Abstract
In the mouse embryo and differentiating embryonic stem cells, the hematopoietic, endothelial and cardiomyocyte lineages are derived from Flk1+ mesodermal progenitors. Here we report that surface expression of Podocalyxin (Podxl), a member of the CD34 family of sialomucins, can be used to subdivide the Flk1+ cells in differentiating embryoid bodies at day 4.75 into populations that develop into distinct mesodermal lineages. Definitive hematopoietic potential was restricted to the Flk1+Podxl+ population, while the Flk1-negative Podxl+ population displayed only primitive erythroid potential. The Flk1+Podxl-negative population contained endothelial cells and cardiomyocyte potential. Podxl expression distinguishes Flk1+ mesoderm populations in mouse embryos at days 7.5, 8.5 and 9.5 and is a marker of progenitor stage primitive erythroblasts. These findings identify Podxl as a useful tool for separating distinct mesodermal lineages.
Keywords: mouse embryonic stem cells, pluripotent cells, mesoderm induction, gastrulation, hematopoiesis, endothelial cell, vascular development, Flk1, Podocalyxin
Introduction
During gastrulation, ectodermal cells of the epiblast ingress through the primitive streak to generate the mesoderm [1–4]. Mesodermal cells exiting the posterior and middle primitive streak, including extraembryonic, hematopoietic, vascular, and cardiac mesoderm, express the vascular endothelial growth factor (VEGF) receptor Flk1 (also known as Kdr or Vegfr2), suggesting that these committed mesodermal lineages are derived from Flk1+ progenitors [5–7]. Due to the restricted accessibility and small size of gastrulation stage mammalian embryos, the mechanisms that control the diversification of Flk1+ progenitors to distinct mesodermal lineages are not well understood. The in vitro differentiation of embryonic stem (ES) cells is a useful surrogate for the developing embryo, owing to their relatively faithful recapitulation of early embryonic developmental processes, the ability to grow them in large numbers, and the ease of their genetic manipulation [8, 9]. Using a Brachyury-GFP transgenic reporter mouse line to follow nascent mesoderm [10], two sequential waves of Flk1 expression were observed in differentiating embryoid bodies (EBs) that contained distinct mesodermal progentors. The early (day 3.25) population had enhanced potential to develop into hematopoietic, endothelial and vascular smooth muscle cells while the later (day 4.25) population had cardiovascular potential [6, 11]. Other markers have also been used to characterize Flk1+ mesodermal progenitors in differentiating EBs. ES-derived cells expressing both platelet-derived growth factor receptor a (PDGFR-a) and Flk1 represent an earlier (“primitive”) mesodermal lineage from which a Flk1+PDGFRa-negative lineage with hematopoietic and endothelial potential is derived [12–14]. Differentiating EB cells expressing both Flk1 and Endoglin (Eng), a receptor for the TGFβ superfamily, are more highly enriched for hemangioblast activity than those expressing Flk1 alone [15]. In the work reported here, we asked whether the distinct potentials of the Flk1+ mesodermal population are reflected by other distinct surface protein profiles.
Conditional (doxycycline, DOX) induction of the homeobox transcription factor gene Mixl1 in i-Mixl1 ES cells accelerated the mesodermal developmental program and increased numbers of hemangioblastic and hematopoietic progenitors [16]. Mixl1 activates the mesendoderm marker gene Gsc by direct binding to its promoter [17], thereby controlling the commitment of differentiating ES cells to mesodermal lineages. In a microarray analysis of genes expressed during the differentiation of induced versus uninduced i-Mixl EBs, we found that Podocalyxin (Podxl or Pclp1) was the most strongly upregulated gene. Podxl is a transmembrane glycoprotein that is closely related to CD34 and Endoglycan (reviewed in ref. [18]). Initially identified on adult kidney, where it regulates podocyte development [19], it was also found on cells of the early mouse embryo [20] and, later, on hemangioblasts, hematopoietic stem and progenitor cells, endothelial cells, and circulating embryonic erythroblasts [20–24].
Having detected the upregulation of Podxl in induced i-Mixl EBs, in which mesoderm formation was accelerated and expanded [16], we systematically examined the expression of Podx1 during ES cell differentiation and asked whether it can be used as a marker for separating mesodermal progenitors. We found that Podxl protein is expressed prior to and then overlapping with Flk1 expression on differentiating EB cells and in the mouse embryo. Furthermore, Podxl expression can be used to subdivide Flk1+ mesoderm into two populations (Flk1+Podxl-negative and Flk1+Podxl+) with partially overlapping but distinct developmental potentials. While the Flk1+Podxl+ population was enriched for hematopoietic potential, the Flk1+Podxl-negative population contained predominantly endothelial and cardiac potentials. The Podxl+Flk1-negative population displayed unexpectedly high primitive erythroid potential. Moreover, Podxl is expressed much earlier on primitive erythroid cells than previously believed, marking not only circulating erythroblasts at embryonic day (E)10–12 but also their progenitors at E7.5–8.5. These results indicate that expression of Podxl is a useful marker for separating Flk1+ mesoderm cells with distinct developmental potentials.
Materials and Methods
Mouse ES cell lines and transgenic mice
E14 ES cells were differentiated through the formation of embryoid bodies (EBs) essentially as described [25], with minor modifications. The ES cells were plated at 20,000 cells/ml in Iscove’s Modified Dulbecco’s Medium (IMDM) containing 15% fetal bovine serum (FBS; CellGro), 2 mM glutamine (Gibco), 50 mg/ml ascorbic acid (Sigma), 5% protein-free hybridoma medium II (PFHM-II; Gibco) and 4.5 × 10−4 M monothioglycerol (MTG; Sigma). The differentiation of EBs was carried out for up to 8d and the EBs were harvested at different time points for flow cytometric analysis or for FACS sorting. To test developmental potential, sorted cells were reaggregated for 20 hr in differentiation medium [10] in 24-well low-cluster plates (Costar) or re-cultured for 2-3d on collagen type IV-coated 6-well plates[26] in differentiation medium containing VEGF (5 ng/ml; R&D Systems) and/or the hematopoietic cytokines erythropoietin (EPO; 2 units/ml; Amgen), Interleukin 3 (IL-3; 100 ng/ml; R&D Systems) and stem cell factor (SCF; 100 ng/ml; R&D Systems).
For embryo studies, the e-globin-H2B-EGFP transgenic mouse line, which expresses a histone H2B-GFP fusion protein under the control of the human e-globin promoter and 3′-UTR and a mLCR enhancer [27–29], was used.
Microarray analysis of differentiating i-Mixl ES cells
Gene expression changes were profiled in differentiating i-Mixl1 ES cells cultured in the presence or absence of DOX (0.1 μg/ml, added 24 hr post differentiation [16], 3 replicates per treatment/time point). Total RNA was isolated from EBs harvested at d2, 3 and 4 (DOX added 1d after plating of ES cells). RNA (1 μg) was subjected to one round of linear amplification (RiboAmp System) to yield 10 μg of RNA. RNA was indirectly labeled using amino allyl-dUTP [30], then conjugated with Cy3 or Cy5. Labeled RNAs were used to screen a 15K mouse developmental cDNA microarray [31]. Pairwise analysis of hybridization results for EBs cultured with or without DOX was performed for samples harvested on each day. Spotfire® software was used for data management and filtering. Gene expression ratios were normalized after filtering the data to remove low-intensity and poor quality spots. Data obtained for replicate samples were in excellent statistical agreement (low adjusted p-value). RNA labeling, microarray hybridization, and initial filtering of data were performed. The data files generated by the array analyses have been submitted to Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40703) at accession code GSE40703.
