Summary
Our finding that LINE-1 undermethylation was present in blood collected from women before clinical detection of their tumor provides evidence from a prospective study that lower global methylation is associated with increased risk of breast cancer.
Abstract
Global decrease in DNA methylation is a common feature of cancer and is associated with genomic and chromosomal instability. Retrospective case–control studies have reported that cancer patients have lower global methylation levels in blood DNA than do controls. We used prospectively collected samples and a case–cohort study design to examine global DNA methylation and incident breast cancer in 294 cases and a sample of 646 non-cases in the Sister Study, a study of 50 884 women aged 35–74 years who had not been diagnosed with breast cancer at the time of blood draw. Global methylation in DNA from peripheral blood was assessed by pyrosequencing of the LINE-1 repetitive element. Quartiles of LINE-1 methylation levels were associated with the risk of breast cancer in a dose-dependent fashion (P, trend = 0.002), with an increased risk observed among women in the lowest quartile compared with those in the highest quartile (hazard ratio = 1.75; 95% confidence interval 1.19, 2.59). We also examined 22 polymorphisms in 10 one-carbon metabolism genes in relation to both LINE-1 methylation levels and breast cancer. We found three single-nucleotide polymorphisms in those genes associated with LINE-1 methylation: SLC19A1 (rs1051266); MTRR (rs10380) and MTHFR (rs1537514), one of which was also associated with breast cancer risk: MTHFR (rs1537514). PON1 (rs757158) was associated with breast cancer but not methylation.
Introduction
Breast cancer is one of the most common cancers in women, with an estimated 226 870 new cases and 39 510 deaths in the USA in 2012 (www.seer.cancer.gov). Both decreased and increased methylation is common in cancer tissue depending on genomic location. Although some gene promoter regions become overmethylated and transcriptionally silenced, the level of global methylation in the cancer genome decreases by 20–60%. This loss of methylation, particularly at long interspersed repeat sequences (LINE-1) and other repetitive elements, is associated with increased genomic instability (1–4). The ~500 000 copies of LINE-1 throughout the human genome (5,6) are normally highly methylated (7,8), and these sequences have been used as a surrogate for estimating global DNA methylation levels (6). In breast tumor tissue, LINE-1 undermethylation is correlated with tumor size, histologic grade and stage of disease (9) and is associated with decreased survival and increased risk of recurrence in breast cancer patients (10). Global undermethylation has been observed even in low-grade tumors, suggesting that demethylation is an early event in breast carcinogenesis (11).
Global methylation differences between cancer cases and controls have been demonstrated using DNA extracted from blood or serum (12). Studies of bladder (13,14) and head and neck cancers (15) have shown that blood DNA from cancer cases has reduced LINE-1 DNA methylation. Although breast cancer case–control studies have not shown differences in LINE-1 methylation of blood DNA (16–19), undermethylation has been shown using other assays of global methylation (16,19,20).
DNA methylation is dependent on DNA methyltransferases that make use of methyl groups donated by S-adenosyl-l-methionine (21). Methyl group availability is dependent on one-carbon metabolism from available folate stores, and polymorphisms in one-carbon metabolism genes affect levels of both folate and DNA methylation (22). Common polymorphisms in genes related to one-carbon metabolism are associated with both breast cancer risk (23–29) and DNA methylation levels in breast tissue of healthy women (24,26).
Retrospective case–control designs are potentially problematic because methylation is measured in samples collected after case diagnosis, so that any observed differences in methylation levels may be due to the disease itself or to treatment for it. Use of a prospective design in which samples are collected before diagnosis is advantageous because it avoids possible postdiagnosis effects. We examined global methylation and incident breast cancer using DNA from prospectively collected blood samples of women enrolled in a large, national cohort study. Global methylation was assessed by pyrosequencing of LINE-1. We also assessed associations between polymorphisms in one-carbon metabolism genes and both LINE-1 methylation levels and breast cancer risk.
Materials and methods
Study population
The Sister Study is a prospective cohort study of environmental and genetic risk factors for breast cancer among 50 884 US and Puerto Rican women (www.SisterStudy.niehs.nih.gov). Eligible participants were between the ages of 35–75 years and had a full or half-sister with breast cancer but had not been diagnosed with the disease themselves. Women completed extensive baseline interviews including family history of cancer, reproductive and medical history and lifestyle and environmental factors. Trained examiners obtained blood samples during a home visit. Enrollment took place in 2003–09 and active follow-up will continue for at least 10 years to ascertain breast cancer and other diagnoses. The study was approved by the Institutional Review Boards of the National Institute of Environmental Health Sciences, National Institutes of Health and the Copernicus Group. All women provided written informed consent.
