Abstract
Background
To clarify the involvement of HIVEP3 and SOX9 coexpression in prostate cancer (PCa).
Methods
A small interfering RNA was used to knockdown SOX9 expression in a PCa cell line and to analyze the effects of SOX9 inhibition on the expression of HIVEP3 in vitro. Then, HIVEP3 and SOX9 expression patterns in the human PCa tissues were detected using quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis and immunohistochemistry.
Results
We found that the downregulation of SOX9 could inhibit the expression of HIVEP3 in the PCa cells in vitro. In addition, both HIVEP3 and SOX9 messenger RNA expression levels in the PCa tissues were significantly higher than those in the noncancerous prostate tissues (P=0.006 and P<0.001, respectively). Moreover, the immunohistochemical staining scores of HIVEP3 in the PCa tissues with PSA failure were significantly higher than those without (P=0.042); the increased SOX9 protein expression was more frequently found in the PCa tissues with a high Gleason score (P=0.045) and a high clinical stage (P=0.012). The tumors showing the HIVEP3-high/SOX9-high expression more frequently had PSA failure (P=0.024). When the patients with an HIVEP3 overexpression combined with the SOX9 overexpression, this group had a worse biochemical recurrence-free survival (P<0.001). Furthermore, the multivariate analysis showed that the HIVEP3/SOX9 coexpression was an independent predictor of an unfavorable biochemical recurrence-free survival.
Conclusion
Our data offer the convincing evidence for the first time that a combined analysis of HIVEP3 and SOX9 may help to predict the tumor progression and prognosis of PCa patients. In particular, the overexpression of HIVEP3 in PCa might partly explain the poor prognosis of patients with an upregulation of SOX9.
Keywords: prostate cancer, HIVEP3, SOX9, clinicopathological feature, biochemical recurrence-free survival
Introduction
Prostate cancer (PCa) is one of the most prevalent malignancies in men and the second most frequent cause of male cancer-related death.1 It is a clinically heterogeneous-multifocal disease, and the incidence is continuously rising. Carcinogenesis and the mechanisms influencing the progression and prognosis of PCa is a multistep process, involving both genetic insults to epithelial cells and changes in the epithelial–stromal interactions.2 The advent of prostate-specific antigen (PSA) screening has led to the earlier detection of clinically localized PCa.3 However, to date, there are no reliable predictors of PCa behavior and aggressiveness. In spite of current therapeutic methods, many patients develop metastases. Androgen suppression therapies (AST) have already been considered as the mainstay of systemic treatment for advanced PCa, especially for metastatic disease.4 Most patients respond to AST and their clinical symptoms could be relieved, although they eventually experience disease relapse and develop more aggressive tumors, commonly termed “hormone-refractory prostate cancer” or “castration-resistant prostate cancer”.5 The clinical benefits of AST for various PCa patients are different. Even among patients with metastatic PCa, for which AST is the established standard of care, the length of clinical remission induced by AST can range from months to more than 10 years.6 Since the carcinogenesis of PCa is initiated when androgen signaling induces epithelial proliferation, invasion, and bud formation, it is of great significance to clarify the precise androgen-initiated programs of early prostate growth to identify more sensitive and more specific PCa markers, which can provide valuable information for the diagnosis and treatment of the disease.
The human immunodeficiency virus type I enhancer binding protein 3 (HIVEP3), also known as KBP-1, KBP1, KRC, SHN3, Schnurri-3, ZAS3, and ZNF40C, is a member of the HIVEP family, which contains multiple zinc finger and acid-rich (ZAS) domains and serine/threonine-rich regions.7 HIVEP3 acts as a transcription factor and is able to regulate nuclear factor kappa B-mediated transcription by binding the kappa B motif in target genes.8 Recent studies have demonstrated that this protein plays a role in various processes, including the immunoglobulin (Ig) gene rearrangement, cell survival, tumor necrosis factor (TNF) signaling in macrophages, IL-2 gene expression in helper T lymphocytes, osteoblastic bone formation, and tumor formation.9–12 For example, mice with a germline deletion of HIVEP3 displayed a massive increase in bone mass, revealing an unexpected role for this protein in the skeletal system;10 in cervical cancer HeLa cells, HIVEP3 deficiency has resulted in anchorage-independent growth and promoted cell cycle progression without an intervening cytokinesis, culminating in the formation of multinucleated giant cells.11 However, the involvement of this protein in PCa carcinogenesis is still unclear.
