Abstract
Regulators of G-protein Signaling (Rgs) proteins are the members of a multigene family of GTPase-accelerating proteins (GAP) for the Galpha subunit of heterotrimeric G-proteins. Rgs proteins play critical roles in the regulation of G protein couple receptor (GPCR) signaling in normal physiology and human diseases such as cancer, heart diseases and inflammation. Rgs12 is the largest protein of the Rgs protein family. Some in vitro studies have demonstrated that Rgs12 plays a critical role in regulating cell differentiation and migration; however its function and mechanism in vivo is largely unknown. Here, we generated a floxed Rgs12 allele (Rgs12flox/flox) in which the exon 2, containing both PDZ and PTB_PID domains of Rgs12, was flanked with two loxp sites. By using the inducible Mx1-cre and Poly I:C system to specifically delete Rgs12 at postnatal 10 days in interferon-responsive cells including monocyte and macrophage cells, we found that Rgs12 mutant mice had growth retardation with the phenotype of increased bone mass. We further found that deletion of Rgs12 reduced osteoclast numbers and had no significant effect on osteoblast formation. Thus, Rgs12flox/flox conditional mice provide a valuable tool for in vivo analysis of Rgs12 function and mechanism through time- and cell-specific deletion of Rgs12.
Keywords: Cre, loxP, FRT, conditional inactivation, Regulator of G protein signaling protein
Introduction
“Regulators of G-protein signaling”, or Rgs proteins, are a highly diverse family of proteins containing the conserved structure of ~120 amino acids, designated “Rgs domain”, which is responsible for accelerating the deactivation of heterotrimeric G-proteins, thus modulating signaling initiated by GPCRs (Popov et al., 1997; Soundararajan et al., 2008). To date, more than 30 kinds of Rgs or Rgs-like proteins have been identified (Manzur and Ganss, 2009). Those proteins vary in their molecular structure, tissue distribution, and intracellular localization, and play divergent roles in different tissues through multiple signaling pathways. Some studies have shown that Rgs proteins are involved in the initiation and progression of cancer, heart diseases and inflammation (Hurst and Hooks, 2009; Shankar et al., 2012; Stewart et al., 2012).
Rgs12 is the largest protein in the Rgs protein family based on its protein molecular weight. It was first identified by Snow et al (Snow et al., 1997). Rgs12 mRNA is expressed in rat spleen, lung, prostate, testis, ovary, kidney and brain regions, including cortex, hippocampus, striatum, thalamus and substantia nigra (Snow et al., 1997). The expression of Rgs12 is also detected at different embryonic stages during mouse development (Martin-McCaffrey et al., 2005). Due to its multi-domain architecture, Rgs12 protein has the potential to regulate multiple signaling pathway components. It contains a Rgs domain, which is responsible for GAP activity (Snow et al., 1998), and another Gα-interaction region, the GoLoco motif, which has guanine nucleotide dissociation inhibitor (GDI) activity toward Gαi subunits (Kimple et al., 2002; Willard et al., 2004). Rgs12 also has a pair of Ras-binding domains (RBDs) (Ponting, 1999), suggesting that Rgs12 may integrate signaling pathways involving both heterotrimeric and monomeric G-proteins. The long Rgs12 splice variant has an N-terminal PDZ (PSD-95/Dlg/ZO-1) domain capable of binding the interleukin-8 receptor B (CXCR2) or its own C-terminal (Snow et al., 1998) and a phosphotyrosine-binding (PTB) domain that associates with tyrosine-phosphorylated N-type calcium channel (Schiff et al., 2000). Further study (Anantharam and Diverse-Pierluissi, 2002) proved that interaction between Rgs12 and tyrosine kinase-phosphorylated calcium channel is direct. Additionally, some evidence showed that Rgs12 facilitates the formation of a Ras/Raf/MEK/ERK multiprotein complex in vitro and suggested that Rgs12 promotes a differentiated phenotype by organizing a TrkA, Ras, Raf, MEK and ERK signal transduction complex in glial cell differentiation (Willard et al., 2007). Our previous study has shown that Rgs12 plays a critical role in regulating calcium oscillations and osteoclast differentiation and activation (Yang and Li, 2007b). Despite such important findings, the role and mechanism of Rgs12 in vivo has hitherto not been addressed.
In Mx1-cre transgenic mice, Cre recombinase is under the control of a type I interferon inducible promoter (Mx1) (Kuhn et al., 1995). This promoter is silent in healthy mice, but it can be induced to the high level of transcription and expression in interferon-responsive cells including monocyte and macrophage cells at any time by the administration of poly I: C (Aliprantis et al., 2008; Ruocco et al., 2005). The inducible Mx1-cre and Poly I:C system has been widely used for inducible deletion of target genes in mouse model (Aliprantis et al., 2008; Hu et al., 2012; Tomita et al., 2000; Wang et al., 2009; Wells et al., 2004). Therefore, to investigate Rgs12 function in vivo, we for the first time generated a conditional allele of Rgs12 and confirmed that this allele is efficient for analyzing Rgs12 in vivo function by breeding with cre transgenic mice.