Study sample
Women in this analysis were selected from the 29 026 participants fully enrolled in the Sister Study by June 2007. Our case–cohort design initially included 328 women who developed incident breast cancer between the time of their blood draw and May 2008 and a random sample of 717 women. Of the random sample, 12 women were among those who developed breast cancer during the study period, resulting in 705 non-cases. LINE-1 methylation assays were conducted on DNA extracted from 1015 whole blood samples and 18 blood clots. Six low-quality samples were excluded, resulting in 1027 samples with high-quality measures. Methylation levels differ by race and ethnicity (30), so we restricted analyses to non-Hispanic white women, resulting in the exclusion of 87 women and yielding an analysis sample of 294 cases and 646 non-cases. Medical records and pathology reports were used to verify breast cancer diagnosis. When medical records were unavailable (n = 39), self-reported data were used; the accuracy of self-report in our data was high for cancer verification (98%).
DNA extraction and bisulfite conversion
Genomic DNA was extracted using an automated process (Autopure LS; Gentra Systems) and DNA concentrations were determined using a Nanodrop ND-1000 spectrophotometer. Three DNA aliquots from each participant were bisulfate converted in independent “batches” of bisulfate conversion reactions for a total of 49 independent bisulfite conversion batches. Samples of DNA (500ng) were denatured by adding 5.5 μl 2M NaOH and incubating at 37°C for 10 minutes. The cytosine deamination reaction occurred by adding 30 μl freshly prepared 10mM hydroquinone, followed by 520 μl 3M sodium bisulfite, pH 5.0, and incubating the sample at 50°C overnight. The samples were desalted using the Wizard DNA Clean-Up System (Promega) and the resin was washed twice with 80% isopropanol. Column-purified DNA was desulfonated by adding 5.5 μl 3M NaOH for 5 minutes. Samples were neutralized by the addition of 33 μl 10M NH4Ac and the DNA precipitated in ethanol overnight at −20°C. DNA was pelleted by centrifuging for 30 minutes at 4°C, followed by a wash with 70% ethanol and centrifugation at 4°C for 10 minutes. The bisulfite-converted DNA was air-dried and resuspended in 10 μl diethylpyrocarbonate H2O. At least three different members of a set of 10 control samples were included in each batch to facilitate adjustment for batch-to-batch variability (batch effects). These control samples included peripheral blood lymphocyte DNA (Promega), the MCF-7 cell line (ATCC) and blood from eight healthy female donors. The Promega peripheral blood lymphocyte DNA was included as a control sample in all batches, whereas other control samples were included in overlapping subsets. Both case and non-case samples were present in each batch, with a relative proportion of roughly 2:1 non-cases to cases.
PCR amplification and pyrosequencing
Bisulfite-converted DNA was amplified with primers designed to recognize a consensus LINE-1 sequence (modified from ref. 6): LINE-1-F, 5′-TTTTTTGAGTTAGGTGTGGG -3′; LINE-1-R, 5′-Biotin-TCTCACTAAA AAATACCAAACAA-3′ and LINE-1-Seq, 5′-AGTTAGGTGTGGGATATA GT-3′ (IDT). The 25 μl PCR reaction mix contained 1× reaction buffer (Qiagen), 1.5mM MgCl2, 800nM dNTPs, 5 pmol of forward and reverse primers, 0.8U HotStarTaq polymerase (Qiagen) and 1 μl bisulfite-converted DNA. PCR cycling parameters were as follows: 95°C hot start for 15 minutes, 45 cycles of amplification with 95°C for 20 s, 50°C for 20 s, 72°C for 20 s and a final extension at 72°C for 5 minutes. Following PCR, the biotin-labeled DNA product was bound to streptavidin-coated Sepharose beads (GE Healthcare), purified and denatured using 0.2M NaOH to a single-stranded template. Pyrosequencing primers at 0.3 μmol/l were annealed to the single-stranded template and pyrosequencing was carried out using PSQ HS 96 System (Biotage). Percentage methylation at four CpG sites within the amplicon was quantified using the PSQ Software (Biotage).
Genotyping
We selected 22 single-nucleotide polymorphisms (SNPs) from 10 genes in the one-carbon metabolism pathway using the SNPinfo GenePipe tool, which prioritizes SNPs based on previous genome-wide association study P values, linkage disequilibrium and functional prediction scores (Xu et al. 31). We used P values from the Cancer Genetic Markers of Susceptibility (http://cgems.cancer.gov/) breast cancer genome-wide association study (32) as input for SNPinfo.
