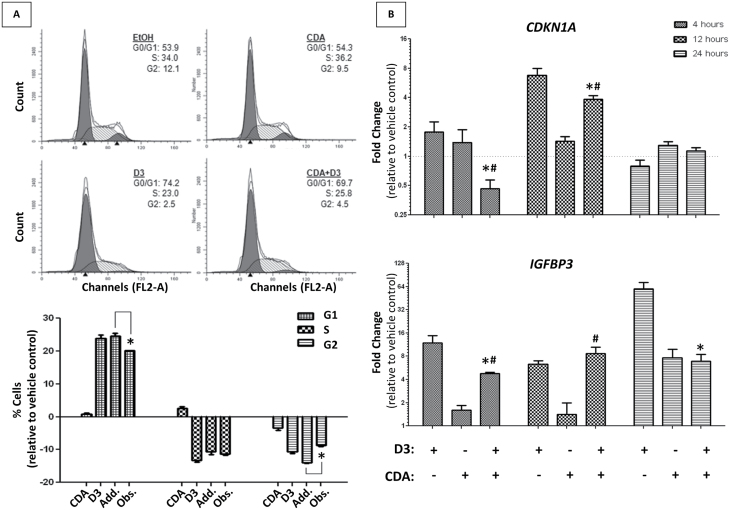
Fig. 3.
The effects of 1,25α(OH)2D3 and CDA on the cell-cycle status and gene regulation. (A) Cell-cycle analysis of RWPE-1 cells measured by FACS analysis. Cells were treated with ligand ED25 concentrations individually and in combination for 24h. Representative cell-cycle distributions are shown (upper panel), with subsequent quantification of three biological replicate populations (lower panel). Significant changes (determined by t-test) of cell population at any given cell cycle are noted, comparing additive results to those observed experimentally. (B) VDR candidate gene expression profile in RWPE-1 cells. Expression of known VDR-regulated genes (IGBFP3, CDKN1A) at different time points of treatment (4, 12, 24h) determined via Q-RT–PCR. The 1α,25(OH)2D3 (D3) and CDA were dosed individually and in combination at ED25 concentrations and were compared with respective vehicle controls. Significant changes of gene expression between combination treatment and individual treatments (*D3, #CDA) are noted.
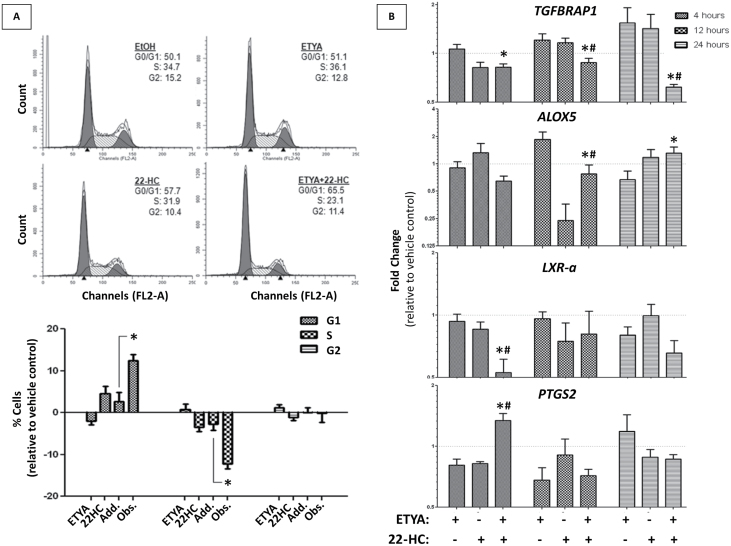
Fig. 4.
The effects of 22-HC and ETYA on the cell-cycle status and gene regulation. (A) Cell-cycle analysis of RWPE-1 cells measured by FACS analysis. Cells were treated with ligand ED25 concentrations individually and in combination for 72h. Representative cell-cycle distributions are shown (upper panel), with subsequent quantification of three biological replicate populations (lower panel). Significant changes (determined by t-test) of cell population at any given cell cycle are noted, comparing additive results to those observed experimentally. (B) PPARγ candidate gene expression profile in RWPE-1 cells. Expression of candidate PPARγ-regulated genes (LXR-a, PTGS-2, TGFBRAP1, ALOX 5) at different time points of treatment (4, 12, 24h) determined via Q-RT–PCR. ETYA and 22-HC were dosed individually and in combination at ED25 concentrations and were compared with respective vehicle controls. Significant changes of gene expression between combination treatment and individual treatments (*ETYA, #22-HC) are noted.