SOX9, a member of the SOX (SRY [sex-determining region Y] box) gene superfamily, is a developmental transcription factor which plays an important role in the regulation of sex determination, cartilage development, intestinal differentiation, and adult progenitor cell pool maintenance.13 Schaeffer et al14 have reported that SOX9 may be one of the earliest molecules expressed in the primordial prostate, predating even the expression of NKX3.1, a transcription factor and classical marker of prostate lineage, suggesting the implication of SOX9 in prostate development. Similarly, Huang et al15 also determined that SOX9 may be required for prostate development and prostate cancer initiation. Accumulating studies showed the emerging role of SOX9 in cancer. Wang et al16,17 reported the important functions of SOX9 in both the development and maintenance of the normal prostate and indicated that these functions contribute to the PCa tumor growth and invasion. Our previous study reported that the SOX9 upregulation was significantly associated with aggressive tumor progression and poor prognosis in PCa, suggesting a key role of SOX9 in the carcinogenesis of PCa.18
To date, there has been no investigation of the relationship between HIVEP3 and SOX9, and the clinicopathologic relevance of combined HIVEP3 and SOX9 expression in PCa tissues. In the present study, we first used the small interfering RNA (siRNA) to knockdown SOX9 expression in a PCa cell line and to analyze the effects of SOX9 inhibition on the expression of HIVEP3 in vitro. Then, we evaluated the potential association of the coexpression of HIVEP3 and SOX9 in PCa tissues with clinicopathologic findings and postresectional survival by an immunohistochemical analysis. Our results suggest that the combined HIVEP3 and SOX9 expression can be used as an independent marker for the prognosis in patients with PCa.
Materials and methods
Patients and tissue samples
The study was approved by the Research Ethics Committee of the Central Hospital of Panyu District, Guangzhou Medical University, and Southern Medical University, Guangzhou, Guangdong, People’s Republic of China. Informed consent was obtained from all patients. All specimens were handled and made anonymous according to the ethical and legal standards.
The Guangzhou First People’s Hospital provided 26 fresh PCa tissues (approximately 52–86 years old, mean ± standard deviation [SD] =70.41±1.09 years, tumor-nodes-metastasis [TNM] staging from I to III) and paired 26 adjacent benign tissues of prostate for quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis.
For semiquantitative immunohistochemistry, a tissue microarray was provided by the Massachusetts General Hospital in Boston, MA, USA. Also, 98 PCa tissues (from patients aged approximately 37–83 years old, mean ± SD =57.83±0.746 years, TNM staging from I to III) and 26 paired adjacent benign tissues were included. All the tissues were obtained immediately during the operation of the transurethral resection prostate and suprapubic prostatectomy. None of the patients recruited in this study had chemotherapy or radiotherapy before the surgery. The pathological diagnosis was performed preoperatively and confirmed postoperatively. All patients were reviewed, and all specimens were reexamined in March 2010. The TNM grade and the Gleason score were examined by the same group of two senior pathologists who were experienced in PCa diagnosis. Detailed information on the clinical features of all patients in this study is shown in Table 1. The patients were given a follow-up exam, ranging from 3–12 years. For the analysis of survival and follow-up, the date of the prostatectomy was used to represent the beginning of the follow-up period. The primary analysis endpoint was biochemical recurrence-free survival. Others analysis endpoints were overall survival and metastasis-free survival. Patients who received adjuvant or neoadjuvant hormonal or radiation treatment prior to cancer recurrence were excluded.
Table 1.
Clinical features of all patients
| Sample type and clinical features | Experiment type (cases)
|
|
|---|---|---|
| qRT-PCR | IHC | |
| Prostate cancer | 26 | 98 |
| Mean age (years) | ||
| <60 | 20 | 62 |
| ≥60 | 6 | 36 |
| Serum PSA levels (ng/mL) | ||
| <4 | 5 | 16 |
| ≥4 | 21 | 82 |
| Gleason score | ||
| <8 | 24 | 87 |
| ≥8 | 2 | 11 |
| Clinical stage | ||
| <T2A | 17 | 59 |
| ≥T2A | 9 | 39 |
| Metastasis | 0 | 5 |
| Adjacent benign prostate tissue | 26 | 26 |
Abbreviations: qRT-PCR, quantitative reverse transcription polymerase chain reaction; IHC, immunohistochemistry; PSA, prostate-specific antigen.