Genotyping was conducted by the National Institute of Environmental Health Sciences Molecular Genetics Core Facility using a custom-designed Illumina GoldenGate Genotyping Assay. A total of 20 HapMap trios were genotyped to evaluate parent-parent-child error. Illumina BeadStudio genotyping software (version 1.6.3) was used to call genotypes. Individual genotypes with an Illumina GenCall score <0.25 were assigned as missing. The overall call rate was 0.998. Both averaged parent-parent-child genotype error and averaged replication error were zero. The concordance between genotype results and HapMap data for the 20 HapMap trios was 0.998.
Statistical analyses
To model the case–cohort data and fully control for batch and age effects on methylation levels, we conducted the analysis in two stages. First, we employed a linear mixed model that included batch as a random factor to estimate an average control methylation level for each batch. To remove batch-to-batch variability, we adjusted raw methylation values within each batch by subtracting the estimated average control methylation level for that batch so that all batches had the same average for lab control samples after adjustment. These batch-adjusted LINE-1 methylation values were categorized into the following quartile groups (Q1–Q4, with Q1 the lowest) based on the distribution among non-cases: 74.8–75.8, 75.9–76.2, 76.3–76.5 and 76.6–78.0%. We then used a modified proportional hazards model for case–cohort design to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) for the associations between quartiles of LINE-1 methylation and the risk of breast cancer (33). Parameters and asymptotic variances were estimated using the method described by Prentice and Self (34). To enable full adjustment for age effects on risk, the primary time scale used was age, with women having delayed entry into risk sets (left censored) at the age when they had their blood drawn. Women were followed until breast cancer diagnosis or until censored at the earlier of the age at loss to follow-up or the cutoff date of 15 May 2008. The following variables were assessed as possible confounders: number of first-degree relatives with breast cancer, having a mother with breast cancer, age at menarche, ever having had a full-term pregnancy and age at menopause. None of these variables substantially altered the estimates, so they were not retained in the models. A test for linear trend of HRs was calculated by treating quartiles of methylation as a four-level ordinal variable in the model.
To evaluate how time elapsed since blood draw may influence the risk relevance of measured methylation values, we employed a linear mixed model to test the association between methylation level and time between blood draw and diagnosis of breast cancer in cases, adjusting for batch as a random effect. This analysis assesses possible diminution of the relationship over time, either due to the relationship being secondary to prediagnostic disease or due to covarying drift in risk-related methylation levels and risk over time.
Associations between one-carbon metabolism genotypes and the risk of breast cancer were examined using modified proportional hazard models for case–cohort design, as described above. Associations between one-carbon metabolism genotypes and LINE-1 methylation levels were assessed using a linear mixed effect model adjusting for batch and subject as random effects in non-cases. All statistical analyses were performed using SAS 9.1.3 (Cary, NC).
Results
Cases were more likely than non-cases to be aged 60 years or older and postmenopausal at enrollment, but the two groups were similar with regard to education level, body mass index, ever use of oral contraceptives and number of pregnancies (Table I). By design, all participants had at least one sister diagnosed with breast cancer, but participants who developed breast cancer were more likely to have had their mother or an additional sister diagnosed as well. For cases, the mean age at diagnosis was 57.9 years (standard deviation = 9.3 years) and the average time between blood draw and breast cancer diagnosis was 487 days (range: 5–1389). For non-cases, the average time between blood draw and the end of the follow-up period (May 2008 for this analysis) was 902 days (range 351–1675).
Table I.