Flow cytometry
EBs were dispersed to single cells using non-enyzmatic Cell Dissociation Buffer (Gibco). Cells were re-suspended in phosphate buffered saline (PBS) containing 10% fetal bovine serum (FBS) and incubated with antibody (Table S1) on ice for 15 min. Cells were then washed once with PBS containing 10% FBS and incubated with allophycocyanin (APC)-conjugated streptavidin (eBioscience) on ice for 15 min. After washing with PBS containing 10% FBS, cells were then resuspended in PBS containing 3% FBS and 3 mM 4′,6-diamidino-2-phenylindole (DAPI) and sorted using a FACSAria III or FACS Influx instrument (BD). For analysis of additional surface markers, dissociated EB cells or re-cultured cells were incubated on ice for 15 min with anti-Flk1 and -Podxl and antibodies against CD144 (VE-cadherin), CD117 (c-Kit), CD31 (Pecam1), and CD41 (Table S1). The cells were washed once with PBS containing 10% FBS and incubated on ice for 15 min with APC-conjugated (eBioscience), FITC-conjugated (BioLegend) or PE/Cy7-conjugated (BD Pharmingen) streptavidin. The cells were then washed with PBS containing 10% FBS and resuspended in PBS containing 3% FBS and 3 mM DAPI for flow cytometric analysis using a BD LSRII system. The data were analyzed using FlowJo software (v8.8.6; TreeStar).
Quantitative real-time RT-PCR (QRT-PCR)
Total RNA was isolated from ES cells, EBs and sorted cells using the Qiagen RNeasy mini kit (Qiagen) as per manufacturer’s instructions. cDNA was synthesized using Superscript III first-strand synthesis system (Invitrogen) or iScript cDNA sysnthesis kit (Bio-Rad) as per the manufacturer’s instructions, with 1 mg of total RNA template in a 20 ml reaction. The TaqMan protocol and primers and probes were described previously [16]. Primer sequences for the SYBR Green method are shown in Table S2. For SYBR Green QRT-PCR, the 20-ml reaction contained 1 ml of cDNA, 5 mM each primer and 10 ml of SYBR Green PCR master mix (Applied Biosystems) or iQ SYBR Green Supermix (Bio-Rad). The reactions were run in a 7900HT fast real-time PCR instrument (Applied Biosystems) using the following program: 95°C, 2 min.; 40 cycles of: 95°C, 15 sec.; 55°C, 15 sec.; 72°C, 30 sec.; 95°C, 15 sec.; and 60°C, 15 sec. The Gapdh gene served as an internal control and the relative expression level of each gene was calculated from the Ct value of the gene normalized to that of Gapdh, as described [29]. The arbitrary value in the formula was set at 100,000.
Hematopoietic progenitor assays
For the primitive erythroid colony-forming cell (EryP-CFC) assay, sorted cells were combined with a methylcellulose mixture for EryP colonies [32, 33] at 20,000 cells/ml and incubated at 37°C, and EryP colonies were scored after 4d. For multi-lineage hematopoietic progenitor colony assays, sorted cells were combined with a methylcellulose mixture (10,000 or 25,000 cells/ml) for multi-lineage hematopoietic colonies, modified from ref. [25] by an increase in ascorbic acid and omission of transferrin [1% methylcellulose (w/v), 15% plasma-derived serum, 10% PFHMII, 2 mM glutamine, VEGF (10 ng/ml), bFGF (10 ng/ml), hSCF (100 ng/ml), hIL-3 (20 ng/ml), EPO (4 units/ml), TPO (20 ng/ml), hIL-11 (50 ng/ml), hIGF-1 (25 ng/ml), GM-CSF (1 ng/ml) and IMDM] and incubated at 37°C. Cytokines were purchased from R&D Systems; PFMII was from GIBCO®/Invitrogen. Hematopoietic colonies were scored at d5–10, depending on the lineage.
Assays for cardiac potential
Sorted cells (105 cells/ml) were reaggregated in StemPro-34 serum-free medium (Invitrogen) supplemented with 2 mM glutamine, transferrin (200 mg/ml), 0.5 mM ascorbic acid and 4.5 × 10−4 M MTG in ultra-low-attachment 24-well plates (Costar) for 24 hr, essentially as described [34]. Individual aggregates were re-cultured in StemPro-34 containing VEGF (5 ng/ml), bFGF (30 ng/ml) and 2 mM glutamine in 96-well plates for 4d and the fraction of beating (contracting) colonies was scored.
Immunofluorescence analyses
Sorted cells were re-cultured on collagen type IV-coated 8-chamber slides (Millipore) at 30,000 cells per chamber in differentiation medium in the presence or absence of VEGF (5 ng/ml) for 2-3d. Immunofluorescence analysis was performed as described [35].
DiI-Ac-LDL uptake
The sorted cells were re-cultured on collagen type IV-coated 8-chamber slides at 30,000 cells per chamber in differentiation medium containing VEGF (5 ng/ml) for 3d. The cells were then incubated with acetylated low density lipoprotein labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindo-carbocyanine perchlorate (DiI-Ac-LDL; Biomedical Technologies) for 4 hr followed by fixation and mounting as per manufacturer’s instructions.
Immunofluorescence analyses
Cells were fixed by incubation in 3% (v/v) formaldehyde in PBS followed by incubation with blocking buffer containing 5% horse serum in PBS. The cells were then incubated at 4°C overnight with rat anti-mouse CD31 (Southern Biotech). The cells were then washed with 0.5% (v/v) Triton X-100 in PBS and incubated at room temperature for 1 hr with AlexaFluor 568 goat anti-rat IgG (H+L) (Invitrogen, A-11077) at a dilution of 1:1000. After washing with 0.5% (v/v) Triton X-100 in PBS, the cells were mounted with VECTASHIELD HardSet mounting medium with DAPI (Vector Laboratories) and viewed using an Axioplan 2 fluorescence microscope (Carl Zeiss).
Angiogenesis/Tube Formation Assay
120,000 cells were resuspended in medium (90% DMEM, 10% FBS, 50 ng/mL VEGF and 2.5 ng/mL bFGF) at a concentration of 600,000 cells/mL and cultured on a 48-well plate pre-coated with Matrigel (BD) for one hour at room temperature. Tube formation was visually assessed every 1 hr for up to 6 hr. Phase-contrast images were acquired using a Canon EOS Rebel T2i camera mounted on a Ziess AxioVert25 using a Plan-NEOFLUAR 5X/NA0.15 objective.
Results
Expression of Podocalyxin distinguishes populations of Flk1+ mesoderm during EB differentiation
To examine the transcriptional changes that occur in response to increases in Mixl1, we screened a 15K mouse developmental cDNA microarray [31] using RNAs isolated from uninduced versus DOX-induced i-Mixl1 EBs [16] at different times (d2, 3 or 4) after initiation of differentiation (DOX was added on d1, as described [16]). A cluster analysis revealed dynamic changes in gene expression in the DOX-treated EBs (Fig. S1). Significant changes in gene expression were observed for numerous developmentally regulated genes, including transcription factors, regulators of signaling pathways, growth factors, and cell adhesion molecules. As early as day 2, transcription of genes involved in mesoderm formation, hematopoiesis and the development of blood vessels and the heart was activated (Table S3 and Fig. S1) and the most strongly upregulated gene (~12-fold) was Podxl. Flk1 is the earliest known surface marker for nascent mesoderm [5]. We hypothesized that the Flk1+ population in d3-4 EBs (peak mesoderm induction) is heterogeneous and multipotent. Therefore, it should be possible to subdivide the Flk1+ cells into different subpopulations with more restricted potential, using additional markers.