Cell culture
The lymph node carcinoma of the prostate (LNCaP), the human prostate carcinoma cell line, was purchased from the American Type Culture Collection (Manassas, VA, USA) and was cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (#SH30027.01, HyClone™, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (#10438-026, Gibco® = product line of Life Technologies, Carlsbad, CA, USA), 2 mM L-glutamine, and antibiotics. All cell lines were maintained at 37°C in a humidified chamber supplemented with 5% CO2.
RNA interference and cell transfection
The LNCaP cells were seeded into 6-well plates and incubated overnight, and then they were transfected using Lipofectamine® 2000 (Life Technologies, Carlsbad CA, USA), according to the manufacturer’s instructions. The siRNA against SOX9 (siSOX9) and the control siRNA were synthesized by the Shanghai Invitrogen Biotechnology Co, Ltd Shanghai, People’s Republic of China. For every 105 cells, 0.5 μg SOX9 siRNA or control siRNA was diluted and mixed with 3 μL transfection reagent. After mixing and incubating for 30 minutes, the transfection mixture was added to the cells. After 6 hours, the medium was changed to growth medium. The LNCaP cells were used for functional assay 24 hours after transfection.
qRT-PCR
The quantitative RT-PCR was used to examine the expression status of HIVEP3 and SOX9 messenger RNA (mRNA) in the LNCaP cells with and without the siSOX9 transfection (LNCaP-S and LNCaP-NC cells), in 26 PCa tissues, and in the paired 26 adjacent benign tissues.
The cDNA (complementary DNA) templates for qRT-PCR were synthesized from RNA samples. The primers 5′-AGC GAA CGC ACA TCA AGA C-3′ and 5′-GCT GTA GTG TGG GAG GTT GAA-3′ were used to amplify 110 base pair (bp) transcripts of SOX9. The primers 5′-TTTTGGCAGGATTCGAGGCA-3′ and 5′-GAAGGCACCACTTCCGATGA-3′ were used to amplify the 201-bp transcripts of HIVEP3. The primers 5′-GGT GGC TTT TAG GAT GGC AAG-3′ and 5′-ACT GGA ACG GTG AAG GTG ACA G-3′ were used to amplify the 161-bp transcripts of β-actin. Gene expression was determined using the SYBR Green® Realtime PCR Master Mix (#QPK-201, Toyobo Co, Ltd Osaka, Japan) and 10 μg of template. qRT-PCR was performed on a MyiQ.2 Two-Color Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA), using the following amplification conditions: 5 minutes at 95°C, followed by 40 cycles of 10 seconds at 95°C; 40 cycles of 20 seconds at 60°C; and 40 cycles of 20 seconds at 72°C. All assays were carried out in triplicate. The cycle threshold values were determined using the IQ5™ Optical System Software version 2 (Bio-Rad). Gene expression in each sample was normalized with the housekeeping gene (β-actin) expression. The relative quantification of target gene expression was evaluated using the comparative cycle threshold method.
Immunohistochemistry analysis
The specimens were fixed in 10% neutral buffered formalin (MP Biomedicals, Solon, OH, USA) and subsequently embedded in paraffin. The paraffin-embedded tissues were cut at 3 μm and then deparaffinized with xylene and rehydrated for further hematoxylin and eosin or peroxidase–diaminobenzidine immunohistochemistry staining, employing the EnVision™ System (Dako, Carpinteria, CA, USA). Briefly, following a brief proteolytic digestion and a peroxidase blocking of tissue slides, the slides were incubated overnight with the primary antibodies against respective target proteins (SOX9 antibody, [SC0093, Santa Cruz Biotechnology, Inc, Dallas, TX, USA]; HIVEP3 antibody, [PAB21918, Abnova, Taipei, Taiwan]) at a dilution of 1:100 at 4°C. After washing, the peroxidase-labeled polymer and substrate-chromogen were then employed to visualize the staining of the interested proteins. In each immunohistochemistry run, negative controls were carried out by omitting the primary antibody; whereas, the overexpression of respective target proteins confirmed by Western blotting were used as positive controls.