Cell-cycle analysis was undertaken to assess if the interactions between NR ligands reflected altered patterns of cell-cycle progression. Specifically, these approaches revealed that 1α,25(OH)2D3-mediated G1 arrest was significantly attenuated in the presence of CDA, reflecting the proliferation data (Figure 3A). Intriguingly, VDR target gene expression was also found to be differentially expressed in the ligand combination compared with individual treatments in a manner that reflected this attenuation, indicating strong evidence of gene-regulatory overlap between activated NRs. For instance, VDR-mediated induction of CDKN1A (encodes the cell-cycle regulating protein p21(waf1/cip1)) was squelched in the presence of CDA (Figure 3B), coinciding with the subadditive inhibitory nature of this combination treatment on RWPE-1 growth. Although 1α,25(OH)2D3-induced VDR regulation of IGFBP3 was similarly squelched in the presence of CDA, CYP24A1 was not (Supplementary Figure S5, available at Carcinogenesis Online), suggesting that the presence of CDA may not alter VDR signaling homogeneously.
To investigate the impact of ETYA and 22-HC, we also examined cell-cycle status and observed a synergistic G1 block of cells treated in combination with ETYA/22-HC, compared with individual treatments (Figure 4A). Again, these findings are consistent with the superadditive growth inhibitory nature of this ligand combination.
It has been proposed in several studies that PPARs and LXRs regulate one another’s expression in a positive-feedback loop regulating both cholesterol and fatty acid metabolism (29). Therefore, we examined the expression of LXR-α and observed a significant downregulation upon co-treatment. Similarly, the combination treatment suppressed expression of the putative PPARγ target TGFBRAP1 (1) at all time points examined relative to individual treatments, and squelched ETYA-mediated induction of ALOX5 at 12 h. Conversely, the combination treatment enhanced expression of PTGS2 as early as 4 h posttreatment. Combinatorial responses were also observed for CDKN1A and IGFBP3 (Supplementary Figure S5, available at Carcinogenesis Online). These candidate gene analyses suggest that gene-regulatory patterns induced by combining PPAR and LXR ligands are not predictable from the individual agents alone and reflect more accurately the cell biology.
Expression of the NR superfamily is significantly deregulated in PCa
These results, along with abundant evidence from the literature, support the concept that NRs in the prostate act cooperatively to sense and respond a wide range of lipophilic compounds. Therefore, we thought it logical to examine if NRs are commonly deregulated in malignant prostate. To investigate the status of the NR superfamily in malignancy, we undertook in silico approaches. RWPE-2 cells are transformed with the v-Ki-ras oncogene and a malignant isogenic clone of RWPE-1 cells (22,23). RWPE-2 microarray datasets, available through ArrayExpress database, were queried for NR expression (30) (Supplementary Figure S6, available at Carcinogenesis Online). Compared with RWPE-1, several NR expression levels were downregulated, including LXR-a, LXR-b (−1.38, −1.45), VDR (−1.5) and retinoid receptors RARG and RXRA (-1.71, -1.74). Meanwhile, others were elevated, notably the glucocorticoid receptor (GR) (2.14).
Mining of a human PCa dataset made available by Taylor et al. (26,27) assessed NR expression in clinical PCa (Figure 5A). Gene expression data were considered from 98 white, non-Hispanic patients. NR expression was considered significantly altered in patients whose tumor showed differential expression relative to a pool of matched adjacent normal samples with a defined threshold of Z = ±2. Fourteen NRs were differentially expressed in over 18% of PCa patients, including LXR-a, LXR-b (18.4%, 31.6%), PPARA (31.6%), RXRA (41.8%) and RARB, RARG (38.8%, 61.1%) (Supplementary Table S3, available at Carcinogenesis Online). Of these, only RORG was predominantly elevated (22.5%) while the remaining NRs were largely lost. Of interest, RARG and RXRA were two most downregulated NRs in RWPE-2 compared with RWPE-1 (Supplementary Figure S6, available at Carcinogenesis Online) and also represented the most commonly underexpressed NR transcripts in the primary tumors, occurring in 40.8% and 59.18% of cases, respectively.