Baseline characteristics of participants
| Characteristics | Non-cases (n = 646), n (%) | Cases (n = 294), n (%) |
|---|---|---|
| Age | ||
| 35–39 | 26 (4.0) | 8 (2.7) |
| 40–49 | 176 (27.2) | 75 (25.5) |
| 50–59 | 246 (38.1) | 101 (34.3) |
| 60–69 | 162 (25.1) | 86 (29.3) |
| 70–74 | 36 (5.6) | 24 (8.2) |
| Education | ||
| High school or less | 80 (12.4) | 44 (15.0) |
| Some college | 210 (32.6) | 76 (25.9) |
| Bachelor’s degree or higher | 355 (55.0) | 174 (59.1) |
| Missing | 1 | 0 |
| Body mass index | ||
| Normal or underweight (<24.9) | 275 (42.6) | 120 (41.0) |
| Overweight (25.0–29.9) | 212 (32.9) | 84 (28.7) |
| Obese (≥30.0) | 158 (24.5) | 89 (30.4) |
| Missing | 1 | 1 |
| Ever used oral contraceptives | 538 (83.3) | 240 (81.6) |
| Number of births (including stillbirths) | ||
| 0 | 114 (17.7) | 62 (21.1) |
| 1 | 96 (14.9) | 39 (13.3) |
| 2 | 251 (38.9) | 102 (34.7) |
| 3 or more | 184 (28.5) | 91 (31.0) |
| Missing | 1 | 0 |
| Number of sisters diagnosed with breast cancer | ||
| 1 | 586 (90.7) | 255 (86.7) |
| 2 or more | 60 (9.3) | 39 (13.3) |
| Mother diagnosed with breast cancer | ||
| Yes | 126 (20.0) | 79 (27.6) |
| Missing | 16 | 8 |
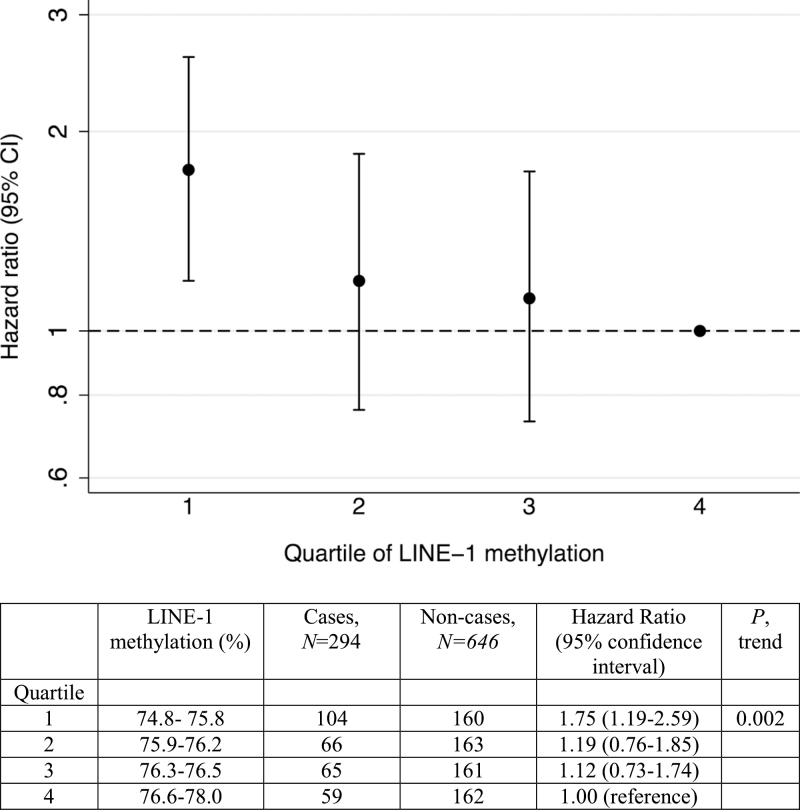
The modified proportional hazard analysis revealed an inverse association between quartile of LINE-1 methylation and the risk of breast cancer in a dose-dependent fashion (P, trend = 0.002), with an increased risk observed among women in the lowest quartile of methylation compared with those in the highest quartile (HR = 1.75; 95% CI: 1.19, 2.59) (Figure 1). LINE-1 methylation levels were associated with the timing of blood collection relative to diagnosis; for each year closer to diagnosis, methylation levels decreased by an average of 0.20% (P = 0.03)
Fig. 1.
Associations between quartile of LINE-1 methylation and the risk of breast cancer. Methylation levels were categorized into quartiles based on the distribution among controls. HRs are adjusted by attained age.
Of 22 SNPs in 10 genes involved in one-carbon metabolism, 3 were significantly associated with LINE-1 methylation: a non-synonymous polymorphism in exon 2 SLC19A1 (rs1051266, P = 0.013), a gene involved in folate transport; a non-synonymous polymorphism in MTRR (rs10380, P = 0.045) (Table II) and a 3′-untranslated region polymorphism in MTHFR (rs1537514, P = 0.029). This latter SNP was also one of two SNPs found to be associated with breast cancer risk: MTHFR (rs1537514) was associated with a reduced risk of breast cancer (HR = 0.69; 95% CI: 0.48, 0.99), whereas a synonymous SNP in PON1 (rs757158) was associated with an increased risk of breast cancer (HR = 1.25; 95% CI: 1.01, 1.54).
Table II.