Following a hematoxylin counterstaining, immunostaining was scored by two independent experienced pathologists, who were blinded to the clinicopathological data and clinical outcomes of the patients. The scores of the two pathologists were compared, and any discrepant scores were scored through reexamining the staining by both pathologists to achieve a consensus score. The number of positive-staining cells in ten representative microscopic fields was counted and the percentage of positive cells was calculated. Given the homogenicity of the staining of the target proteins, the tumor specimens were scored in a semiquantitative manner. The percentage scoring of immunoreactive tumor cells was as follows: 0 (0%); 1 (1%–10%); 2 (11%–50%); and 3 (>50%). The staining intensity was visually scored and stratified as follows: 0 (negative); 1 (weak); 2 (moderate); and 3 (strong). A final immunoreactivity score (IRS) was obtained for each case by multiplying the percentage and the intensity score. Protein expression levels were further analyzed by classifying IRS values as low and as high, based on a median of IRS value.
Statistical analysis
The software programs SPSS version 13.0 for Windows (IBM Corporation, Armonk, NY, USA) and SAS 9.1 (SAS Institute, Cary, NC, USA) were used for statistical analysis. Continuous variables were expressed as Statistical analyses were performed with Fisher’s exact test for any 2×2 tables, the Pearson’s chi-square test for non-2×2 tables, the Kaplan–Meier method for the question of survival, the Chiquest trend test for ordinal data, and the Cox regression analysis for the multivariate analysis. The Spearman correlation was calculated between the expression levels of HIVEP3 and SOX9 in PCa tissues. Differences were considered statistically significant when P<0.05.
Results
Downregulation of SOX9 expression by siRNA interference inhibits HIVEP3 expression in PCa cells
To clarify the relationship between HIVEP3 and SOX9, the siRNA interference technique was used to knockdown SOX9 expression in PCa cell line LNCaP. The qRT-PCR assay showed that the expression level of SOX9 mRNA was obviously downregulated in LNCaP-S cells compared with LNCaP-NC cells (P<0.001, Figure 1). In addition, the expression level of HIVEP3 in LNCaP-S cells with the inhibition of SOX9 by SOX9 siRNA was significantly lower than that treated by control siRNA (LNCaP-S versus LNCaP-NC, P<0.001, Figure 1), suggesting that the HIVEP3 expression in the PCa cells was closely related to the SOX9 expression.
Figure 1.
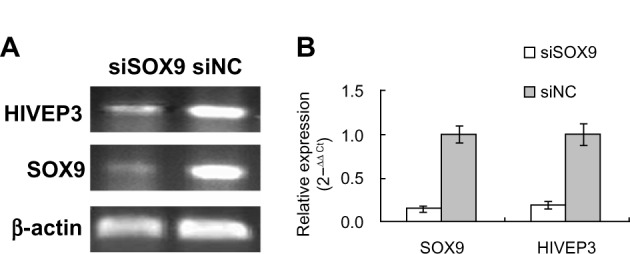
Downregulation of SOX9 expression by siRNA interference inhibits HIVEP3 expression in PCa cells.
Notes: (A) Electrophoresis of qRT-PCR production. (B) Relative expression of HIVEP3 and SOX9 in PCa cells.
Abbreviations: PCa, prostate cancer; qRT-PCR, quantitative reverse transcription polymerase chain reaction; siRNA, small interfering RNA; siSOX9, siRNA for SOX9; siNC, normal control siRNA.
HIVEP3 and SOX9 mRNA and protein expression in PCa and adjacent benign prostate tissues
We examined the HIVEP3 and SOX9 mRNA expression in 26 PCa tissues relative to their pair-matched adjacent benign prostate tissues using qRT-PCR and found that both HIVEP3 and SOX9 mRNA expression in PCa tissues was significantly higher than that in adjacent benign prostate tissues (P=0.006 and P<0.001, respectively, Figure 2).
Figure 2.
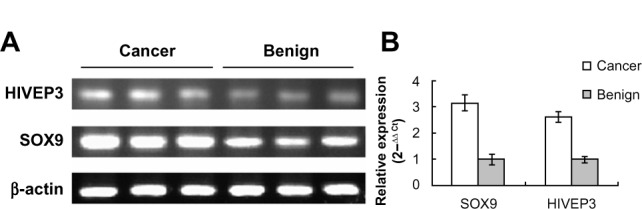
HIVEP3 and SOX9 mRNA expression in PCa and adjacent benign prostate tissues.