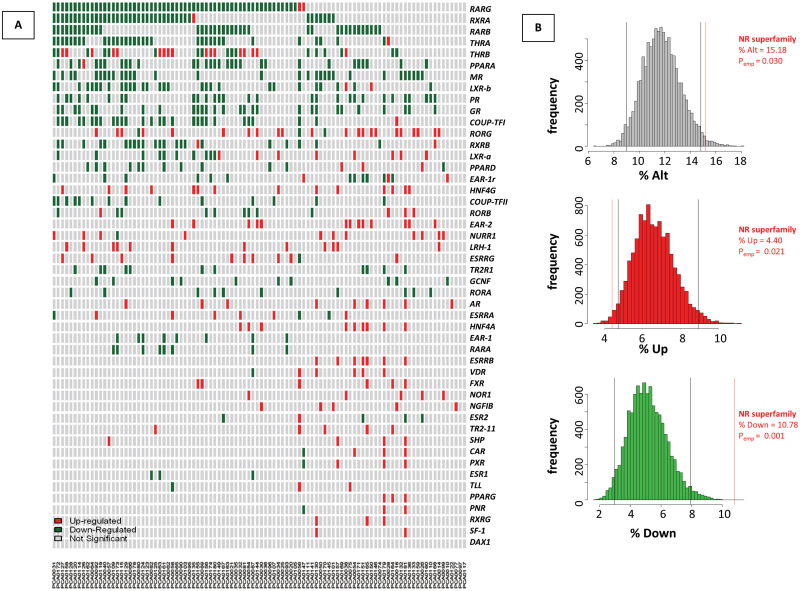
Fig. 5.
Expression alterations of NR in 98 primary tumors from the MSKCC Prostate Oncogenome Project. (A) The MSKCC cBio Cancer Genomics Portal (http://cbioportal.org) PCa dataset (27) was queried to assess significant changes in NR expression among a cohort of 98 white, non-Hispanic primary PCa patients compared with a pool of normal adjacent prostate samples. The significance threshold for the expression of a given gene to be significantly altered was set to Z = ±2 (red/green, respectively). The total percent of patients in which a given NR expression is significantly deregulated (% alt), downregulated (% down) or upregulated (% up) are shown, along with the means for each type of deregulation across all 48 NRs. (B) Bootstrapping results assessing significance of NR deregulation observed from dataset available from the PCa patient cohort. Histograms depict the distribution of significant gene alterations [total alterations (upper), upregulated (middle), downregulated (lower)], for 10 000 random samples of 48 genes from 26 440 gene set list. Black lines indicate 95% confidence intervals of each distribution, and red line represents the average deregulation of the 48 NR superfamily.
Considering all of the 48 members of the NR superfamily together revealed that each gene member was significantly altered on average in 15.2% of patients: 10.8% underexpressed and 4.4% overexpressed (Supplementary Table S3, available at Carcinogenesis Online). To test whether this deregulation is more than would be predicted by chance, we applied a bootstrapping approach. We sampled 10 000 replicates of 48 random genes within the 26 446 gene set and determined the distribution of significant gene expression changes among the patient cohort. We found that NR superfamily expression was significantly altered (P emp = 0.03) in these patients, and more specifically that NR expression was lost in primary PCa (P emp = 0.001) (Figure 5B). Our results also show that NR expression was upregulated less than would be expected by random chance (P emp = 0.021), further indicating that NR expression is actively selected against in PCa. Taken together, these observations suggest that expression of the NR superfamily as a whole is lost in primary PCa. Supporting that the deregulation of this critical transcription factor family is important in PCa, we performed a comparable analysis to determine the deregulation of another transcription factor family with members implicated in PCa progression, the ETS transcription factor family, and found that its expression is not significantly altered by similar criteria (Supplementary Figure S7, available at Carcinogenesis Online). To verify this finding further, NR expression was queried in another PCa patient cohort available from TCGA. This dataset confirmed expression patterns found in data from MSKCC, indicating significant loss of expression of many NRs including RARs, RXRs, LXRs and PPARs, whereas RORG remained elevated (Supplementary Figure S8, available at Carcinogenesis Online).