Results and discussion
Generation of Rgs12flox/flox conditional knockout mouse
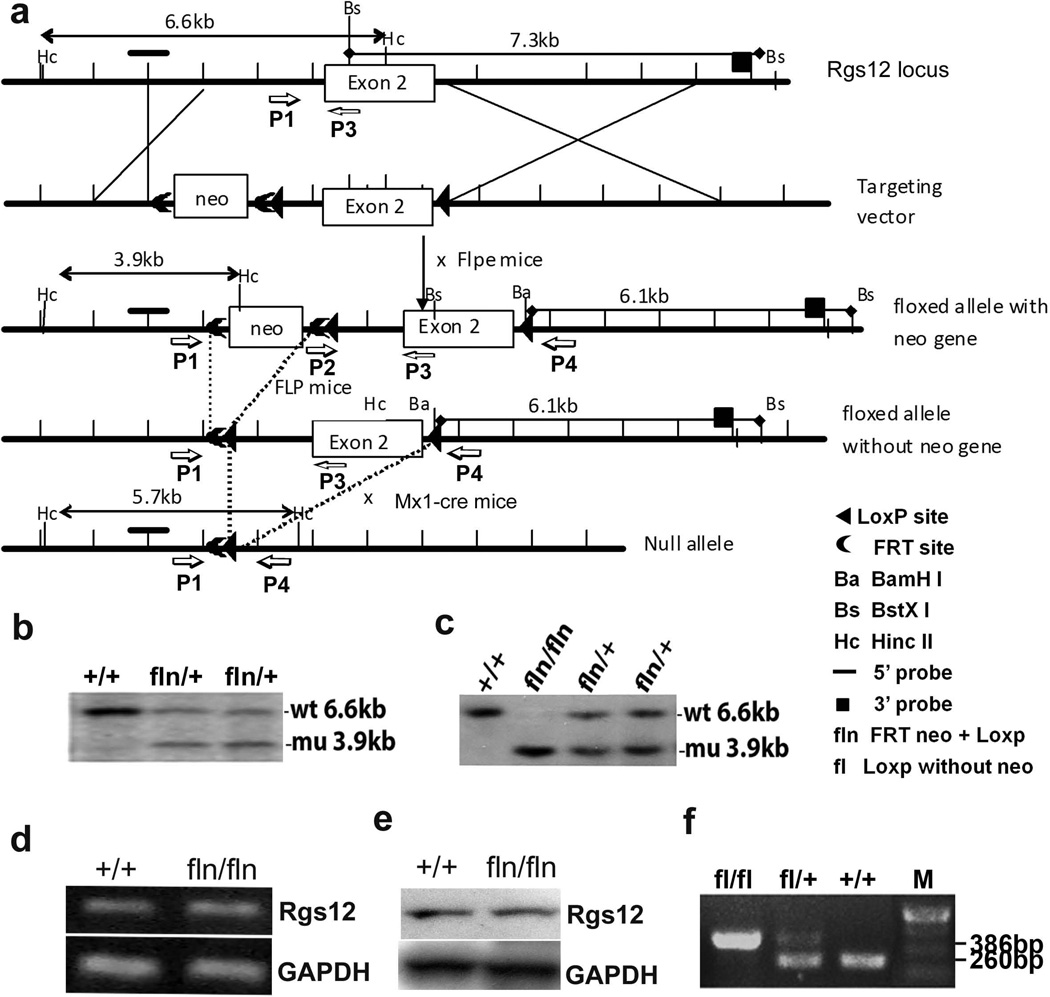
The mouse Rgs12 gene spans 84155 base pairs of genomic DNA and contains 17 exons, which are located within the chromosome 5. We isolated a Rgs12 genomic DNA fragment and constructed a Rgs12 conditional target vector. The targeting vector contains 1.1 kb and 4.92 kb of homologous genomic DNA (Fig. 1a). The linearized targeting vector was electroporated into SV129 embryonic stem (ES) cells. The mutant ES clones were screened and confirmed by southern blot analysis. Thirteen colonies demonstrated the 6.6-kb wild-type (Wt) and 3.9-kb targeted bands by southern blotting with the 5’ external probe (Fig. 1b). Two independent clones were used for generation of chimeric mice and resulted in germ line transmission. F1 mice were genotyped by southern blotting analysis using tail genomic DNA samples for the presence of the targeted Rgs12floxneo allele. Heterozygous mice were intercrossed, and the result of genotyping F2 progeny at 1 week after birth showed homozygous Rgs12floxneo/floxneo mice at the expected mendelian ratio. Southern blotting analysis of genomic DNA from the Rgs12floxneo/floxneo mutant mice and Wt mice showed that the bands corresponding to the Wt are 6.6 kb and that the bands corresponding to targeted mutation with the neo gene (fln: FRTneo+LoxP) are 3.9 kb as expected (Fig. 1c). The homozygous Rgs12floxneo/floxneo mice exhibited normal lifespan and fertility, and did not display any apparent phenotypic abnormality. Reverse transcriptase PCR (RT-PCR) analysis detected Rgs12 expression in mouse bone marrow cells from both Wt and Rgs12floxneo/floxneo mice (Fig. 1d). Western blot analysis using antibody against Rgs12 further confirmed that Rgs12 was expressed in both Wt and Rgs12floxneo/floxneo mice (Fig. 1e), indicating that the insertion of LoxP and Frt-Neo-Frt cassette does not alter Rgs12 gene expression. Some homozygous Rgs12floxneo/floxneo mice were bred to FLPeR transgenic mice (Farley et al., 2000) to remove the Frt-Neo-Frt expression cassette in the germ line to generate Rgs12flox/+. Rgs12flox/+ were mated with Rgs12flox/+ to generate Rgs12flox/flox alleles. The bands corresponding to the neo gene-deleted mutation were 386bp, and the bands corresponding to Wt alleles were 260bp, as determined by PCR analysis (Fig. 1f).
Fig. 1. Generation of Rgs12 conditional null allele.
a. The targeting vector contains 1.1 kb and 4.92 kb of homologous genomic DNA fragments and 4.22kb of excised DNA flanked with LoxP. The 4.22 kb fragment which contains exon 2 (encoding 627 aa that contains both PDZ and PTB domains) of Rgs12 gene was flanked with LoxP and excised. The targeting vector-pRgs12N was constructed by placing a PGK-neo selectable marker flanked by two FRT sites at 3887bp and 2060bp upstream of exon 2. One LoxP site was introduced at 2041bp upstream of exon 2, and the other one was introduced at 201bp downstream of exon 2. b. Identification of Rgs1floxneo/+ mutant ES clones by southern blot. The bands corresponding to the Wt were 6.6 kb and the bands corresponding to targeted mutation with neo gene (floxneo: FRTneo+LoxP) were 3.9 kb as expected. c. Identification of Rgs12floxneo/floxneo alleles by southern blot. The bands corresponding to the Wt were 6.6 kb and the bands corresponding to Rgs12floxneo/floxneo alleles with neo gene (floxneo: FRTneo+LoxP) were 3.9 kb as expected. d. RT-PCR analysis of Rgs12 expression in mouse bone marrow cells from both Wt and Rgs12floxneo/floxneo mice. e. Western blot analysis of Rgs12 expression in both Wt mice and Rgs12floxneo/floxneo mice. f. Identification of Rgs12flox/flox and Rgs12flox/+ alleles by PCR analysis. Rgs12floxneo/floxneo mice were mated with FLPeR transgenic mice to delete the neo gene to generate Rgs12flox/flox and Rgs12flox/+ alleles (fl: LoxP without neo). The bands corresponding to the targeted mutation without neo gene (fl) were 386bp and the bands corresponding to Wt were 260bp.