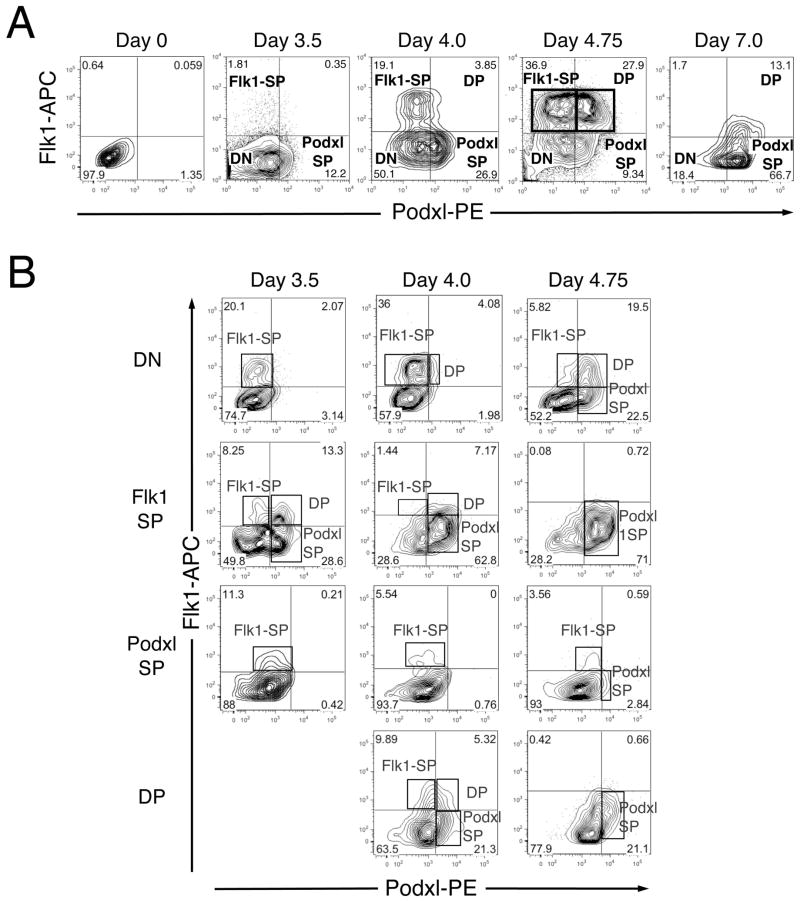
We examined the expression pattern of Podxl on undifferentiated and differentiating mouse ES cells using flow cytometry (Fig. 1A). Podxl is not expressed on undifferentiated mouse ESC but is upregulated during their differentiation, preceding the expression of Flk1 protein and RNA (Figs. 1A, S2 and S3), and divides the Flk1+ population into two distinct Flk1+ subpopulations (Fig. 1, boxed). By d4.75, four distinct populations (Flk1-SP, Flk1-single positive; Podxl-SP, Podxl-single positive; DP, Flk1 and Podxl double positive; DN, double negative) were identified. We, therefore, focused on this time point for subsequent studies.
Figure 1. Expression of Podxl segregates populations of Flk1+ mesoderm during EB differentiation.
E14 ES cells were induced to differentiate as EBs and harvested at the days indicated. For sorting, EBs were dispersed to single cells and stained with anti-Flk1 and anti-Podxl antibodies. (A) Flow cytometric analysis showing subdivision of Flk1+ differentiating ES cells based on expression of Podxl. Boxes highlight the clear separation of Flk1-SP and DP populations at d4.75. (B) Representative flow cytometric analysis of sorted populations after re-aggregation and culture. Differentiating EBs were harvested at the indicated times, dissociated and subjected to antibody staining and FACS. Sorted populations (DN, Flk1-SP, Podxl1-SP and DP) were reaggregated and cultured for 20hr and then analyzed using flow cytometry. The new cell populations formed during re-culture are boxed. The d3.5 DP population was too small to analyze and is, therefore, not represented in this figure. This experiment was repeated three times, with comparable results.
Given the early emergence of Podxl-SP cells in differentiating EBs, we asked whether they could give rise to Flk1+ cells and thus represent a novel population of mesodermal progenitors. The various populations were isolated from EBs at d3.5, 4.0 and 4.75 using fluorescence-activated cell sorting (FACS), allowed to reaggregate for 20 hr [10], and then analyzed for surface expression of Flk1 and Podxl. The purity of the sorted populations was confirmed by flow cytometry immediately after sorting (Figure S4). A representative experiment (out of 3) is shown in Fig. 1B. The largest numbers of new Flk1+ cells arose from reaggregated DN cells (up to ~40%). Only about half as many new Flk1+ cells formed from reaggregated Flk1-SP cells (~22%), while the DP and Podxl-SP populations gave rise to still fewer (~15% and ~12%, respectively).
In contrast, new Podxl+ cells arose largely from reaggregated Flk1-SP (up to ~71%), followed by DN (~42%) and DP (26%) cells (Fig. 1B). Reaggregated Podxl-SP cells gave rise almost exclusively to Flk1+ cells and were also the smallest population at d4.75 (Fig. 1A). Reaggregated Podxl-SP did not give rise to any new DP cells. Only DN and Flk1-SP cells generated all four subpopulations after reaggregation. Together, these findings suggest different developmental potentials for the four subpopulations from d4.75 EBs.
Mesodermal character of the sorted cell populations
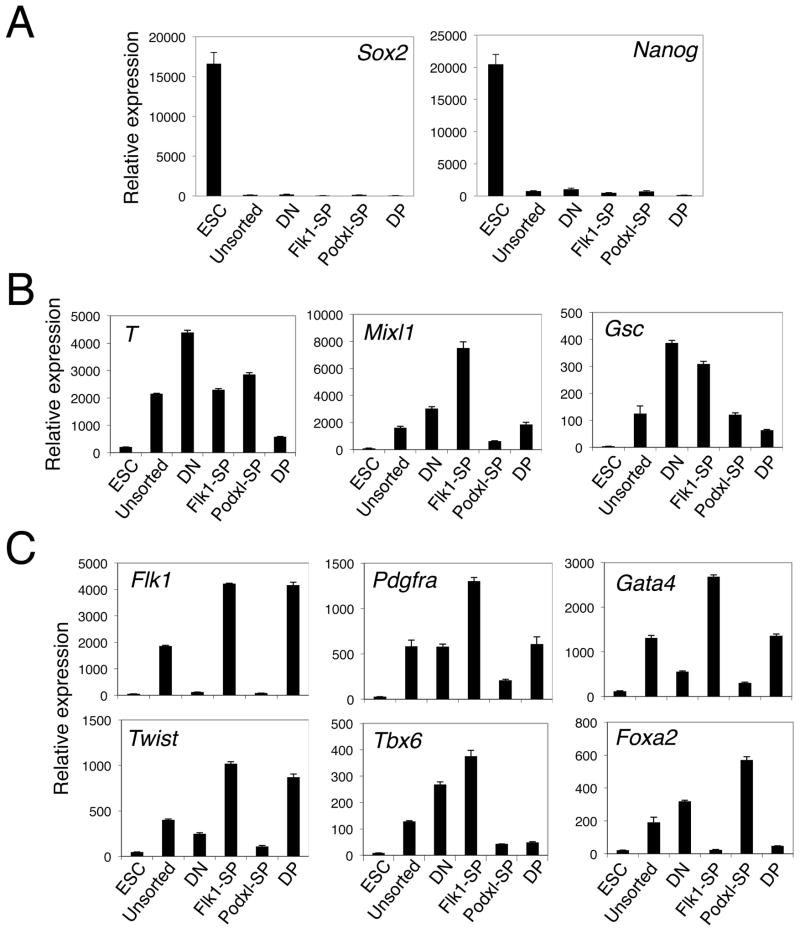
To evaluate the mesodermal character of the cell populations sorted from d4.75 EBs, QRT-PCR analyses were performed. None of these populations expressed the pluripotency markers Sox2 and Nanog (Fig. 2A). The highest levels of expression of Brachyury (T), a marker of the primitive streak and nascent mesoderm, were detected in the DN, Flk1-SP and Podxl-SP populations, with very low level expression in the DP population (Fig. 2B). In contrast, within the Flk1-SP population, markers of primitive streak, mesoderm and mesendoderm (T, Mixl1 and Gsc, Fig. 2B) were expressed at significant levels. The Flk1-SP population also transcribed markers of lateral plate (Twist), paraxial (Pdgfra and Tbx6), and pre-cardiac (Gata4) but not axial (Foxa2) mesoderm (Fig. 2C). Together, these results suggest that the Flk1-SP population contains early Flk1+Pdgfra+ mesoderm progenitors and/or their derivative paraxial, pre-cardiac and vascular mesodermal cells [14]. The DP population showed an expression profile most like lateral plate mesoderm (Twist, Fig. 2C). These results suggest that the Flk1-SP and DP populations separated by expression of Podxl represent distinct mesodermal populations.