Expression of NR-targeting miRNAs reflect altered NR expression
After establishing a significant repression of the NR superfamily in non-malignant prostate epithelial cells and its distortion in a cohort of PCa patients, we next assessed whether deregulated miRNA expression in the PCa cohort could play a role in NR network deregulation. To form a list of miRNAs predicted to target deregulated NRs, we utilized miRWalk, a comprehensive software providing miRNA target prediction results for up to 10 commonly utilized prediction algorithms (31). Those NRs with significant deregulation in at least 18% of primary tumors (Figure 5A, Supplementary Table S3, available at Carcinogenesis Online) were examined further. For each deregulated NR, a list of predicted miRNAs was assembled with the criteria requiring at least 5 out of 10 prediction algorithms producing positive predictions for the interaction. This miRNA list was then consolidated to examine which predicted miRNAs targeted multiple deregulated NRs. For our final analysis, miRNAs targeting at least four deregulated NRs were considered (Supplementary Figure S9, available at Carcinogenesis Online). From these filters, 29 miRNAs were compiled, several of which have recently been implicated in PCa [i.e. mir-106b (32), mir-20a (33), mir-141 (34)]. Seventy-eight patients from the MSKCC cohort examined for NR expression had matched miRNA microarray data, and expression of the NR-targeting miRNA was assessed in a similar manner as described previously for the NR superfamily (Supplementary Figure S10, available at Carcinogenesis Online). The NR-targeting miRNA subset was expressed at a higher level than the mean of all miRNAs across the array (8.42%) but did not lie outside of 2 SD of the bootstrap distribution. However, the expression of these miRNAs was lost significantly less than would be predicted by random chance (4.25%, P emp = 0.044), indicating that their expression is actively maintained in PCa and may play a role in the deregulation of NR transcripts in these patients.
Discussion
The current study aimed to examine how NRs may operate in the prostate as a network to integrate hormonal and environmental signals to cell fate decisions; functions that may be distorted in malignancy. Non-malignant prostate epithelial cells were used to examine the expression of NRs, how this varied through the cell cycle and to the combinatorial actions of binary NR ligand treatments. In this manner, we were able to establish a subset of NR interactions that display emergent behavior patterns that were not predictable when considering each NR alone.
We exploited different platforms and approaches and identified 35 NRs detectable by different methods, with a consensus cohort of 15 robustly expressed NRs (detected by all methods) in normal prostate cells (Table I). Importantly, we were able to infer biological meaning to this expression by interrogating their binary responsiveness to a panel of ligands that target 12 well-studied NRs. These approaches identified superadditive and subadditive interactions within the NR network and revealed convergent control of cell proliferation by means of combined gene-regulation and cell-cycle control.
The map of these interactions suggests that dietary-derived factors modify hormonal signaling. Notably, the LXRs, sensing cholesterol metabolites, emerged as strongly cooperating with a range of receptors including the PPARs that sense fatty acids. Similarly, another dietary-derived and regulated factor, the primary bile acid CDA, suppressed the actions of several receptors including steroidal sensing ESRα and AR, and the VDR that binds the secosteroid 1α,25(OH)2D3. Taking key interactions that were indicative of these subadditive (i.e. 1α,25(OH)2D3 and CDA), and superadditive (i.e. ETYA and 22-HC) behaviors, we investigated their impact upon cell-cycle status and candidate gene regulation and found underlying cooperative regulation supportive of proliferative responses.
The biology of the prostate cannot be examined fully without considering the role of the AR. In the normal prostate, the AR regulates gene expression programs that exert a profound control on cell growth and survival (35). Blockade of the AR in androgen-sensitive PCa via androgen deprivation therapy (ADT) yields significant clinical benefit by triggering apoptosis and other extreme stresses, even in disseminated disease. PCa cells that survive and traverse this crucible adapt rapidly in association with distorted histone modifications and DNA methylation to generate the ADT-recurrent AR transcriptome. Consequently, the ADT-recurrent PCa cells display altered activity of chromatin remodeling enzymes, such as the oncogene EZH2 (36,37), and changes in H3K27me3 (38) and DNA methylation patterns (39–41). ‘Opening’ of different enhancer regions allows the AR to govern a cistrome that controls genes that promote cell survival and proliferation in ADT-RCaP cells, whereas other AR target genes become transcriptionally resistant associated with increased genome-wide DNA methylation (42). However, despite its central biological role, the AR interactions with other NRs appear to be fewer in number than other NRs and the extent of changes in expression in primary tumors is less than observed than other receptors such as those responsive to retinoids.