Inducible deletion of Rgs12 causes growth retardation
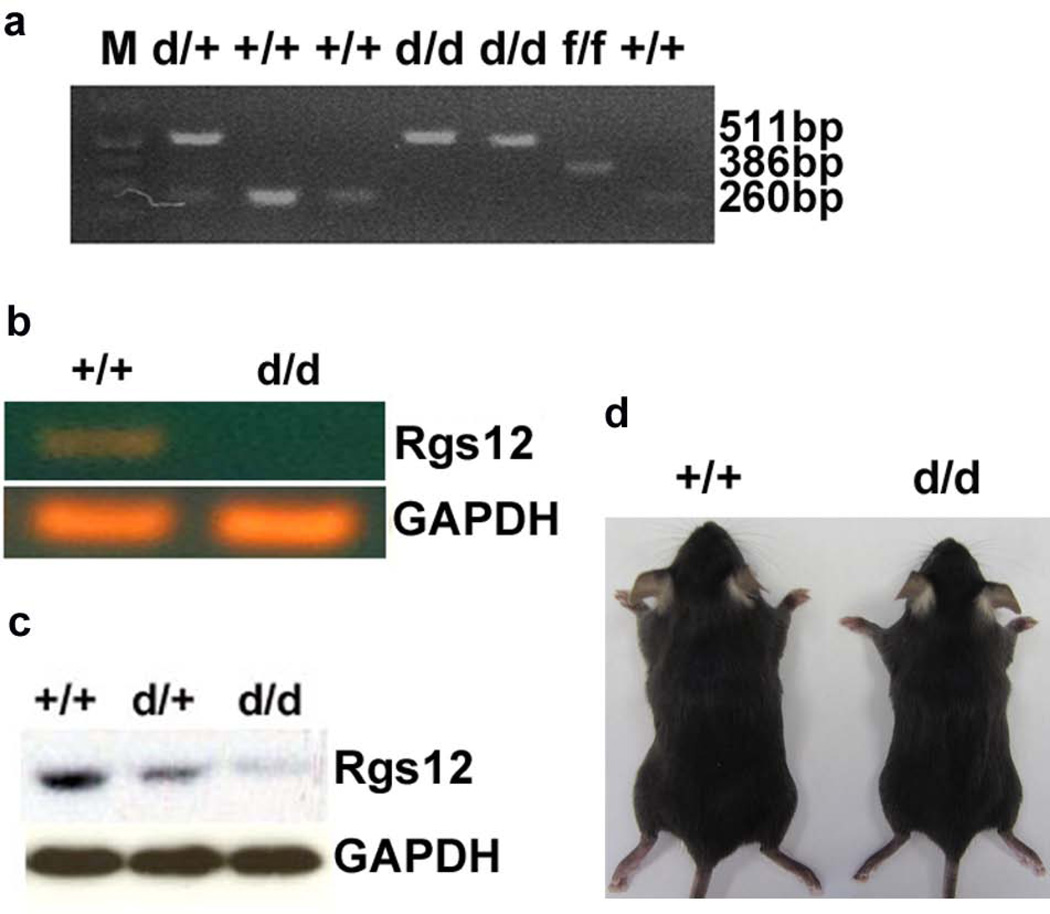
To demonstrate whether the LoxP-flanked exon 2 of Rgs12 gene can be efficiently deleted in vivo, a Mx1-cre inducible transgenic line and Poly I:C system was used to delete Rgs12 gene at postnatal 10 days as described in Materials and Methods (Aliprantis et al., 2008). The deletion of exon 2 in Rgs12del/del/cre mice was confirmed by PCR analysis (Fig. 2a). The 511bp bands representing the targeted alleles were observed in Rgs12del/+ and Rgs12del/del/cre mice. The 260bp bands representing the Wt alleles were detected in Rgs12del/+ and Wt mice. The 386bp bands represented the neo-deleted alleles (fl) as a control. Rgs12del/+ mice were viable, fertile, and did not display any visible abnormalities. To confirm that the deletion of Rgs12 exon2 results in deficiency of Rgs12 expression, we performed RT-PCR and Western blot analysis using cell lysates extracted from mouse bone marrows. The 225bp bands indicating Rgs12 expression were observed in the extracts from Rgs12del/+/cre and Wt/cre mice, but not from Rgs12del/del/cre mutants (Fig. 2b). Western blot analysis showed that Rgs12 protein was not expressed in Rgs12del/del/cre mouse bone marrows (Fig. 2c). Additionally, Rgs12del/del/cre mice were born with the expected Mendelian ratios, but they showed 10–25% reductions in the body weight compared with Wt/cre mice as shown in Fig. 2d. These data demonstrated that Rgs12 was deleted in Rgs12del/del/cre mutant mice.
Fig. 2. Rgs12 expression is disrupted in Rgs12del/del/cre embryos.
Rgs12flox/flox/cre and Wt/cre mice were injected with Poly I: C every other day for three times from E14.5 to E18.5 to delete exon 2 of Rgs12 gene. a. PCR genotype of Rgs12del/del/cre and Rgs1del/+/cre embryos. The primer pair P1/P3 was for amplifying a fragment of 386bp in the flox allele, and a fragment of 260bp in Wt alleles. The primer pair of P1/P4 was for amplifying a fragment of 511bp in Rgs12del/del/cre allele. b. RT-PCR analysis of RNA isolated from BMMs. Bone marrow mRNA from Rgs12del/del/cre and Wt/cre mice was for detecting Rgs12 transcription, which was detectable in Wt/cre BMMs, but undetectable in Rgs12del/del/cre BMMs. c. Western blotting analysis of Rgs12 expression from Rgs12del/del/cre, Rgs12Del/+/cre and Wt/cre BMMs. Total protein extracts from BMMs were blotted for Rgs12 and GAPDH (the loading control). Rgs12 expression was undetectable in Rgs12del/del/cre mice. d. Appearance of Rgs12del/del/cre mice.