Figure 2. Mesodermal character of the sorted cell populations.
EBs were dissociated, stained with antibodies against Flk1 and Podxl, and FACS-sorted to isolate each of the four populations (DN, Flk1-SP, Podxl-SP and DP). Total RNA was isolated and analyzed for expression of the marker genes indicated using QRT-PCR. The expression of each gene was normalized to that of Gapdh. ESC, undifferentiated ES cells; unsorted, unsorted d4.75 EBs. (A) The sorted cell populations did not express pluripotency markers Sox2 and Nanog. (B) Differential expression of the nascent mesodermal marker T and mesendodermal markers Mixl1 and Gsc. (C) Differential expression of mesodermal markers Flk1, Twist (lateral plate mesoderm), Tbx6 and Pdgfra (paraxial mesoderm), Foxa2 (axial mesoderm), and Gata4 (pre-cardiac mesoderm). All QRT-PCR assays were performed at least three times; a representative result is shown for each marker.
Podxl marks the first population with primitive erythropoietic potential in EBs
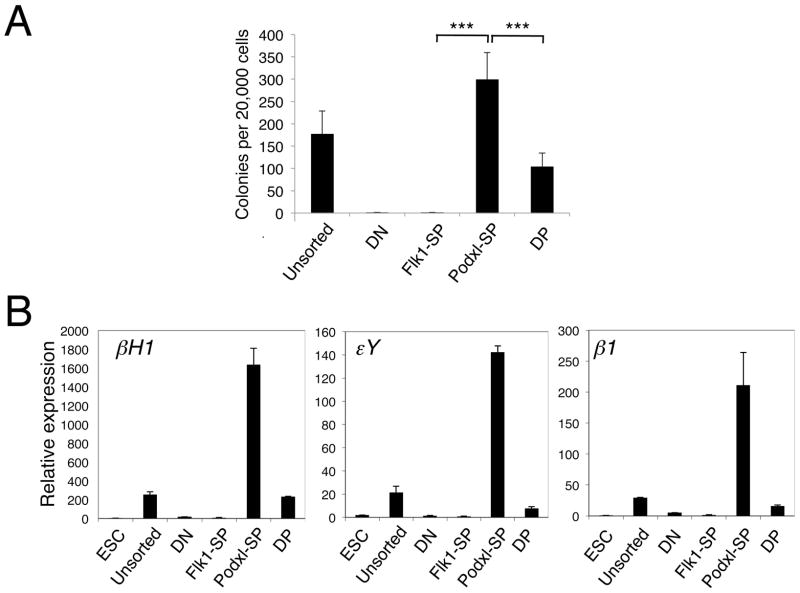
Primitive erythroid cells (EryP) are the first hematopoietic lineage to arise in the gastrulating embryo [36] or in differentiating ES cells [25]. To determine which of the sorted cell populations from d4.75 EBs contain EryP potential, we performed clonogenic progenitor assays in methylcellulose and scored EryP colonies (representing EryP-colony forming cells or EryP-CFC). Most of the EryP potential (75%) was detected in the Podxl-SP population, with the remaining 25% in the DP population (Fig. 3A). Embryonic (βH1 and εY) and adult β1 globins were expressed at significant levels in Podxl-SP cells and at much lower levels in DP cells (Fig. 3B). Essentially no expression of globin genes was detected in the DN or Flk1-SP population. Together with the EryP-CFC data, these findings indicate that the earliest cell population with primitive erythropoietic potential is marked by Podxl expression.
Figure 3. The Podxl-SP population contains primitive erythropoietic potential.
(A) EryP-CFC assay was performed to assess the primitive erythropoietic potential of the d4.75 EB populations. Sorted cells (20,000) were plated in methylcellulose supplied with EPO and EryP colonies were scored after 4d. Primitive erythroid potential was present predominantly in the Podxl-SP population. The data from 3 independent experiments were analyzed using an unpaired two-tailed Student’s t-test (*** p < 0.001). (B) QRT-PCR analysis of embryonic (βh1 and εY) and adult (β1 or βmaj) β-like globin gene expression in the d4.75 EB populations. The expression of each gene was normalized to that of Gapdh.
The Flk1+Podxl+ (DP) population is enriched for definitive hematopoietic potential
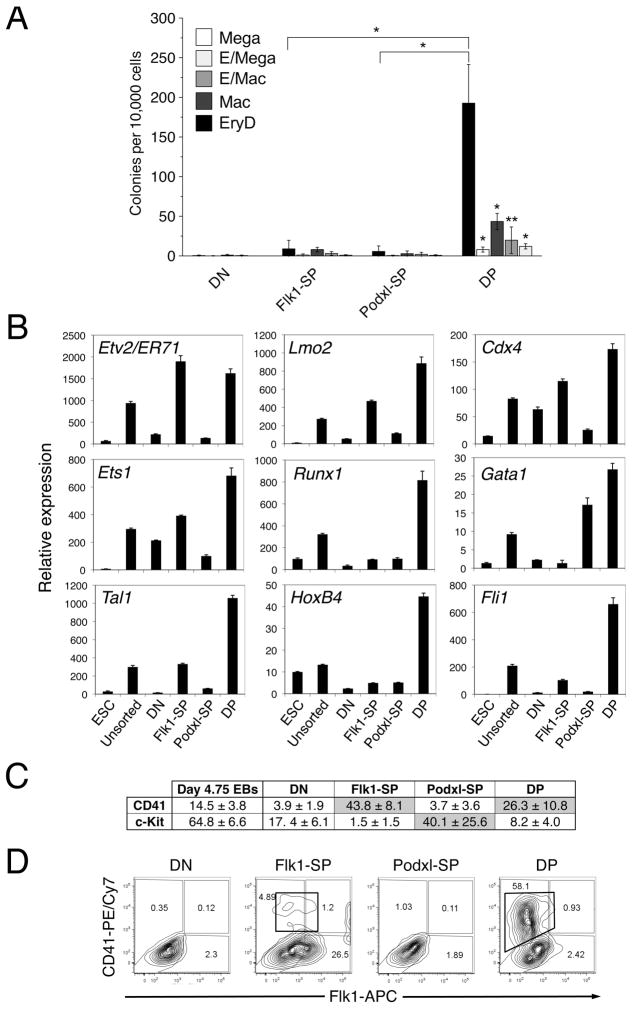
To evaluate the definitive hematopoietic potential of the four populations, we performed multilineage hematopoietic progenitor assays. Only the DP population generated significant numbers of definitive hematopoietic colonies (EryD, macrophage, megakaryocyte, mixed erythroid/macrophage or erythroid/megakaryocyte; Fig. 4A). Hematopoietic transcription factor genes (Lmo2, Cdx4, Ets1, Runx1, Gata1, Tal1, HoxB4 and Fli1) were also expressed largely in the DP population (Fig. 4B). The DP population at d4.75 expressed both c-Kit and CD41 (Fig. 4C), two surface markers of definitive hematopoietic progenitors [37, 38]. However, these markers were not co-expressed (Fig. 4C and data not shown). These data are consistent with results published by others which show significant expression of c-Kit but low levels of CD41 in day 4.75 EBs [37]. CD41 was expressed on the surface of a majority (58%) of DP cells that were sorted and re-cultured for 3d on collagen type IV [26, 39] but on few, if any, cells from the other three recultured populations (Fig. 4D). Taken together, these results suggest that the DP population contains definitive hematopoietic potential.
Figure 4. The DP population contains definitive hematopoietic potential.