Roles for bile acids such as CDA are well established and are maintained at 5–15 µM in the serum, the level of which is proportional to dietary fat consumption. It is therefore of interest that a number of epidemiological and in vivo studies have found implications of bile acid concentrations to cancers of the esophagus, liver, small intestine and colon (17). In fact, mice lacking the FXR required for proper export of bile acids maintain elevated hepatic bile acid concentrations and display a high propensity for hepatocarcinogenesis (43). Similarly, children with deficiencies in bile acid export transporters have increased incidence of hepatocellular carcinoma (44).
Although the implication of bile acids to gastrointestinal carcinogenesis is well documented, the contribution of prolonged exposure of high serum bile acids to systemic tissues, such as the prostate, is less understood. CDA can only signal and bind to intracellular NRs if those cells express solute carrier member transporter proteins on their surface and indeed these are altered in PCa by genetic and epigenetic mechanisms (45–47). Furthermore, during ADT in men with PCa, circulating levels of bile acids are elevated, potentially suggesting that AR signaling systemically may alter bile acid levels, highlighting further cross talk between these receptors (48). The potential role of bile acids, mediated through the FXR, in modulating steroid signaling has recently been reviewed (49).
It has been hypothesized that bile acids may allow organs to preemptively sense when there will be a fresh supply of triglycerides after a meal (50). In this manner, CDA may act to squelch antiproliferative responses from other external signals in order to allow unabated cell division while there is a sufficient supply of raw materials to build more cells. The recognized receptor of CDA, FXR, is associated with induction of detoxifying enzymes and xenobiotic transporter pumps. Similarly, VDR and pregnane X receptor act as bile acid sensors (51). Thus, through the NR network, the presence of CDA prepares tissue for cellular recycling by deferring its antiproliferative signaling capacities and provides for its own detoxification if concentrations are too high. Chronic, elevated serum bile acid levels due to high dietary fat intake might then overwhelm detoxifying pathways and provide a favorable tumor environment. Given the absence of FXR in RWPE-1, it is intriguing that the role of CDA in this context may be to antagonize the VDR directly, as described for the structurally related secondary bile acid LCA (52), resulting in squelched ability of 1α,25(OH)2D3 to elicit cell-cycle arrest and antimitotic signals.
22-HC cooperates with other factors to inhibit proliferation potently. Endogenously, 22-HC is a metabolic intermediate in the production of steroid hormones from cholesterol. The LXRs are responsive to 22-HC and in turn activate mechanisms of cholesterol efflux, including upregulation of the ABCG1 transporter, resulting in reduced intracellular and membrane-associated cholesterol (53). Cholesterol metabolism is critical in deriving many of the compounds sensed by the NRs, and thus it is not surprising that LXRs reside as an integral node in the prostate NR network. Via the LXRs, 22-HC sensing has been implicated in dampening androgen-dependent proliferation in PCa cells and has been proposed as a therapeutic target in this setting (54). The LXRα is itself inhibited by FXR via small heterodimer partner in the liver, suggesting that the dampening of NR-network-mediated antiproliferative signals by CDA may work through inactivating basal LXR function.
The interactive nature of LXRs and PPARs indicates that lipid and cholesterol metabolism is regulated cooperatively through these NRs in the prostate. Several reports have suggested a PPAR–RXR–LXR axis that cooperatively controls lipid and cholesterol homeostasis in hepatocytes (55) and macrophages (29). Our binary studies revealed gene-regulatory overlap of PPAR and LXR activation, corroborating that this axis may be active in normal prostate epithelia. Providing strong evidence of these observations, it was recently determined by chromatin immunoprecipitation-sequencing that PPARα binds to 71–88% of all identified LXR–RXR response elements in mouse liver, and that this binding occurs in a mutually exclusive manner (20). These molecular understandings might provide rationale for the confounding epidemiological evidence linking serum cholesterol to PCa (56,57).