Deletion of Rgs12 significantly increases bone mass
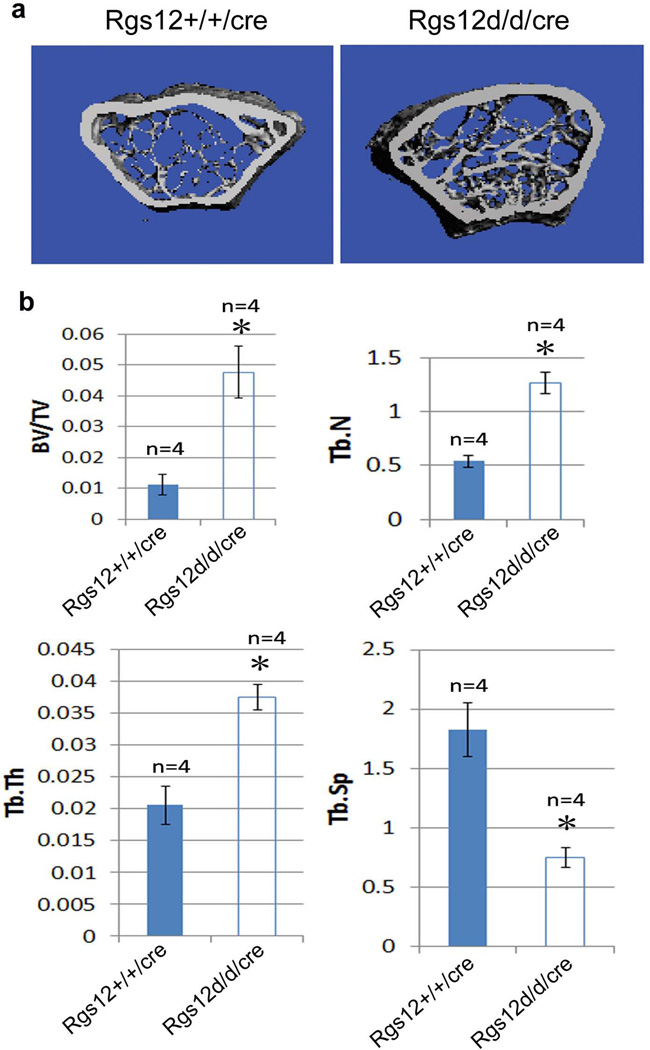
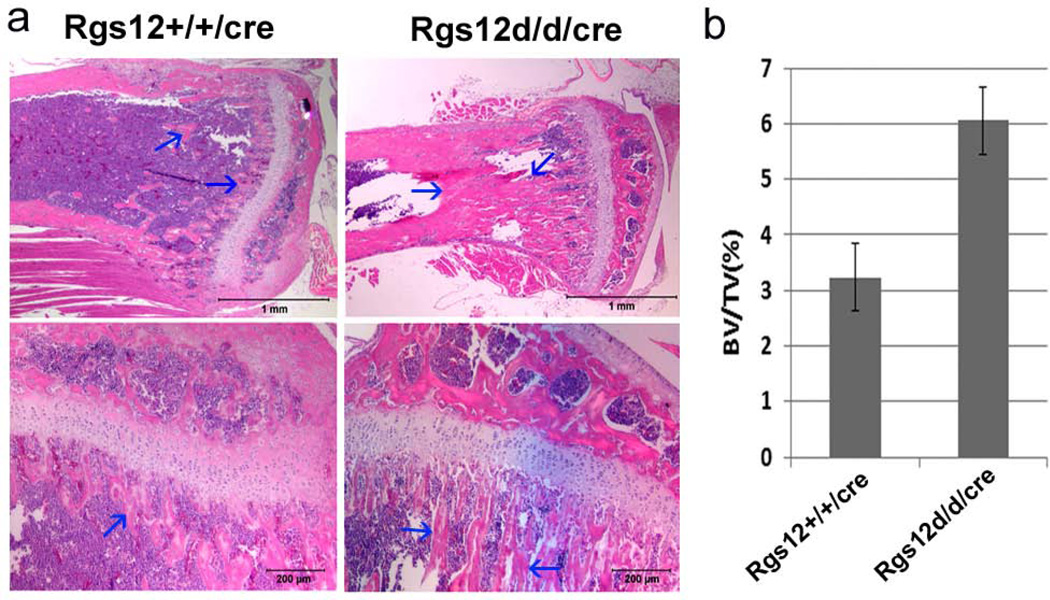
Our previous in vitro study has shown that RNA-interference (RNAi)-mediated knockdown of the Rgs12 transcript inhibits osteoclast differentiation (Yang and Li, 2007b). Additionally, Mxl-cre can efficiently delete the target gene in interferon-responsive cells including monocytes and macrophage cells which are the osteoclast progenitor cells. Hence, to determine whether disruption of Rgs12 leads to the actual changes in bone physiology, 10-day-old Wt/cre and Rgs12del/del/cre mice were injected with Poly I:C as described in Materials and Methods. Two months later following the injection, mice were harvested for the bone phenotype analysis. Micro-CT results showed an apparent increase in the bone mass in Rgs12del/del/cre mice compared with Wt/cre mice (Fig. 3a). The percentage of bone volume to total bone volume (BV/TV), trabecular thickness (Tb.Th) and trabecular number (Tb. N) in the tibia of Rgs12del/del/cre mice were respectively 4.28, 2.34 and 1.82-folds of that in Wt/cre mice (Fig. 3b). Trabecular spacing (Tb.Sp) in the tibia of Rgs12del/del/cre mice was 0.40-fold of that in Wt/cre mice, suggesting an increased bone mass phenotype. To further confirm the phenotype, histological H&E staining analysis were performed in the long bones of Rgs12del/del/cre mice. We found that the long bones in Rgs12del/del/cre were osteopetrotic in appearance (Fig. 4a), with abundance of bone and cartilage trabeculae. The percentage of bone area to total marrow space in long bones of Rgs12del/del/cre mice was 1.9 folds of that in Wt/cre mice (Fig. 4b). The growth plates of Rgs12del/del/cre mice were relatively irregular and slightly extended compared with Wt/cre growth palates (Fig. 4a). These results suggest that Rgs12 likely affects osteoclast and/or osteoblast formation.