Notes: (A) Electrophoresis of qRT-PCR production. (B) Relative expression of HIVEP3 and SOX9 in tissue.
Abbreviations: mRNA, messenger RNA; qRT-PCR, quantitative reverse transcription polymerase chain reaction; PCa, prostate cancer.
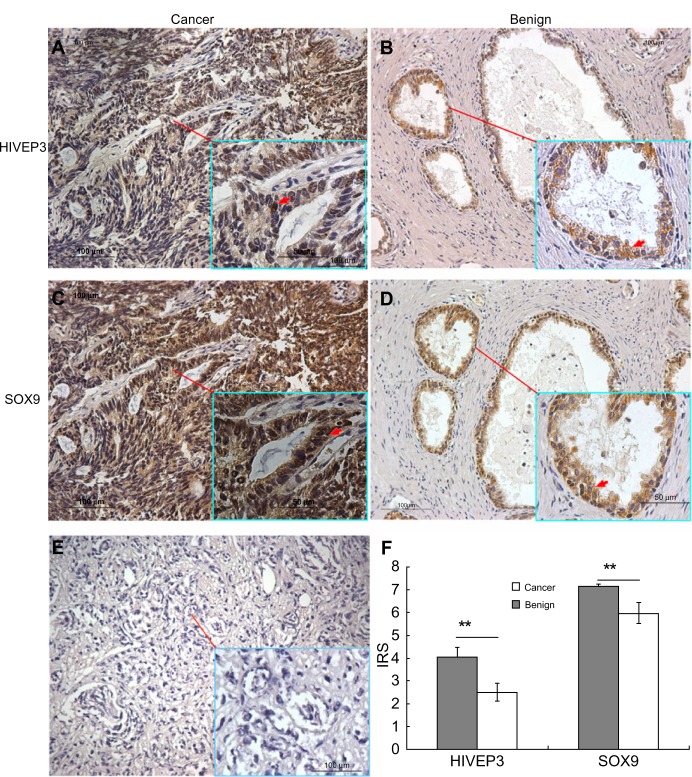
Then, the expression and localization of HIVEP3 and SOX9 proteins in 98 PCa and 26 adjacent benign prostate tissues were examined using immunohistochemical analysis. The immunohistochemical staining of HIVEP3 in PCa tissues was localized in cell membranes and cytoplasm, while the SOX9 protein was localized in the nucleus of the tumor cells in the PCa tissues (Figure 3A and B). Consistent with the data of the qRT-PCR assay, both the expression levels of HIVEP3 (IRS, PCa =4.05±0.41 versus benign =3.49±0.38, P=0.006) and SOX9 (IRS, PCa =7.16±0.041, versus benign =5.96±0.461, P<0.001) in the PCa tissues were significantly higher than those in the adjacent benign prostate tissues (Figure 3C). Of the 98 PCa patients investigated, 49 cases (50.0%) were regarded as the HIVEP3-high expression group, and 49 cases (50.0%) were regarded as the SOX9-high expression group, based on the median value of the IRS. The Spearman rank correlation analysis showed that the expression level of the HIVEP3 protein in the PCa tissues had a significantly positive correlation with that of SOX9 protein (Spearman correlation coefficient [rs] =0.51, P<0.001, Table 2).
Figure 3.
Immunohistochemical staining for HIVEP3 and SOX9 proteins in PCa and adjacent benign prostate tissues.
Notes: (A) HIVEP3 strongly positive staining was found in the cytoplasm and nucleus of the PCa cell; (B) HIVEP3 weakly positive staining in the PCa was found in the cytoplasm and the nucleus of adjacent benign prostate gland cell; (C) SOX9 strongly positive staining was found in the cytoplasm and the nucleus of the PCa cell; (D) SOX9 moderately positive staining in the PCa was found in the cytoplasm and the nucleus of the adjacent benign prostate gland cell; (E) Negative immunostaining of negative controls with the primary antibody omitted in the PCa tissues; (F) Immunoreactivity scores (IRS) of SOX9 and HIVEP3 in PCa and adjacent benign prostate tissues. Inset areas in A–E is 400X magnification. The arrows in A–D show positive staining cells. **P<0.001 in comparison between PCa and adjacent benign prostate tissues.