(A) Multilineage progenitor assay to assess hematopoietic potentials of sorted d4.75 EB cell populations. Cells (10,000) were plated in methylcellulose supplied with cytokines. EryD colonies were scored on d5 and other colonies on d10. Data from 3 independent experiments were analyzed using an unpaired two-tailed Student’s t-test. The DP population generated significantly larger numbers of definitive hematopoietic colonies than did any of the other populations (* p < 0.0001; ** p < 0.05). EryD, definitive erythroid; Mega, megakaryocyte; Mac, macrophage; E/Mac, mixed erythroid/macrophage; and E/Mega, mixed erythroid/megakaryocyte colonies. (B) QRT-PCR analysis of the expression of genes encoding hematopoietic and endothelial transcription factors. Total RNA was isolated from the sorted d4.75 EB populations (DN, Flk1-SP, Podxl-SP and DP) and analyzed using QRT-PCR. The expression of each gene was normalized to that of Gapdh. This experiment was performed three times, with comparable results. ESC, undifferentiated ES cells; unsorted, unsorted d4.75 EBs. (C) The sorted d4.75 EB populations were analyzed by flow cytometry for expression of c-Kit and CD41. This experiment was repeated three times, with comparable results. (D) Representative flow cytometric analysis of Flk1 and CD41 on d4.75 EB populations after reculturing on collagen type IV for 3d. This experiment was performed 4 times, with comparable results. The CD41+ cells formed from the DP and Flk1-SP populations are boxed.
The Flk1-SP and DP populations contain endothelial potential
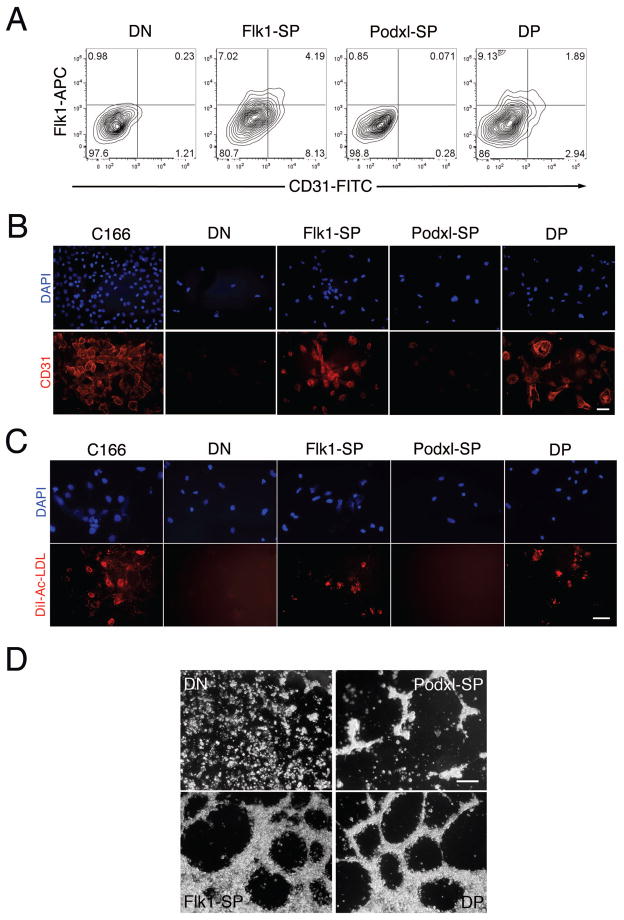
The expression of the endothelial transcription factor genes Etv2/ER71, Tal1, Lmo2, Fli1 and Ets1 [40] in the Flk1-SP and DP populations at d4.75 (Fig. 4B) led us to evaluate their endothelial potential. Expression of the endothelial markers Flk1 and CD31 on sorted cells after re-culture on collagen IV in the presence of VEGF was measured using flow cytometry. Both the Flk1-SP and DP populations, but not the Podxl-SP or DN populations, expressed Flk1 and CD31 (Fig. 5A) and displayed uptake of DiI-Ac-LDL (Fig. 5C). CD31 expression was further confirmed by immunofluorescence (Fig. 5B). Finally, the Flk1-SP and DP cells formed capillary-like structures on matrigel (Fig. 5D).
Figure 5. The Flk1-SP and DP populations display endothelial potential.
(A) The sorted d4.75 cell populations (DN, Flk1-SP, Podxl-SP and DP) were cultured on collagen type IV for 3d and analyzed for expression of the endothelial marker CD31 (Pecam1) using flow cytometry. This experiment was performed 4 times, with comparable results. (B) Representative immunostaining for Pecam1. The sorted d4.75 cell populations (DN, Flk1-SP, Podxl-SP and DP) were recultured on collagen type IV in the presence of VEGF (5 ng/ml) for 3d and then stained with an antibody against Pecam1 (primary) and an Alexa Fluor 568 goat anti-rat secondary antibody. Scale bar, 50 microns. (C) Representative DiI-Ac-LDL uptake assay. The sorted d4.75 cell populations (DN, Flk1-SP, Podxl-SP and DP) were recultured on collagen type IV in the presence of VEGF for 3d and then incubated with DiI-Ac-LDL (10 mg/ml) for 4 hr. Scale bar, 50 microns. (D) Formation of capillary like structures on Matrigel. The sorted d4.75 populations (DN, Flk1-SP, Podxl-SP and DP) were cultured on Matrigel in the presence of VEGF and bFGF. Images were acquired 6 hr post-culture. The analyses in (B) and (C) were performed 3 times, with comparable results. The analysis shown in (D) was repeated twice, with comparable results. C166 is a mouse endothelial cell line used as a positive control.
Cardiac mesoderm potential is restricted to the Flk1-SP population
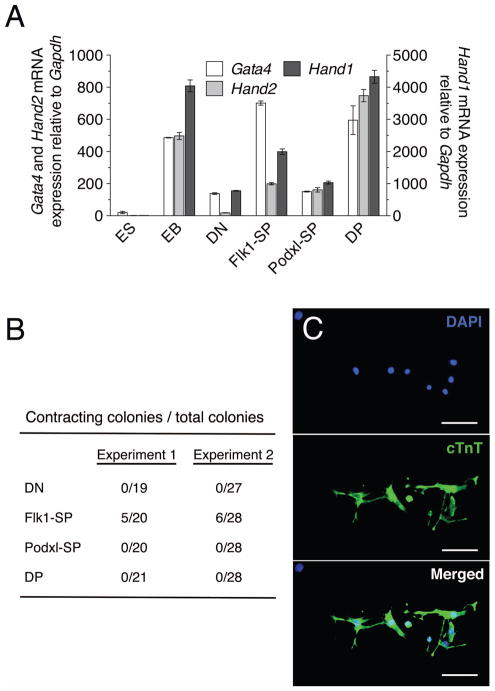
The high levels of expression of Gata4 (Fig. 2C), a marker of pre-cardiac mesoderm [41], in Flk1-SP cells prompted us to ask whether this population possesses cardiac potential. We examined cardiac markers Gata4, Hand1, Hand2, and Nkx2.5 in sorted d4.75 populations. Gata4, Hand1 and Hand2 were all expressed in both Flk1-SP and DP populations at higher levels than in DN and Podxl-SP cells (Fig. 6A). Nkx2.5 expression was not detectable at d4.75 in our analysis; others have reported that it is activated ~d6 of EB differentiation [42, 43]. To examine the potential of the sorted populations to form beating colonies, cells sorted from d4.75 EBs were allowed to reaggregate for 24 hr and were then cultured for 4d in the presence of VEGF and bFGF [34]. Beating colonies were found only in the cultured Flk1-SP aggregates (Fig. 6B). Expression of the cardiac marker troponin T (cTnT) was analyzed using immunofluorescence and was detected only in cells from beating colonies from Flk1-SP cells (Fig. 6C). Collectively, these data suggest that the expression of Flk1 alone is indicative of the segregation of mesodermal cells to the myocardial lineage.