Fig. 3. Deletion of Rgs12 causes a significant increase of bone mass.
a. Micro-CT of the femurs from 12-week-old Wt/cre and Rgs12del/del/cre mice. An apparent increase in the bone mass was observed in Rgs12del/del/cre mice compared with Wt/cre mice. b. Quantitative analysis of the percentage of BV/TV, Tb.Th, Tb. N and Tb.Sp in the tibia of Rgs12del/del/cre and Wt/cre mice. N=4. P<0.01, Rgs12del/del/cre vs. Wt/cre (student’s t-test).
Fig. 4. Deletion of Rgs12 causes increased bone mass phenotype.
a. Histological H&E staining analysis of 12-week-old mice tibia. Upper panel: magnification 1.5X, lower panel: magnification 10X. Blue arrows indicate bone tissue. b. Quantitative analysis of the percentage of bone area to total bone marrow space in femur bones of Rgs12del/del/cre mice and in Wt/cre mice. N=4, P<0.01 (student’s t-test).
Increased bone mass in Rgs12-deleted mice results from decreased osteoclast differentiation
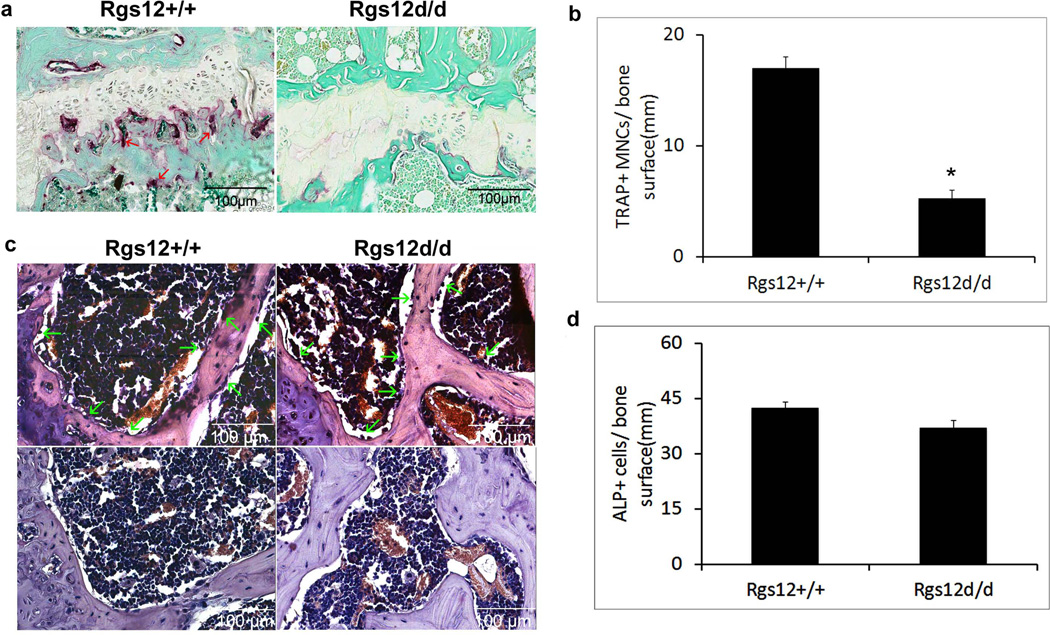
To examine whether deletion of Rgs12 affect osteoclast differentiation and activity, Histochemical stains of 12-wk-old mouse tibiae for the osteoclast enzyme, tartrate-resistant acid phosphatase (TRAP), were performed. The result showed that few TRAP+ osteoclasts in Rgs12del/del/cre tibiae (Fig. 5a). Consistent with this result, quantitative analysis of TRAP+ osteoclast number per millimeter (mm) of bone surface showed a 3.23 fold decrease in Rgs12del/del/cre tibiae compared with Wt/cre tibiae (Fig. 5b), indicating that the deletion of Rgs12 impaired osteoclast differentiation. Most recently, it was reported that Mx1 is also expressed in a subset of mesenchymal stem cells (MSCs) (Park et al., 2012). Those Mx1+ MSCs cells are osteoblast lineage-restricted stem/progenitor cells. They respond to tissue stress and migrate to sites of injury, supplying new osteoblasts during fracture healing. To define whether the deletion of Rgs12 also affects osteoblast differentiation, we performed alkaline phosphatase (ALP) staining for analyzing osteoblasts in the same-age bones. The result showed that there were the ALP+ osteoblasts in both Wt/cre and Rgs12del/del/cre tibiae (Fig. 5c, upper panel). Quantitative analysis further showed that there was no significant difference in osteoblast numbers per mm of bone surface between Rgs12del/del/cre and Wt/cre mice (Fig. 5d), indicating that the increased bone mass phenotype most likely resulted from defective osteoclastogenesis.
Fig. 5. Increased bone mass in Rgs12-deleted mice results from decreased osteoclastogenesis.
12-wk-old mouse tibiae from Rgs12del/del/cre and Wt/cre mice were stained for TRAP and ALP activity. a. TRAP staining for analyzing osteoclasts. Red arrows: osteoclasts. b. Quantitative analysis osteoclast number per mm of the bone surface. *P<0.05 (student’s t-test). c. ALP staining for analyzing osteoblasts. Upper panel: ALP staining, lower panel: negative control. Green arrows: osteoblasts. d. Quantitative analysis osteoblast number per mm of the bone surface. P>0.05 (student’s t-test).