Associations between one-carbon metabolism SNPs and LINE-1 methylation and breast cancer
| Gene | SNP | Chromosome | Position | Major/minor allele | Amino acid | MAF | LINE-1 methylation | Breast cancer | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Coefficient | P value | HR | 95% CI | |||||||
| MTHFR | rs3737967 | 1 | 11770036 | G/A | 0.04 | 0.24 | 0.12 | 0.81 | 0.48–1.36 | |
| MTHFR | rs1537514 | 1 | 11770655 | G/C | — | 0.1 | 0.23 | 0.03 | 0.69 | 0.48–0.99 |
| MTHFR | rs1801131 | 1 | 11777063 | T/G | E429A | 0.32 | 0.03 | 0.66 | 1.03 | 0.82–1.28 |
| MTHFR | rs1801133 | 1 | 11778965 | G/A | A222V | 0.35 | 0.03 | 0.69 | 0.99 | 0.80–1.23 |
| MTR | rs12354209 | 1 | 235025875 | A/G | — | 0.39 | −0.11 | 0.1 | 0.9 | 0.72–1.11 |
| MTR | rs1805087 | 1 | 235115123 | A/G | D919G | 0.18 | −0.11 | 0.22 | 1.19 | 0.91–1.55 |
| DNMT3A | rs13036246 | 2 | 25386473 | T/C | — | 0.47 | −0.1 | 0.14 | 1.02 | 0.84–1.25 |
| DNMT3A | rs7575625 | 2 | 25402406 | A/G | — | 0.43 | −0.07 | 0.27 | 1.05 | 0.85–1.29 |
| DNMT3A | rs17745484 | 2 | 25404634 | C/T | — | 0.33 | 0.02 | 0.8 | 1.06 | 0.86–1.32 |
| MTRR | rs1801394 | 5 | 7923973 | G/A | I22M | 0.45 | −0.11 | 0.1 | 1.09 | 0.88–1.35 |
| MTRR | rs1532268 | 5 | 7931179 | C/T | S175L | 0.38 | 0.04 | 0.57 | 1.07 | 0.86–1.33 |
| MTRR | rs10380 | 5 | 7950191 | C/T | H595Y | 0.1 | −0.22 | 0.05 | 0.89 | 0.63–1.28 |
| BHMT | rs3733890 | 5 | 78457715 | G/A | R239Q | 0.30 | 0.04 | 0.56 | 0.93 | 0.74–1.18 |
| PON1 | rs662 | 7 | 94775382 | T/C | Q192R | 0.3 | 0.07 | 0.36 | 0.97 | 0.77–1.22 |
| PON1 | rs854560 | 7 | 94784020 | A/T | L55M | 0.36 | 0.06 | 0.41 | 0.96 | 0.78–1.20 |
| PON1 | rs757158 | 7 | 94793464 | C/T | — | 0.43 | −0.08 | 0.23 | 1.25 | 1.01–1.54 |
| MTHFD1 | rs2236225 | 14 | 63978598 | G/A | R653Q | 0.44 | 0 | 0.94 | 0.83 | 0.68–1.03 |
| DNMT3B | rs742630 | 20 | 30814325 | C/G | — | 0.41 | 0.11 | 0.13 | 1 | 0.80–1.25 |
| CBS | rs234706 | 21 | 43358419 | G/A | — | 0.36 | −0.05 | 0.49 | 0.99 | 0.80–1.23 |
| SLC19A1 | rs1051266 | 21 | 45782222 | C/T | H27R | 0.41 | 0.17 | 0.01 | 0.98 | 0.79–1.23 |
| TCN2 | rs9606756 | 22 | 29336860 | A/G | I23V | 0.11 | 0.13 | 0.21 | 0.83 | 0.59–1.16 |
| TCN2 | rs1801198 | 22 | 29341610 | C/G | R259P | 0.45 | 0.03 | 0.61 | 1.09 | 0.90–1.34 |
Bold indicates P value ≤0.05. MAF, minor allele frequency; MTRR, methionine synthase reductase; PON1, paraoxonase 1; SLC19A1, solute carrier family 19.
Discussion
Using a prospective case–cohort study of 940 women, we found statistically significant LINE-1 undermethylation in the peripheral blood DNA of women who developed breast cancer compared with those who remained breast cancer-free throughout our study period. Undermethylation of LINE-1 was present in blood samples collected before clinical detection, consistent with the idea that global demethylation is an early event in breast carcinogenesis. Among cases, undermethylation was greater in women who provided blood closer to the time of diagnosis, indicating that undermethylation of LINE-1 may be a marker of active breast cancer. Our use of samples collected and stored before cancer diagnosis is a particular strength of this study because it avoids a number of potential biases inherent in case–control designs including case–control differences in sample collection and processing and the possibility that the changes observed in cases were introduced by responses to the disease itself or by biopsy, surgery, treatment or other postdiagnostic life events. Pyrosequencing has been established as a precise assay for assessing LINE-1 methylation levels in peripheral blood (35) and a particular strength of our study is that three independent replicate measures were made on each sample, with three independent bisulfite conversions, PCR and pyrosequencing reactions.