Figure 6. The Flk1-SP population contains cardiac potential.
Sorted d4.75 populations (DN, Flk1-SP, Podxl-SP and DP) were reaggregated for 24 hr in StemPro-34 serum-free medium followed by reculture of the aggregates for 4d in the presence of VEGF and bFGF. Total and contracting colonies formed by the aggregates were scored. (A) QRT-PCR analysis of cardiac mesoderm marker expression. Total RNA was isolated from the sorted d4.75 EB populations (DN, Flk1-SP, Podxl-SP and DP) and analyzed using QRT-PCR. The expression of each gene was normalized to that of Gapdh. This experiment was performed two times, with comparable results. ESC, undifferentiated ES cells; unsorted, unsorted d4.75 EBs. (B) Results of two independent experiments are shown as number of beating colonies/number of total colonies. (C) Immunostaining of colonies formed from the Flk1-SP cell aggregates using rat anti-mouse cTnT (primary) and Alexa Fluor 488 goat anti-rat (secondary) antibodies. (Scale bars, 100 microns)
Podxl expression distinguishes Flk1+ mesoderm populations in mouse embryos
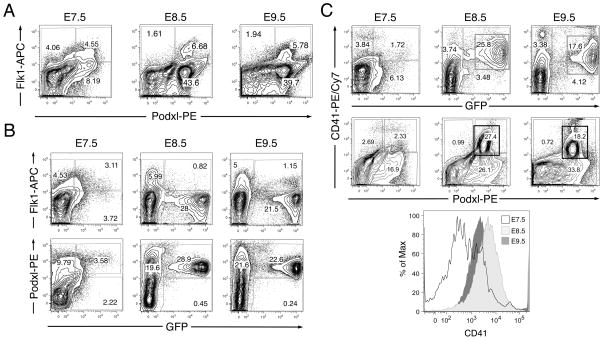
To determine whether Podxl expression distinguishes Flk1+ mesoderm populations in vivo, mouse embryos were harvested at E7.5, E8.5 and E9.5, dispersed to single cells, and analyzed for surface expression of Flk1 and Podxl using flow cytometry. As observed for differentiating EBs (Fig. 1), developing mouse embryos contain Flk1-SP, Podxl-SP and DP populations (Fig. 7A) at each stage analyzed (E7.5 and E8.5, prior to the onset of circulation, when EryP are restricted to the yolk sac, which contains progenitor activity; and E9.5, when EryP are found in the bloodstream but progenitor activity has disappeared [29, 36]).
Figure 7. Podxl marks the primitive erythroid population in mouse embryos.
Transgenic e-globin::H2B-GFP embryos (E7.5, 8.5 and 9.5) were dissociated to single cells for flow cytometric analysis. (A) Expression of Podxl and Flk1 in E7.5-E9.5 mouse embryos. As observed for differentiating ES cells (Fig. 1), developing mouse embryos contain Flk1-SP, Podxl-SP and DP populations at each stage. (B) Flk1 was expressed on the surface of a significant subset of EryP/GFP+ cells at E7.5 but, at E8.5 and E9.5, was absent from the vast majority of EryP. In contrast, the expression of Podxl on GFP+ EryP increased dramatically from E7.5 to E8.5, when nearly all EryP were Podxl+. (C) Expression of the hematopoietic marker CD41 correlated well with GFP fluorescence at each stage of development, indicating that most EryP express CD41. The Podxl+CD41+ populations are boxed. The histogram at the bottom displays changes in expression of CD41 from E7.5 to E9.5. The experiment shown in panels A–C has been repeated twice, with comparable results.
A previous report had demonstrated that Podxl is expressed on embryonic peripheral blood cells at E10 and on Ter119+ CD71+ cells from E12 yolk sac [20]. To determine more directly whether this protein is expressed on EryP, we used a transgenic mouse line in which a histone H2B-GFP reporter is expressed in the nuclei of primitive erythroid cells (EryP) under the control of a human e-globin promoter and a mLCR enhancer [27–29]. GFP+ (EryP) cells were stained with antibodies against Podxl and Flk1 and analyzed using flow cytometry. As expected [29], Flk1 was expressed on the surface of a subset of EryP/GFP+ cells at E7.5 but was absent from most EryP at E8.5 and E9.5 (Fig. 7B). In contrast, the expression of Podxl on GFP+ EryP increased sharply from E7.5 to E8.5, when nearly all EryP were Podxl+ (Fig. 7B). Conversely, a significant proportion (27–60%) of Podxl+ cells in the embryo were EryP (Fig. 7B). Expression of the hematopoietic marker CD41 correlated well with GFP fluorescence at each stage of development (Fig. 7C). CD41 expression increased from E7.5 to E8.5 (with increased numbers of EryP progenitors [29]) and then began to fall by E9.5, a time when EryP are circulating in the blood and progenitor activity is absent [36, 44].
Discussion
Our knowledge of hematopoietic development and the hierarchical relationships among multipotent progenitors has benefitted enormously from the use of cell surface markers to prospectively identify, isolate and characterize populations of cells at different stages of commitment (e.g. see refs. [45, 46]). The ability to subdivide mesodermal progenitors on the basis of surface protein expression, in an analogous approach, would advance our understanding of the lineage specification of mesoderm. Here, we report that ES cell-derived mesoderm identified by surface Flk1 expression can be separated, according to Podxl expression, into distinct populations with distinguishable potentials.
The Podxl-SP subpopulation sorted from EBs at d4.75 (a time when four subpopulations were clearly identified) gave rise only to primitive erythroid cells, and this potential was significantly higher than that of the other three sorted populations. These findings were unexpected, as lineage tracing studies previously established that both primitive and definitive hematopoietic cells are derived from Flk1+ mesoderm [7] and Flk1 marks a subset of E7.5 (progenitor stage) EryP (ref. [29] and this work).
The expression of Podxl was detected earlier than that of Flk1 in differentiating ES cells, raising the possibility that at least some Flk1+ mesoderm cells are derived from an earlier Podxl+ cell population. Consistent with this idea, reaggregated Podxl-SP cells sorted from EBs at various times gave rise almost exclusively to Flk1-SP cells. Therefore, Flk1 expression may segregate Podxl-expressing mesoderm into subsets before Podxl expression segregates Flk1-expressing mesoderm. In contrast, reaggregated Flk1-SP and DP cells produced Flk1-SP, DP and Podxl-SP cells. These observations suggest that the Podxl-SP population contains early mesoderm cells comparable to those that form the extraembryonic mesoderm and primitive erythroid progenitors of the yolk sac. These data, therefore, suggest that Podxl is potentially an earlier marker of the commitment of the Flk1+ mesodermal cells to the primitive erythroid fate.
In mouse embryos, Podxl was expressed on more than 50% of GFP+ transgenic EryP at E7.5 (around the time progenitor activity is first detected using colony assays [29]) and on essentially all EryP at E8.5 and E9.5. Moreover, a large proportion of all Podxl+ cells in the embryo are EryP (~27%, 60% and ~50% at E7.5, E8.5 and E9.5, respectively). Therefore, expression of Podxl marks not only circulating EryP but also their progenitors and provides a new tool to identify the first hematopoietic cells of the embryo. Podxl has anti-adhesive properties [47] and may play a role in the mobilization of erythroid progenitors from bone marrow [48]. It is tempting to speculate that this anti-adhesion also contributes to the release of primitive erythroblasts from the yolk sac as circulation initiates in the embryo.
In contrast with the Podxl-SP subpopulation, DP cells contained essentially all of the definitive hematopoietic potential. The DP population also expressed the lateral plate mesoderm marker Twist and all hematopoietic genes tested at the highest levels. It contained a minor fraction of primitive and endothelial potential. The primitive and definitive hematopoietic lineages have previously been distinguished based on temporal patterns of Flk1+ cell emergence in EBs [49]. Here, we have separated these lineages at the same temporal stage of EB differentiation, based on cell surface markers.