In summary, our study is potentially important and clinically relevant to bone loss diseases. The availability of conditional Rgs12flox/flox mice will provide us with a valuable in vivo model for tissue-specific genetic analysis of the molecular pathways involving Rgs12 in early embryogenesis, postnatal development and diseases.
Materials and methods
Generation of Rgs12floxneo/floxneo allele
Rgs12 genomic DNA fragments from a 129/Sv genomic library (Stratagene) were screened with a 32P-labeled 1.0 kb Rgs12 cDNA fragment as a probe and confirmed by restriction mapping and sequencing. The probe of 1.0 kb Rgs12 cDNA fragment is located at 860bp-1880bp of Rgs12 full length cDNA sequence (Genbank accession no. NM_173402). Targeting vector was constructed by inserting a 16.1 kb Rgs12 genomic DNA fragment into a Bluescript KS vector. A replacement targeting vector was constructed with the 1.1 kb fragment of 5’ homologous arm, the 1.9 kb Frt-flanked Neo-LoxP positive selection cassette, the 4.22 kb fragment excised DNA flanked with LoxP which contains exon 2 (encoding 627 aa that contains both PDZ and PTB domains), and 4.92 kb of homologous genomic 3’ homologous arm. The targeting vector-pRgs12 was constructed by placing a PGK-neo selectable marker flanked by FRT sites at 3887bp and 2060bp upstream of exon 2. One LoxP site was introduced at 2041bp upstream of exon 2, and the other one was introduced at 201bp downstream of exon 2.
For embryonic stem cell (ES) transfection, the targeting vector was linearized with Not I and electroporated into SV129 mouse ES cells (Nagy et al., 1993), which were selected with 200 mg/ml of G418. G418-resistant ES clones were screened by Southern blot analysis of Hinc II-digested genomic DNA with a 5’ external probe. Briefly, 10ug of genomic DNA from mouse tails was digested with Hinc II restriction enzymes, fractionated in 0.8% agarose gels, transferred to nylon membranes, and hybridized with 32P-labeled 865bp 5’ external probe. The targeting frequency was 13 of 144. Two independently targeted ES clones were microinjected into C57BL/6J host blastocysts. Male chimeric mice were bred with C57BL/6J females to obtain germ line transmission of the Rgs12floxneo targeted allele. The genotype of this allele was confirmed by southern blot as described above and PCR genotyping. Briefly, Rgs12floxneo and Rgs12floxneo/floxneo mice were genotyped using two primers P2 (5’-TAAAATGAGGAAATTGCATCGC-3’) and P3 (5’-CACCGCACACACAAAATAAATATCA-3’) and the product length is 375bp. Rgs12floxneo/+ or Rgs12floxneo/floxneo mice were mated with the FLPeR mice expressing the Flpe recombinase (Farley et al., 2000) to excise the neo cassette and generate the Rgs12flox/+ allele. Male Rgs12flox/+ were mated with female Rgs12flox/+ to generate Rgs12flox/flox allele. Excision of the neo cassette was determined by PCR using the primers P1 (5’–CAGTTATTGGAAACTATCTCATGAC-3’) and P3 (5’-CACCGCACACACAAAATAAATATCA-3’), which yielded a 385bp band for the flox allele without neo gene and a 260bp band for Wt allele. All studies and procedures performed on mice were carried out at the Laboratory Animal Facility of the University at Buffalo (UB) and were approved by the UB Institutional Animal Care and Use Committee. The mice were maintained in mixed genetic backgrounds. Rgs12flox/+ or Rgs12flox/flox alleles will be available to the research community upon acceptance of manuscript.
Generation of the Rgs12del/del/cre Alleles
Rgs12flox/+ heterozygous mice were mated with the Mx1-cre line (Kuhn et al., 1995) to generate Rgs12flox/+/cre allele. Then male Rgs12flox/+/cre mice were crossed with female Rgs12flox/+ mice for generating Rgs12flox/flox/cre, Rgs12flox/flox, Rgs12+/+/cre (Wt/cre) and Rgs12+/+ mice. Rgs12flox/flox/cre and Wt/cre mice at 10-day-old were i.p. injected with 250 µg of poly I: C/PBS or PBS in every other day for a total of three doses to eliminate Rgs12 gene and generate Rgs12del/del/cre and Wt/cre mice (Xu et al., 2001). Excision of Rgs12 genomic sequences from tails was determined by PCR using the primers P1 (5’–CAGTTATTGGAAACTATCTCATGAC-3’) and P4 (5’–TCCCCAAGCCTCTTACTTCA–3’), yielding a 511-bp mutant band.
Preparation of bone marrow derived monocyte/macrophage cells (BMMs)
Non-adherent bone marrow cells were obtained from the bone marrow of Rgs12del/del/cre and Wt/cre mice as described (Clohisy et al., 1987; Clohisy et al., 1989). Briefly, the ends of freshly harvested femurs were excised. The bone marrow was collected by flushing the medullary cavity with ice-cold DPBS through a 25-gauge needle. The marrow plug was dispersed by several passages through 18-gauge needles, and the cells were pelleted and incubated in tissue culture plates, at 37°C in 5% CO2, in the presence of colony stimulating factor-1 (20µg/ml). After 24h in culture, the non-adherent cells were layered on a Ficoll-Hypaque gradient (Invitrogen) and the cells at the gradient interface were collected and used for RT-PCR and western blot analyses.
Western blotting
Western blots were performed as described (Yang and Li, 2007a). Briefly, bone marrows or spleens from Poly I :C induced mice were isolated and homogenized in a homogenizer with RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 1% sodium deoxycholate) with protease inhibitor cocktail (Sigma). Protein concentrations were measured using BCA Protein Assay kit (Pierce). About 10–20 micrograms protein samples were separated in a denaturing 10% SDS–PAGE gel and transferred to nitrocellulose membrane. The membranes were washed, blocked (5% milk in PBS-T) and incubated in primary chicken antibody of Rgs12 (1:500, Abcam) or rabbit antibody of GAPDH (1:2000, Genscript) overnight at 4°C. Secondary antibodies were conjugated with horseradish peroxidase. Visualization was done with ECL Western blotting detection reagents (Pierce).