Disruption of the usually heavily methylated LINE-1 sites increases the chance of undesired mitotic recombination. Such recombination can lead to the reactivation of transposable elements and their integration at random sites in the genome, resulting in mutagenesis and genomic instability (36). Several studies of global methylation in blood DNA for other cancer types including colon, head and neck, bladder and stomach have found an increased risk of cancer for those in the lowest quantile of global methylation compared with those in the highest (12). Studies of breast cancer; however, have yielded mixed results. Choi et al. (16) found decreased methylation of 5-methyldeoxycytosine in the blood DNA of women with breast cancer compared with controls; likewise both Wu et al. (20) and Cho et al. (19) found decreased methylation of Satellite2. However, Xu et al. (17) found increased global methylation among cases using the luminometric methylation assay. Previous studies of LINE-1 (16–19) did not find an association with breast cancer, but some studies measured LINE-1 in only a small number of cases (<50) (16,19). In a separate epigenome-wide study of Sister Study blood DNA samples, we have also reported that cases were relatively undermethylated at CpG islands near gene transcription start sites (37).
This study contributes to a growing literature examining methylation in blood DNA showing global undermethylation in the blood DNA of those who develop cancer compared with controls (12). However, little is known about the correlation between global methylation in blood and that in target tissues (17). Evidence based on limited studies supports small tissue-specific differences in global methylation levels (38). In a study comparing DNA global methylation in 40 breast cancer patients, tumor tissue and white blood cells were significantly correlated for LINE-1 methylation and both were relatively undermethylated relative to control blood DNA, although the methylation difference between case and control blood cells was not statistically significant (19).
Blood may be more susceptible than other tissues to methylation changes in response to environmental exposures such as chemical pollutants, diet and other lifestyle factors, which may in turn be associated with an increased risk of cancer. These epigenetic alterations may be acquired gradually, resulting in observable differences only after long-term exposure (39). In cross-sectional studies, LINE-1 methylation in blood DNA was lower among smokers (40), inversely associated with arsenic levels in toenails (40) and higher among women with greater physical activity over the life course (41). DNA methylation is a plausible mechanism through which the cumulative effects of environmental exposures may influence cancer risk or it may be a marker of these effects.
One concern is the possibility that differences in the cell populations making up peripheral blood may account for methylation differences between cases and controls. However, LINE-1 measurements in purified granulocyte and lymphocyte samples were not significantly different in one study (40). Fifty CpGs shown previously to be differentially methylated according to leukocyte cell type (42) were not found to be significant predictors of breast cancer in a case–cohort analysis of our samples (37), providing some reassurance that our findings were not due to shifts in leukocyte subpopulations.
We examined four polymorphisms of methylenetetrahydrofolate reductase (MTHFR) in relation to LINE-1 methylation levels and the risk of breast cancer. MTHFR plays a key role in one-carbon metabolism by converting 5,10-methylenetetrahydrofolate to 5-methyl tetrahydrofolate, the primary circulating form of folate. MTHFR is a rate-limiting enzyme in the pathway and thus regulates the levels of methyl groups available for methylation reactions. We found that the SNP in the 3′-untranslated region of MTHFR (rs1537514) was associated with increased LINE-1 methylation and reduced risk of breast cancer. Based on predictions in SNPinfo (31), this SNP is also a putative microRNA-binding site. We did not find significant associations for the C/T nucleotide polymorphism at position 677 that was previously associated with lower genome-wide methylation levels (22) and altered MTHFR enzyme activity (43,44) nor the A/C polymorphism at nucleotide 1298 that was previously associated with MTHFR enzyme activity (45).
We saw a significant association in non-cases between LINE-1 methylation and rs10308, a missense polymorphism (His595Tyr) in methionine synthase reductase (MTRR), a gene involved in regeneration of methionine synthase required for one-carbon metabolism; this SNP has been associated with risk of pancreatic cancer (46). We also found a significant association between LINE-1 methylation and rs1051266, a missense polymorphism (His27Arg) in the solute carrier family 19 (SLC19A1), a gene involved in folate uptake and transport; this SNP has been associated with risk of colorectal adenomas (47). Finally, we found a significant association between breast cancer risk and a SNP (rs757158) in a putative transcription factor-binding site upstream of the paraoxonase 1 (PON1) gene although previous studies found no association between this SNP and breast cancer risk (48–50).