Flk1-SP cells gave rise to small numbers of definitive hematopoietic cells and to endothelial cells and cardiomyocytes, while the DP population gave rise to primitive and definitive hematopoietic and endothelial cell lineages. The expression of endothelial and hematopoietic markers by Flk1-SP and DP cells was consistent with their respective developmental potentials. The Flk1-SP cells expressed high levels of genes required for endothelial development but very low levels of hematopoietic genes, were capable of Ac-LDL uptake, and generated CD31+ cells. These observations suggest that this population contains greater endothelial than hematopoietic potential. The Flk1-SP population also expressed high levels of the cardiac maker Gata4 and gave rise to contracting cTnT+ cardiomyocytes. They are likely similar to the Brachyury-GFP/Flk1+ cardiac progenitors reported previously [6, 11].
As noted in the Introduction, populations of cells expressing both Flk1 and Eng are enriched for hemangioblast activity compared with Flk1+ populations alone [15]. After submission of this work, it was reported that expression of Eng marks the first hematopoietic progenitors in the early embryo [50] and the hematopoietic and endothelial lineages can be distinguished on the basis of the levels of Eng expression at later developmental stages [51]. Interestingly, inducible overexpression of Eng may influence a switch that determines hematopoietic versus endothelial and cardiac potential [52]. It will be of interest to examine Eng expression on the various cell populations we have analyzed here. CD40 and Icam2 are expressed sequentially during the earliest stages of blood specification both in vitro and in vivo [53]. Their temporal relationship to Podxl and Eng remain to be determined.
Conclusions
Podxl expression provides a new tool for separating distinct mesodermal cell populations in differentiating EBs without the need for a transgenic reporter ES cell line and should, therefore, be translatable to human pluripotent cell systems. More precise segregation of early mesodermal cell populations with unique developmental potentials will become possible as Podxl and other markers are employed in combination and will ultimately facilitate the isolation of lineage-specific progenitors from differentiating pluripotent cells for use in regenerative medicine.
Supplementary Material
Figure S1. Clusters and patterns of gene expression in uninduced and DOX-induced EBs. A 15K developmental cDNA microarray was screened using RNAs isolated from uninduced versus DOX-induced i-Mixl1 EBs. The heat map shows expression of genes during differentiation under Dox(+) and Dox(−) conditions. Averaged expression for each time point for each cluster is shown on the right. Gene Ontology (GO) enrichment analysis revealed mesoderm formation genes (Dkk1, Nckap1, Eomes, Hmga2) in cluster #8 (see Table S3). Exploration of 458 genes for distribution of GO terms containing the words “hematopoiesis” and “blood” also suggested selective enrichment of genes associated with these terms in cluster #8 (Cdh2, Amot, Gata2, Gas6).
Figure S2. Expression of Flk1 and Podxl on cells from differentiating EBs. E14 ES cells were induced to differentiate as EBs and harvested at the days indicated. Cells were stained with anti-Flk1 and anti-Podxl1 antibodies and then subjected to flow cytometric analysis.
Figure S3. Expression of Flk1 and Podxl mRNA in differentiating EBs. E14 ES cells were induced to differentiate as EBs and harvested at the days indicated. Total RNA was isolated and analyzed using QRT-PCR. The expression of Flk1 and Podxl was normalized to that of Gapdh.
Figure S4. Flow cytometric analysis confirms purity of sorted populations. (A) Flow cytometric analysis showing subdivision of Flk1+ differentiating ES cells based on expression of Podxl in day 4.75 EBs. Boxes represent the gating schematic used to FACS sort each population. (B) Flow cytometric analysis of each population following sorting. Immediately after sorting, each population was analyzed by flow cytometry to measure sorting efficiency.
Acknowledgments
We thank Ambjuj (Butch) Upadhyay for outstanding technical assistance and Dr. Andrei Vacaru for thoughtful comments on the manuscript. This work was supported by a grant from the National Institutes of Health (RO1 HL62248) to M.H.B.
Footnotes
Disclosure of potential Conflicts of Interest: The authors declare no conflict of interest.
Author contributions: Conception and design: H.Z., S.T.F., J.I., M.H.B.; collection and assembly of data: H.Z., J.L.N., S.T.F., J.I., P.D., D.P., S.L.D’S., M.A.D.; data analysis and interpretation: H.Z., J.L.N., S.T.F., J.I., P.D., D.P., M.A.D., M.H.B.; manuscript writing: H.Z., S.T.F., M.H.B.; financial support: I.R.L., M.A.D., M.H.B.; final approval of manuscript: M.H.B.
References
- 1.Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 2.Kinder SJ, Tsang TE, Quinlan GA, et al. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- 3.Kinder SJ, Loebel DA, Tam PP. Allocation and early differentiation of cardiovascular progenitors in the mouse embryo. Trends Cardiovasc Med. 2001;11:177–184. doi: 10.1016/s1050-1738(01)00091-3. [DOI] [PubMed] [Google Scholar]
- 4.Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 5.Ema M, Takahashi S, Rossant J. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood. 2006;107:111–117. doi: 10.1182/blood-2005-05-1970. [DOI] [PubMed] [Google Scholar]
- 6.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Lugus JJ, Park C, Ma YD, et al. Both primitive and definitive blood cells are derived from Flk-1+ mesoderm. Blood. 2009;113:563–566. doi: 10.1182/blood-2008-06-162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 9.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Fehling HJ, Lacaud G, Kubo A, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 11.Kouskoff V, Lacaud G, Schwantz S, et al. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci U S A. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakurai H, Era T, Jakt LM, et al. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586. doi: 10.1634/stemcells.2005-0256. [DOI] [PubMed] [Google Scholar]
- 13.Era T, Izumi N, Hayashi M, et al. Multiple mesoderm subsets give rise to endothelial cells, whereas hematopoietic cells are differentiated only from a restricted subset in embryonic stem cell differentiation culture. Stem Cells. 2008;26:401–411. doi: 10.1634/stemcells.2006-0809. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka H, Hayashi M, Nakagawa R, et al. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- 15.Perlingeiro RC. Endoglin is required for hemangioblast and early hematopoietic development. Development. 2007;134:3041–3048. doi: 10.1242/dev.002907. [DOI] [PubMed] [Google Scholar]
- 16.Willey S, Ayuso-Sacido A, Zhang H, et al. Acceleration of mesoderm development and expansion of hematopoietic progenitors in differentiating ES cells by the mouse Mix-like homeodomain transcription factor. Blood. 2006;107:3122–3130. doi: 10.1182/blood-2005-10-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Fraser ST, Papazoglu C, et al. Transcriptional activation by the Mixl1 homeodomain protein in differentiating mouse embryonic stem cells. Stem Cells. 2009;27:2884–2895. doi: 10.1002/stem.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen JS, McNagny KM. The role of podocalyxin in health and disease. J Am Soc Nephrol. 2009;20:1669–1676. doi: 10.1681/ASN.2008070782. [DOI] [PubMed] [Google Scholar]
- 19.Doyonnas R, Kershaw DB, Duhme C, et al. Anuria, omphalocele, and perinatal lethality in mice lacking the CD34-related protein podocalyxin. J Exp Med. 2001;194:13–27. doi: 10.1084/jem.194.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyonnas R, Nielsen JS, Chelliah S, et al. Podocalyxin is a CD34-related marker of murine hematopoietic stem cells and embryonic erythroid cells. Blood. 2005;105:4170–4178. doi: 10.1182/blood-2004-10-4077. [DOI] [PubMed] [Google Scholar]