RT-PCR
Total RNA was isolated from bone marrows or spleens of Poly I:C-induced mice using the Trizol reagent (Invitrogen), and cDNAs were synthesized by SuperScriptTM first strand synthesis system for RT-PCR (Invitrogen) according to the manufacturers’ protocols. Subsequently, the cDNAs were analyzed by PCR for Rgs12 gene (NM_173402.2) using the following primer pairs (Rgs12-F1, 5’ GTGACCGTTGATGCTTTCG-3, which is located upstream of exon 2 and at 27–48bp of the Rgs12 gene (NM_173402.2), and Rgs12-R1 5’-CCACTTCGACGCTCCGCACT–3’, which is located in the deleted sequence and at 252-233 of Rgs12 gene (NM_173402.2)). GAPDH (forward primer, 5’-ACCACAGTCCATGCCATCAC-3’; reverse primer, 5’-T CCACCACCC TGTTGCTGTA-3’) was used as a quality and loading control.
Histological analysis
Animals were euthanized by CO2 asphyxiation at 8 weeks or the indicated times after birth. A general pathologic examination of all tissues and organ systems was conducted. Representative bones (femurs and tibiae) were dissected and fixed in 10% formalin buffer, and then decalcified in EDTA. All samples were dehydrated through graded alcohols, embedded in paraffin, sectioned at 5um, and stained with hematoxylin and eosin.
TRAP staining
TRAP staining was performed by a modification of a previously described (Yang and Li, 2007a) method. Briefly, sections were incubated for 15 minutes at 37°C in freshly prepared 0.1 mol/L Tris buffer, pH 5.0, 1.35 mmol/L naphthol AS-MX phosphate (Sigma), 0.362 mol/L N,N-dimethylformamide, 3.88 mmol/L Violet LB salt (Sigma), and 25 mmol/L sodium tartrate. Slides were rinsed for 10 minutes and counterstained with fast green. For quantitation of osteoclasts, five TRAP stained-tibiae slides from Rgs12del/del/cre and Wt/cre mice were analyzed by using both NIH Image J and Adobe Photoshop. In each slides, ten random × 100 objective views located at the proximal ends of tibia were selected for the measurement of the TRAP+ osteoclast number per mm of bone surface as described (Hino et al., 2007).
ALP Staining
ALP staining was performed as described (Miao and Scutt, 2002). Briefly, paraffined bone sections were deparaffinized, hydrated through a xylene and graded alcohol series, and preincubated overnight in 1% magnesium chloride in 100 mm Tris maleate buffer (pH 9.2). Then the sections were incubated in ALP substrate solution (ALP staining Kit, Sigma) for 2 h at room temperature. After washing with distilled water, the sections were counterstained with hematoxylin. The negative control staining was performed on the same bone tissue at the same time with same conditions without the ALP substrate. For quantitation of osteoblasts, five ALP stained-tibiae slides from Rgs12del/del/cre and Wt/cre mice were analyzed by using both NIH Image J and Adobe Photoshop. In each slides, ten random × 100 objective views located at the proximal ends of tibia were selected for the measurement of the ALP+ osteoblast number per mm of bone surface as described (Xian et al., 2012).
Immunohistochemistry
Immunohistochemical staining of paraffin sections was performed as described previously (Yang and Li, 2007a). Paraffin was removed by citrosolv (Fisherbrand), rinsed with tap water. Non-specific labeling was blocked using 10% normal chicken serum, 5% bovine serum albumin, and 0.1% Triton X-100. Excess blocker was removed and the sections were incubated in a combination of primary Rgs12 chicken antibody (Abcam) diluted in block solution for 12 hours at 4 °C. The sections were rinsed in TBS and then incubated with HRP-conjugated anti-chicken IgY secondary antibody (Jackson Imunoresearch laboratories) for 2 hours at room temperature. Sections were rinsed, and further stained with VectaStain Elite ABC kit and DAB enzyme substrates (Vector Laboratories).
Micro-computed Tomography (µCT) determination of bone mass
A quantitative analysis of the bone changes and bone microarchitecture were performed by a Micro-CT system (USDA Grand Forks Human Nutrition Research Center, Grand Forks, ND 58202, USA). The fixed tibiae from Rgs12del/del/cre and Rgs12+/+/cre mice were analyzed and calculated for morphometric indices, including total volume (TV), bone volume (BV), BV/TV as described (Bensamoun et al., 2006; Hawse et al., 2008).
Statistical analysis
Where indicated, experimental data were reported as mean ± SD of triplicate independent samples. Data were analyzed by student’s t-test to determine statistically significant differences between groups. P values <0.05 were considered significant.
Acknowledgments
We thank Mr. David Hadbawnik for the critical reading of the manuscript. We thank Dr. Margaret A. Thompson and the Gene Manipulation Core of the Children’s Hospital, Boston Mental Retardation and Developmental Disabilities Research Center (P30 HD 18655), for technical assistance with the ES cell injections performed for this study. This work was supported by National Institute of Health grant AR055678 (S. Yang), AR061052 (S. Yang) and the USDA CRIS project 5450-51000-046-00D (J. Cao).
Footnotes
Conflict of Interest Disclosure
The authors have no conflict of interest to declare.