Our study had some potential limitations. Although LINE-1 undermethylation was observed in blood collected before breast cancer diagnosis, our relatively short follow-up time (average of 1.3 years before diagnosis for cases) limited our ability to determine how far in advance of clinical detection these differences are observable. Our sample size for the analysis of polymorphisms was small. Because of the number of tests involved, our findings on one-carbon metabolism genotypes are particularly subject to type 1 (false positive) error. We chose to report P values without correction for multiple comparisons because the SNPs in this study were selected based on previous genome-wide association study P values, linkage disequilibrium and functional prediction scores. These SNP findings should be interpreted with caution because none of the associations would have reached the same level of significance after adjustment for multiple comparisons. Our findings were based on women with a family history of cancer and may not generalize to other women; however, such a history would seem unlikely to influence the relationships between SNPs and methylation or methylation and breast cancer.
In conclusion, using prospectively collected blood samples, we found a statistically significant association between LINE-1 undermethylation and an increased risk of breast cancer in non-Hispanic white women in the Sister Study. We also found some evidence that particular SNPs in one-carbon metabolism genes influence both LINE-1 methylation levels and breast cancer status. Undermethylation of LINE-1 was present in blood samples collected before clinical detection, consistent with the idea that that global undermethylation is associated with increased cancer risk.
Funding
Intramural Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES044005, Z01 ES049033).
Acknowledgements
We thank Dr Lauranell Burch, Re Bai and Raymond Thomas of the National Institute of Environmental Health Sciences Molecular Genetics Core for technical assistance and Karen Baldwin, Gleta Carswell and Alexandra J.White of the National Institute of Environmental Health Sciences Laboratory of Molecular Genetics for assistance with the laboratory assays. We also thank Drs Bonnie R.Joubert and Donna D.Baird for helpful comments on an earlier draft of this paper.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CI
confidence interval
- HR
hazard ratio
- MTHFR
methylenetetrahydrofolate reductase
- SNP
single-nucleotide polymorphism.
References
- 1. Dumitrescu R.G. (2012). DNA methylation and histone modifications in breast cancer. Methods Mol. Biol., 863, 35–45 [DOI] [PubMed] [Google Scholar]
- 2. Esteller M., et al. (2001). DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum. Mol. Genet., 10, 3001–3007 [DOI] [PubMed] [Google Scholar]
- 3. Estécio M.R., et al. (2007). LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE, 2, e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chalitchagorn K., et al. (2004). Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene, 23, 8841–8846 [DOI] [PubMed] [Google Scholar]
- 5. Weisenberger D.J., et al. (2005). Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res., 33, 6823–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang A.S., et al. (2004). A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res., 32, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kochanek S., et al. (1993). DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J., 12, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmid C.W. (1998). Does SINE evolution preclude Alu function? Nucleic Acids Res., 26, 4541–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soares J., et al. (1999). Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer, 85, 112–118 [PubMed] [Google Scholar]
- 10. van Hoesel A.Q., et al. (2012). Hypomethylation of LINE-1 in primary tumor has poor prognosis in young breast cancer patients: a retrospective cohort study. Breast Cancer Res. Treat., 134, 1103–1114 [DOI] [PubMed] [Google Scholar]
- 11. Jackson K., et al. (2004). DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol. Ther., 3, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 12. Woo H.D., et al. (2012). Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS ONE, 7, e34615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilhelm C.S., et al. (2010). Implications of LINE1 methylation for bladder cancer risk in women. Clin. Cancer Res., 16, 1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cash H.L., et al. (2012). LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int. J. Cancer, 130, 1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsiung D.T., et al. (2007). Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev., 16, 108–114 [DOI] [PubMed] [Google Scholar]
- 16. Choi J.Y., et al. (2009). Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis, 30, 1889–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu X., et al. (2012). DNA methylation in peripheral blood measured by LUMA is associated with breast cancer in a population-based study. FASEB J., 26, 2657–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flanagan J.M., et al. (2009). Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum. Mol. Genet., 18, 1332–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cho Y.H., et al. (2010). Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res., 30, 2489–2496 [PMC free article] [PubMed] [Google Scholar]