Subsequently, we used tumor archive data to reveal that the expression of the NR network in prostate tumors is significantly more distorted than predicted by chance, and that this may in part be result of targeting by miRNA. The distortion of NR superfamily expression in PCa was significant in a clinical cohort of tumors and suggests that prostate tissue maintenance relies on its ability to sense and respond to a range of dietary lipophilic compounds. For instance, LXR ligand-mediated signaling was strongly suppressive toward cellular proliferation in our study, and thus, selection against its expression would be predicted in PCa patients. Indeed LXR-a and LXR-b are deregulated in a substantial proportion of these patients, with LXR-b alongside PPARA among the most commonly downregulated NRs in the primary tumor cohort. By contrast, VDR and FXR, encoding receptors that mediate bile acid signaling, display infrequent deregulation. It is also interesting to note that the most commonly lost NRs include several of the retinoid receptors (RARG, RARB, RXRA, RXRB) and reflect the squamous metaplasia of the prostate in RARγ null mice (7). Interestingly, retinoids were only additive within the NR network in the current study, yet may reflect limitations in our method to elucidate interactive ligands. By contrast, LXRs were interdependent on PPARs and thus their combined behavior is not apparent when considering each receptor in isolation. The concept that together LXRs and PPARs represent an emergent conduit of NR signaling and gene regulation was hitherto unsuspected in the prostate.
Expression status is not the only manner by which the NR can be disrupted. Mutations in the AR are well established, and with large-scale sequencing efforts it is apparent that mutations and deletions are detected in various NRs. Preliminary screens of all 48 NRs for tissues where DNA sequence data are available in TCGA (i.e. breast, ovarian and colon) suggest that these genes are commonly distorted at the genomic level as well. In parallel, a number of reports have examined how distorted interactions with co-activators or corepressors can distort the transcriptional actions of NRs. Specifically, we have focused on interactions of VDR and PPAR interactions with NCOR1 and NCOR2/SMRT and in both cases, we have established selective suppression of transcriptional actions by these receptors in PCa progression (58).
To this established understanding, we now have also added the concept of distortion to miRNA that target NRs. Our analysis of NR-targeted miRNAs (Supplementary Figure S10, available at Carcinogenesis Online) indicates that global miRNA deregulation plays a significant role in the desensitization of the prostate to dietary constituents. This in turn begs the question as to what processes regulate miRNA expression. Of course, there are likely many events that are critical, but it is interesting to speculate on the interdependent status of NRs and miRNA. For example, VDR expression is commonly retained in this PCa cohort, of which a key target is miR-106b (59), the most upregulated miRNA in our analysis (Supplementary Table S3, available at Carcinogenesis Online). Meanwhile, miR-106b targets several downregulated NRs, including PPARA, suggesting that miRNAs add yet another layer of complexity within the NR network.
Together, the current observations of NR expression and miRNA co-expression combined with our established findings on epigenetic distortion suggest highly intricate processes disrupting the NR superfamily in the prostate. Genomic understanding will only add to this complexity. We also present evidence that this disruption to the NR superfamily is more significant than expected and does not occur for another important transcription superfamily, namely the ETS family. The intricacy within the network provides the cell with a finely tuned homeostatic framework with which to sense and respond to dietary and hormonal cues. In all, we identified a potential homeostatic network of NRs in normal prostate epithelia with the capacity to respond to a range of naturally occurring lipophilic ligands. We observed cooperative control of cellular proliferation, gene transcription and cell-cycle progression upon dual treatments with these compounds. We also identify significant loss of NR expression in a PCa patient cohort, with implicating evidence of the involvement of NR-targeted miRNAs.
Supplementary material
Supplementary Tables 1–3 and Figures 1–10 can be found at http://carcin.oxfordjournals.org/
Funding
M.J.C. acknowledges the support of NucSys, a European Community FP6- Marie Curie Research Training Network ; CanSys, an Atlantis DoE-EU training network [P116J090011]; the Biotechnology and Biological Sciences Research Council, UK; and support in part from National Institute of Health [R01 CA095367-06 and 2R01-CA-095045-06]. M.J.C. also acknowledges support, in part, of the NCI Cancer Center Support Grant to the Roswell Park Cancer Institute [CA016056].
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- 9cRA
9-cis retinoic acid
- 22-HC
22-hydroxycholesterol
- ADT
androgen deprivation therapy
- ATRA
all-trans retinoic acid
- Bez
bezafibrate
- CDA
chenodeoxycholic acid
- DHT
dihydrotestosterone
- EPA
eicosopentanoic acid
- ETYA
eicosotetraenoic acid
- FXR
farnesoid X receptor
- LCA
lithocholic acid
- LXR
liver X receptor
- miRNA
microRNA
- mRNA
messenger RNA
- MSKCC
Memorial Sloan-Kettering Cancer Center
- NR
nuclear receptor
- PCa
prostate cancer
- PPAR
peroxisome proliferator-activated receptor
- Q-RT–PCR
quantitative real-time reverse transcription–polymerase chain reaction
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- SAGE
serial analysis of gene expression
- TCGA
The Cancer Genome Atlas
- VDR
vitamin D receptor.