REFERENCE
- Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, Ostrowski MC, Olsen BR, Glimcher LH. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–3789. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharam A, Diverse-Pierluissi MA. Biochemical approaches to study interaction of calcium channels with RGS12 in primary neuronal cultures. Methods Enzymol. 2002;345:60–70. doi: 10.1016/s0076-6879(02)45007-0. [DOI] [PubMed] [Google Scholar]
- Bensamoun SF, Hawse JR, Subramaniam M, Ilharreborde B, Bassillais A, Benhamou CL, Fraser DG, Oursler MJ, Amadio PC, An KN, Spelsberg TC. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone. 2006;39:1244–1251. doi: 10.1016/j.bone.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Clohisy DR, Bar-Shavit Z, Chappel JC, Teitelbaum SL. 1,25-Dihydroxyvitamin D3 modulates bone marrow macrophage precursor proliferation and differentiation. Up-regulation of the mannose receptor. J Biol Chem. 1987;262:15922–15929. [PubMed] [Google Scholar]
- Clohisy DR, Chappel JC, Teitelbaum SL. Bone marrow-derived mononuclear phagocytes autoregulate mannose receptor expression. J Biol Chem. 1989;264:5370–5377. [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Hawse JR, Iwaniec UT, Bensamoun SF, Monroe DG, Peters KD, Ilharreborde B, Rajamannan NM, Oursler MJ, Turner RT, Spelsberg TC, Subramaniam M. TIEG-null mice display an osteopenic gender-specific phenotype. Bone. 2008;42:1025–1031. doi: 10.1016/j.bone.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K, Nakamoto T, Nifuji A, Morinobu M, Yamamoto H, Ezura Y, Noda M. Deficiency of CIZ a nucleocytoplasmic shuttling protein, prevents unloading-induced bone loss through the enhancement of osteoblastic bone formation in vivo. Bone. 2007;40:852–860. doi: 10.1016/j.bone.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Hu T, Ghazaryan S, Sy C, Wiedmeyer C, Chang V, Wu L. Concomitant inactivation of Rb and E2f8 in hematopoietic stem cells synergizes to induce severe anemia. Blood. 2012;119:4532–4542. doi: 10.1182/blood-2011-10-388231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009;78:1289–1297. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Manzur M, Ganss R. Regulator of G protein signaling 5: a new player in vascular remodeling. Trends Cardiovasc Med. 2009;19:26–30. doi: 10.1016/j.tcm.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Martin-McCaffrey L, Hains MD, Pritchard GA, Pajak A, Dagnino L, Siderovski DP, D'Souza SJ. Differential expression of regulator of G-protein signaling R12 subfamily members during mouse development. Dev Dyn. 2005;234:438–444. doi: 10.1002/dvdy.20555. [DOI] [PubMed] [Google Scholar]
- Miao D, Scutt A. Histochemical localization of alkaline phosphatase activity in decalcified bone and cartilage. J Histochem Cytochem. 2002;50:333–340. doi: 10.1177/002215540205000305. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Spencer JA, Koh BI, Kobayashi T, Fujisaki J, Clemens TL, Lin CP, Kronenberg HM, Scadden DT. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. Raf-like Ras/Rap-binding domains in RGS12- and still-life-like signalling proteins. J Mol Med. 1999;77:695–698. doi: 10.1007/s001099900054. [DOI] [PubMed] [Google Scholar]
- Popov S, Yu K, Kozasa T, Wilkie TM. The regulators of G protein signaling (RGS) domains of RGS4, RGS10, and GAIP retain GTPase activating protein activity in vitro. Proc Natl Acad Sci U S A. 1997;94:7216–7220. doi: 10.1073/pnas.94.14.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruocco MG, Maeda S, Park JM, Lawrence T, Hsu LC, Cao Y, Schett G, Wagner EF, Karin M. I{kappa}B kinase (IKK){beta}, but not IKK{alpha}, is a critical mediator of osteoclast survival and is required for inflammation-induced bone loss. J Exp Med. 2005;201:1677–1687. doi: 10.1084/jem.20042081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff ML, Siderovski DP, Jordan JD, Brothers G, Snow B, De Vries L, Ortiz DF, Diverse-Pierluissi M. Tyrosine-kinase-dependent recruitment of RGS12 to the N-type calcium channel. Nature. 2000;408:723–727. doi: 10.1038/35047093. [DOI] [PubMed] [Google Scholar]
- Shankar SP, Wilson MS, DiVietro JA, Mentink-Kane MM, Xie Z, Wynn TA, Druey KM. RGS16 attenuates pulmonary Th2/Th17 inflammatory responses. J Immunol. 2012;188:6347–6356. doi: 10.4049/jimmunol.1103781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Antonio L, Suggs S, Gutstein HB, Siderovski DP. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochem Biophys Res Commun. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, Siderovski DP. GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain. J Biol Chem. 1998;273:17749–17755. doi: 10.1074/jbc.273.28.17749. [DOI] [PubMed] [Google Scholar]
- Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundstrom M, Doyle DA, Siderovski DP. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits. Proc Natl Acad Sci U S A. 2008;105:6457–6462. doi: 10.1073/pnas.0801508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Huang J, Fisher RA. RGS Proteins in Heart: Brakes on the Vagus. Front Physiol. 2012;3:95. doi: 10.3389/fphys.2012.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol Endocrinol. 2000;14:1674–1681. doi: 10.1210/mend.14.10.0533. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li G, Tse W, Bunting KD. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood. 2009;113:4856–4865. doi: 10.1182/blood-2008-09-181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J Cell Sci. 2004;117:1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Kimple AJ, Johnston CA, Siderovski DP. Fluorescence-based assays for RGS box function. Methods Enzymol. 2004;389:56–71. doi: 10.1016/S0076-6879(04)89004-9. [DOI] [PubMed] [Google Scholar]
- Willard MD, Willard FS, Li X, Cappell SD, Snider WD, Siderovski DP. Selective role for RGS12 as a Ras/Raf/MEK scaffold in nerve growth factor-mediated differentiation. EMBO J. 2007;26:2029–2040. doi: 10.1038/sj.emboj.7601659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian L, Wu X, Pang L, Lou M, Rosen CJ, Qiu T, Crane J, Frassica F, Zhang L, Rodriguez JP, Xiaofeng J, Shoshana Y, Shouhong X, Argiris E, Mei W, Xu C. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat Med. 2012;18:1095–1101. doi: 10.1038/nm.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Li C, Garrett-Beal L, Larson D, Wynshaw-Boris A, Deng CX. Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis. 2001;30:1–6. doi: 10.1002/gene.1025. [DOI] [PubMed] [Google Scholar]
- Yang S, Li YP. RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev. 2007a;21:1803–1816. doi: 10.1101/gad.1544107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Li YP. RGS12 is essential for RANKL-evoked signaling for terminal differentiation of osteoclasts in vitro. Journal of Bone and Mineral Research. 2007b;22:45–54. doi: 10.1359/jbmr.061007. [DOI] [PMC free article] [PubMed] [Google Scholar]