- 20. Wu H.C., et al. (2012). Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis, 33, 1946–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bestor T.H. (2000). The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402 [DOI] [PubMed] [Google Scholar]
- 22. Friso S., et al. (2002). A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl Acad. Sci. USA, 99, 5606–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J., et al. (2005). One-carbon metabolism, MTHFR polymorphisms, and risk of breast cancer. Cancer Res., 65, 1606–1614 [DOI] [PubMed] [Google Scholar]
- 24. Dumitrescu R.G., et al. (2008). DNA hypermethylation phenotypes and one carbon metabolism genetic variants in breast tissues from normal healthy women. Vol. 17 Cancer Epidemiol. Biomarkers Prev., 32nd Annual Meeting, American Society of Preventive Oncology, Bethesda, MD, pp. 460 [Google Scholar]
- 25. Lissowska J., et al. (2007). Genetic polymorphisms in the one-carbon metabolism pathway and breast cancer risk: a population-based case-control study and meta-analyses. Int. J. Cancer, 120, 2696–2703 [DOI] [PubMed] [Google Scholar]
- 26. Marian C., et al. (2008). Global DNA methylation level in normal breast tissue: associations with demographics, lifestyle exposures and one carbon metabolism genes SNPs. Vol. 17 Cancer Epidemiol. Biomarkers Prev., 32nd Annual Meeting, American Society of Preventive Oncology, Bethesda, MD, pp. 463 [Google Scholar]
- 27. Stevens V.L., et al. (2007). Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol. Biomarkers Prev., 16, 1140–1147 [DOI] [PubMed] [Google Scholar]
- 28. Suzuki T., et al. (2008). One-carbon metabolism-related gene polymorphisms and risk of breast cancer. Carcinogenesis, 29, 356–362 [DOI] [PubMed] [Google Scholar]
- 29. Xu X., et al. (2007). Polymorphisms of one-carbon-metabolizing genes and risk of breast cancer in a population-based study. Carcinogenesis, 28, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 30. Terry M.B., et al. (2008). Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol. Biomarkers Prev., 17, 2306–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu Z., et al. (2009). SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res., 37(Web Server issue), W600–W605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hunter D.J., et al. (2007). A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet., 39, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Therneau T.M., et al. (1999). Computing the Cox model for case cohort designs. Lifetime Data Anal., 5, 99–112 [DOI] [PubMed] [Google Scholar]
- 34. Prentice R.L., et al. (1988). Aspects of the use of relative risk models in the design and analysis of cohort studies and prevention trials. Stat. Med., 7, 275–287 [DOI] [PubMed] [Google Scholar]
- 35. Irahara N., et al. (2010). Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J. Mol. Diagn., 12, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kulis M., et al. (2010). DNA methylation and cancer. Adv. Genet., 70, 27–56 [DOI] [PubMed] [Google Scholar]
- 37. Xu Z., et al. (2013). Epigenome-wide association study of breast cancer using prospectively collected sister study samples. J. Natl Cancer Inst., 105, 694–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nelson H.H., et al. (2011). Global methylation in exposure biology and translational medical science. Environ. Health Perspect., 119, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herceg Z., et al. (2010). Introduction: epigenetics and cancer. Adv. Genet., 70, 1–23 [DOI] [PubMed] [Google Scholar]
- 40. Tajuddin S.M., et al. ; Spanish Bladder Cancer/EPICURO Study Investigators (2013). Genetic and non-genetic predictors of LINE-1 methylation in leukocyte DNA. Environ. Health Perspect., 121, 650–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White A.J., et al. (2013). Recreational and household physical activity at different time points and DNA global methylation. Eur. J. Cancer, 49, 2199–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koestler D.C., et al. (2012). Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol. Biomarkers Prev., 21, 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamada K., et al. (2001). Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc. Natl Acad. Sci. USA, 98, 14853–14858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frosst P., et al. (1995). A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet., 10, 111–113 [DOI] [PubMed] [Google Scholar]
- 45. van der Put N.M., et al. (1998). A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am. J. Hum. Genet., 62, 1044–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohnami S., et al. (2008). His595Tyr polymorphism in the methionine synthase reductase (MTRR) gene is associated with pancreatic cancer risk. Gastroenterology, 135, 477–488 [DOI] [PubMed] [Google Scholar]
- 47. Levine A.J., et al. (2011). Variation in folate pathway genes and distal colorectal adenoma risk: a sigmoidoscopy-based case-control study. Cancer Causes Control, 22, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hussein Y.M., et al. (2011). Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Mol. Cell. Biochem., 351, 117–123 [DOI] [PubMed] [Google Scholar]
- 49. Liu C., et al. (2011). Polymorphisms in three obesity-related genes (LEP, LEPR, and PON1) and breast cancer risk: a meta-analysis. Tumour Biol., 32, 1233–1240 [DOI] [PubMed] [Google Scholar]
- 50. Naidu R., et al. (2010). Genetic polymorphisms of paraoxonase 1 (PON1) gene: association between L55M or Q192R with breast cancer risk and clinico-pathological parameters. Pathol. Oncol. Res., 16, 533–5–40 [Google Scholar]