References
- 1. Battaglia S., et al. (2010). Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis, 31, 1650–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baranowski M. (2008). Biological role of liver X receptors. J. Physiol. Pharmacol., 59 (suppl. 7), 31–55 [PubMed] [Google Scholar]
- 3. Abedin S.A., et al. (2009). Elevated NCOR1 disrupts a network of dietary-sensing nuclear receptors in bladder cancer cells. Carcinogenesis, 30, 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kacevska M., et al. (2011). Extrahepatic cancer suppresses nuclear receptor-regulated drug metabolism. Clin. Cancer Res., 17, 3170–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaeding J., et al. (2008). Activators of the farnesoid X receptor negatively regulate androgen glucuronidation in human prostate cancer LNCAP cells. Biochem. J., 410, 245–253 [DOI] [PubMed] [Google Scholar]
- 6. Zhang Z., et al. (2004). Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res., 14, 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lohnes D., et al. (1995). Developmental roles of the retinoic acid receptors. J. Steroid Biochem. Mol. Biol., 53, 475–486 [DOI] [PubMed] [Google Scholar]
- 8. Pommier A.J., et al. (2013). Liver x receptors protect from development of prostatic intra-epithelial neoplasia in mice. PLoS Genet., 9, e1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khanim F.L., et al. (2004). Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene, 23, 6712–6725 [DOI] [PubMed] [Google Scholar]
- 10. Elstner E., et al. (1999). Novel 20-epi-vitamin D3 analog combined with 9-cis-retinoic acid markedly inhibits colony growth of prostate cancer cells. Prostate, 40, 141–149 [DOI] [PubMed] [Google Scholar]
- 11. Campbell M.J., et al. (1998). Expression of retinoic acid receptor-beta sensitizes prostate cancer cells to growth inhibition mediated by combinations of retinoids and a 19-nor hexafluoride vitamin D3 analog. Endocrinology, 139, 1972–1980 [DOI] [PubMed] [Google Scholar]
- 12. Campbell M.J., et al. (1997). Inhibition of proliferation of prostate cancer cells by a 19-nor-hexafluoride vitamin D3 analogue involves the induction of p21waf1, p27kip1 and E-cadherin. J. Mol. Endocrinol., 19, 15–27 [DOI] [PubMed] [Google Scholar]
- 13. Berquin I.M., et al. (2007). Modulation of prostate cancer genetic risk by omega-3 and omega-6 fatty acids. J. Clin. Invest., 117, 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Augustsson K., et al. (2003). A prospective study of intake of fish and marine fatty acids and prostate cancer. Cancer Epidemiol. Biomarkers Prev., 12, 64–67 [PubMed] [Google Scholar]
- 15. Terry P., et al. (2001). Fatty fish consumption and risk of prostate cancer. Lancet, 357, 1764–1766 [DOI] [PubMed] [Google Scholar]
- 16. Holick M.F. (2008). Vitamin D and sunlight: strategies for cancer prevention and other health benefits. Clin. J. Am. Soc. Nephrol., 3, 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bernstein H., et al. (2009). Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol., 15, 3329–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu J., et al. (2011). Dietary cholesterol intake and cancer. Ann. Oncol., 23, 491–500 [DOI] [PubMed] [Google Scholar]
- 19. Perissi V., et al. (2005). Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol., 6, 542–554 [DOI] [PubMed] [Google Scholar]
- 20. Boergesen M., et al. (2012). Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol., 32, 852–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell M.J., et al. (1999). Synergistic inhibition of prostate cancer cell lines by a 19-nor hexafluoride vitamin D3 analogue and anti-activator protein 1 retinoid. Br. J. Cancer, 79, 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sobel R.E., et al. (2005). Cell lines used in prostate cancer research: a compendium of old and new lines–part 2. J. Urol., 173, 360–372 [DOI] [PubMed] [Google Scholar]
- 23. Bello D., et al. (1997). Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis, 18, 1215–1223 [DOI] [PubMed] [Google Scholar]
- 24. Livak K.J., et al. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 25. Lal A., et al. (1999). A public database for gene expression in human cancers. Cancer Res., 59, 5403–5407 [PubMed] [Google Scholar]
- 26. Cerami E., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taylor B.S., et al. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell, 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uhlén M., et al. (2005). A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics, 4, 1920–1932 [DOI] [PubMed] [Google Scholar]
- 29. Chawla A., et al. (2001). A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell, 7, 161–171 [DOI] [PubMed] [Google Scholar]