- 21.McNagny KM, Pettersson I, Rossi F, et al. Thrombomucin, a novel cell surface protein that defines thrombocytes and multipotent hematopoietic progenitors. J Cell Biol. 1997;138:1395–1407. doi: 10.1083/jcb.138.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassetti C, Tangemann K, Singer MS, et al. Identification of podocalyxin-like protein as a high endothelial venule ligand for L-selectin: parallels to CD34. J Exp Med. 1998;187:1965–1975. doi: 10.1084/jem.187.12.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T, Nakano Y, Tanaka M, et al. Identification of podocalyxin-like protein 1 as a novel cell surface marker for hemangioblasts in the murine aorta-gonad-mesonephros region. Immunity. 1999;11:567–578. doi: 10.1016/s1074-7613(00)80132-6. [DOI] [PubMed] [Google Scholar]
- 24.Horvat R, Hovorka A, Dekan G, et al. Endothelial cell membranes contain podocalyxin--the major sialoprotein of visceral glomerular epithelial cells. J Cell Biol. 1986;102:484–491. doi: 10.1083/jcb.102.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy M, Keller GM. Hematopoietic commitment of ES cells in culture. Methods Enzymol. 2003;365:39–59. doi: 10.1016/s0076-6879(03)65003-2. [DOI] [PubMed] [Google Scholar]
- 26.Fraser ST, Ogawa M, Nishikawa S. Embryonic stem cell differentiation as a model to study hematopoietic and endothelial cell development. Methods Mol Biol. 2002;185:71–81. doi: 10.1385/1-59259-241-4:71. [DOI] [PubMed] [Google Scholar]
- 27.Isern J, Fraser ST, He Z, et al. The fetal liver is a niche for maturation of primitive erythroid cells. Proc Natl Acad Sci U S A. 2008;105:6662–6667. doi: 10.1073/pnas.0802032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isern J, Fraser ST, He Z, et al. Dose-dependent regulation of primitive erythroid maturation and identity by the transcription factor Eklf. Blood. 2010;116:3972–3980. doi: 10.1182/blood-2010-04-281196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isern J, He Z, Fraser ST, et al. Single-lineage transcriptome analysis reveals key regulatory pathways in primitive erythroid progenitors in the mouse embryo. Blood. 2011;117:4924–4934. doi: 10.1182/blood-2010-10-313676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.’t Hoen PA, de Kort F, van Ommen GJ, et al. Fluorescent labelling of cRNA for microarray applications. Nucleic Acids Res. 2003;31:e20. doi: 10.1093/nar/gng020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka TS, Jaradat SA, Lim MK, et al. Genome-wide expression profiling of mid- gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron MH, Mohn D. Mouse Embryonic Explant Culture System for Analysis of Hematopoietic and Vascular Development. In: Baron MH, editor. Developmental Hematopoiesis: Methods and Protocols. Chapter 15. Totowa, NJ: Humana Press; 2005. pp. 231–256. [DOI] [PubMed] [Google Scholar]
- 33.Fraser ST, Isern J, Baron MH. Use of transgenic fluorescent reporter mouse lines to monitor hematopoietic and erythroid development during embryogenesis. In: Wasserman PaSPE., editor. Guide to techniques in Mouse Development: Methods in Enzymology. 2010. pp. 403–427. [DOI] [PubMed] [Google Scholar]
- 34.Chen VC, Stull R, Joo D, et al. Notch signaling respecifies the hemangioblast to a cardiac fate. Nat Biotechnol. 2008;26:1169–1178. doi: 10.1038/nbt.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCloskey KE, Stice SL, Nerem RM. In vitro derivation and expansion of endothelial cells from embryonic stem cells. Methods Mol Biol. 2006;330:287–301. doi: 10.1385/1-59745-036-7:287. [DOI] [PubMed] [Google Scholar]
- 36.Palis J, Robertson S, Kennedy M, et al. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 37.Mikkola HK, Fujiwara Y, Schlaeger TM, et al. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 38.Ferkowicz MJ, Starr M, Xie X, et al. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- 39.Nishikawa S-I, Nishikawa S, Hirashima M, et al. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Develop. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- 40.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo CT, Morrisey EE, Anandappa R, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes & Develop. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 42.Chi X, Chatterjee PK, Wilson W, 3rd, et al. Complex cardiac Nkx2–5 gene expression activated by noggin-sensitive enhancers followed by chamber-specific modules. Proc Natl Acad Sci U S A. 2005;102:13490–13495. doi: 10.1073/pnas.0504295102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller-Hance WC, LaCorbiere M, Fuller SJ, et al. In vitro chamber specification during embryonic stem cell cardiogenesis. Expression of the ventricular myosin light chain-2 gene is independent of heart tube formation. J Biol Chem. 1993;268:25244–25252. [PubMed] [Google Scholar]
- 44.McGrath KE, Koniski AD, Malik J, et al. Circulation is established in a stepwise pattern in the mammalian embryo. Blood. 2003;101:1669–1676. doi: 10.1182/blood-2002-08-2531. [DOI] [PubMed] [Google Scholar]
- 45.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pronk CJH, Rossi DJ, Månsson R, et al. Elucidation of the Phenotypic, Functional, and Molecular Topography of a Myeloerythroid Progenitor Cell Hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Takeda T, Go WY, Orlando RA, et al. Expression of podocalyxin inhibits cell-cell adhesion and modifies junctional properties in Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:3219–3232. doi: 10.1091/mbc.11.9.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sathyanarayana P, Menon MP, Bogacheva O, et al. Erythropoietin modulation of podocalyxin, and a proposed erythroblast niche. Blood. 2007;110:509–518. doi: 10.1182/blood-2006-11-056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irion S, Clarke RL, Luche H, et al. Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development. 2010;137:2829–2839. doi: 10.1242/dev.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borges L, Iacovino M, Mayerhofer T, et al. A critical role for endoglin in the emergence of blood during embryonic development. Blood. 2012;119:5417–5428. doi: 10.1182/blood-2011-11-391896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borges L, Iacovino M, Koyano-Nakagawa N, et al. Expression levels of Endoglin Distinctively Identify Hematopoietic and Endothelial Progeny at Different Stages of Yolk Sac Hematopoiesis. Stem Cells. 2013 doi: 10.1002/stem.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baik J, Borges L, Magli A, et al. Effect of endoglin overexpression during embryoid body development. Exp Hematol. 2012;40:837–846. doi: 10.1016/j.exphem.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearson S, Lancrin C, Lacaud G, et al. The sequential expression of CD40 and Icam2 defines progressive steps in the formation of blood precursors from the mesoderm germ layer. Stem Cells. 2010;28:1089–1098. doi: 10.1002/stem.434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Clusters and patterns of gene expression in uninduced and DOX-induced EBs. A 15K developmental cDNA microarray was screened using RNAs isolated from uninduced versus DOX-induced i-Mixl1 EBs. The heat map shows expression of genes during differentiation under Dox(+) and Dox(−) conditions. Averaged expression for each time point for each cluster is shown on the right. Gene Ontology (GO) enrichment analysis revealed mesoderm formation genes (Dkk1, Nckap1, Eomes, Hmga2) in cluster #8 (see Table S3). Exploration of 458 genes for distribution of GO terms containing the words “hematopoiesis” and “blood” also suggested selective enrichment of genes associated with these terms in cluster #8 (Cdh2, Amot, Gata2, Gas6).
Figure S2. Expression of Flk1 and Podxl on cells from differentiating EBs. E14 ES cells were induced to differentiate as EBs and harvested at the days indicated. Cells were stained with anti-Flk1 and anti-Podxl1 antibodies and then subjected to flow cytometric analysis.
Figure S3. Expression of Flk1 and Podxl mRNA in differentiating EBs. E14 ES cells were induced to differentiate as EBs and harvested at the days indicated. Total RNA was isolated and analyzed using QRT-PCR. The expression of Flk1 and Podxl was normalized to that of Gapdh.
Figure S4. Flow cytometric analysis confirms purity of sorted populations. (A) Flow cytometric analysis showing subdivision of Flk1+ differentiating ES cells based on expression of Podxl in day 4.75 EBs. Boxes represent the gating schematic used to FACS sort each population. (B) Flow cytometric analysis of each population following sorting. Immediately after sorting, each population was analyzed by flow cytometry to measure sorting efficiency.