- 30. Galvez A.F., et al. (2011). Differential expression of thrombospondin (THBS1) in tumorigenic and nontumorigenic prostate epithelial cells in response to a chromatin-binding soy peptide. Nutr. Cancer, 63, 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dweep H., et al. (2011). miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform., 44, 839–847 [DOI] [PubMed] [Google Scholar]
- 32. Hudson R.S., et al. (2013). MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene, 32, 4139–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X., et al. (2012). Suppression of CX43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol. Ther., 13, 890–898 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen H.C., et al. (2013). Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate, 73, 346–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kerr J.F., et al. (1972). Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer, 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu J., et al. (2010). An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell, 17, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bachmann I.M., et al. (2006). EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol., 24, 268–273 [DOI] [PubMed] [Google Scholar]
- 38. Wang Q., et al. (2009). Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell, 138, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker A.R., et al. (1988). Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl Acad. Sci. USA, 85, 3294–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yegnasubramanian S., et al. (2004). Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res., 64, 1975–1986 [DOI] [PubMed] [Google Scholar]
- 41. Asatiani E., et al. (2005). Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res., 65, 1164–1173 [DOI] [PubMed] [Google Scholar]
- 42. Mahapatra S., et al. (2012). Global methylation profiling for risk prediction of prostate cancer. Clin. Cancer Res., 18, 2882–2895 [DOI] [PubMed] [Google Scholar]
- 43. Kim I., et al. (2007). Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis, 28, 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knisely A.S., et al. (2006). Hepatocellular carcinoma in ten children under five years of age with bile salt export pump deficiency. Hepatology, 44, 478–486 [DOI] [PubMed] [Google Scholar]
- 45. Yang M., et al. (2011). SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J. Clin. Oncol., 29, 2565–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grisanzio C., et al. (2012). Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc. Natl Acad. Sci. USA, 109, 11252–11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park J.Y., et al. (2007). Candidate tumor suppressor gene SLC5A8 is frequently down-regulated by promoter hypermethylation in prostate tumor. Cancer Detect. Prev., 31, 359–365 [DOI] [PubMed] [Google Scholar]
- 48. Saylor P.J., et al. (2012). Prospective study of changes in the metabolomic profiles of men during their first three months of androgen deprivation therapy for prostate cancer. Clin. Cancer Res., 18, 3677–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baptissart M., et al. (2013). Farnesoid X receptor alpha: a molecular link between bile acids and steroid signaling? Cell. Mol. Life Sci. [Epub ahead of print] (PMID:23784309). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Houten S.M., et al. (2006). Endocrine functions of bile acids. EMBO J., 25, 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xie W., et al. (2001). An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl Acad. Sci. USA, 98, 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Makishima M., et al. (2002). Vitamin D receptor as an intestinal bile acid sensor. Science, 296, 1313–1316 [DOI] [PubMed] [Google Scholar]
- 53. El Roz A., et al. (2012). LXR agonists and ABCG1-dependent cholesterol efflux in MCF-7 breast cancer cells: relation to proliferation and apoptosis. Anticancer Res., 32, 3007–3013 [PubMed] [Google Scholar]
- 54. Lee J.H., et al. (2008). Androgen deprivation by activating the liver X receptor. Endocrinology, 149, 3778–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson S.P., et al. (2004). Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor alpha, retinoid X receptor, and liver X receptor in mouse liver. Mol. Pharmacol., 66, 1440–1452 [DOI] [PubMed] [Google Scholar]
- 56. Hu J., et al. ; Canadian Cancer Registries Epidemiology Research Group (2012). Dietary cholesterol intake and cancer. Ann. Oncol., 23, 491–500 [DOI] [PubMed] [Google Scholar]
- 57. Murtola T.J., et al. (2012). The importance of LDL and cholesterol metabolism for prostate epithelial cell growth. PLoS One, 7, e39445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Battaglia S., et al. (2010). Transcription factor co-repressors in cancer biology: roles and targeting. Int. J. Cancer, 126, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thorne J.L., et al. (2011). Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic Acids Res., 39, 2045–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.