Abbreviations: PCa, prostate cancer; IRS, immunoreactivity scores.
Table 2.
Correlation between HIVEP3 and SOX9 expression in prostate cancer samples
| HIVEP3 expression | SOX9 expression
|
rs | P-value | |
|---|---|---|---|---|
| High (49) | Low (49) | |||
| High (49) | 37 | 12 | 0.51 | <0.001 |
| Low (49) | 12 | 37 | ||
Abbreviation: rs, Spearman correlation coefficient.
Association of HIVEP3 and SOX9 protein expression with the clinicopathological characteristics of PCa
The association of HIVEP3 and SOX9 protein expression with the clinicopathological features of PCa patients is shown in Table 3. The immunohistochemical staining scores of HIVEP3 in the PCa tissues with PSA failure were significantly higher than those without (P=0.042); the increased SOX9 protein expression was frequently found in the PCa tissues with a higher Gleason score (P=0.045) and a higher clinical stage (P=0.012). Moreover, we evaluated the association of the combined expression of HIVEP3 and SOX9, with the clinicopathological characteristics of PCa. Results as shown in Table 4 indicated that tumors showing HIVEP3-high/SOX9-high expression more frequently had PSA failure (P=0.024).
Table 3.
Association of HIVEP3 and SOX9 protein expression with the conventional clinicopathological characteristics in 98 patients with prostate cancer
| Clinical features | Case number | HIVEP3
|
SOX9
|
||
|---|---|---|---|---|---|
| P-value | P-value | ||||
| Age (years) | |||||
| <60 | 62 | 3.63±0.77 | 0.952 | 7.32±0.98 | 0.528 |
| ≥60 | 36 | 3.64±0.83 | 7.19±1.00 | ||
| Serum PSA Levels (ng/mL) | |||||
| <4 | 16 | 3.82±0.92 | 0.318 | 7.52±1.08 | 0.293 |
| ≥4 | 82 | 3.60±0.76 | 7.24±0.96 | ||
| Gleason score | |||||
| <8 | 86 | 3.66±0.79 | 0.189 | 7.13±0.67 | 0.045 |
| ≥8 | 12 | 3.33±0.72 | 8.26±0.82 | ||
| Clinical stage | |||||
| <T2A | 59 | 3.53±0.77 | 0.110 | 6.64±0.71 | 0.012 |
| ≥T2A | 39 | 3.79±0.80 | 7.98±0.88 | ||
| Metastasis | |||||
| No | 93 | 3.64±0.79 | 0.494 | 7.29±0.97 | 0.573 |
| Yes | 5 | 3.39±0.86 | 6.91 ±1.38 | ||
| PSA failure | |||||
| No | 80 | 3.41±0.41 | 0.042 | 7.08±0.97 | 0.001 |
| Yes | 18 | 5.51±0.38 | 8.31 ±0.79 | ||
Note: Bold P-values are statistically significant.
Abbreviation: PSA, prostate-specific antigen.
Table 4.
Association of combined expression of HIVEP3 and SOX9 protein with the conventional clinicopathological characteristics in 98 patients with prostate cancer
| Clinical features | Case number | HIVEP3 and SOX9 expression (n)
|
P-value | ||
|---|---|---|---|---|---|
| HIVEP3(−) SOX9(−) | HIVEP3(+) SOX9(+) | HIVEP3(+) SOX9(−)/HIVEP3(−) SOX9(+) | |||
| Age (years) | |||||
| <60 | 60 | 23 | 20 | 17 | 0.490 |
| ≥60 | 38 | 15 | 16 | 7 | |
| Serum PSA levels (ng/mL) | |||||
| <4 | 12 | 5 | 4 | 3 | 0.938 |
| ≥4 | 86 | 32 | 33 | 21 | |
| Gleason score | |||||
| <8 | 87 | 35 | 31 | 21 | 0.329 |
| ≥8 | 11 | 2 | 6 | 3 | |
| Clinical stage | |||||
| <T2A | 59 | 19 | 28 | 12 | 0.051 |
| ≥T2A | 39 | 18 | 9 | 12 | |
| Metastasis | |||||
| No | 93 | 35 | 34 | 24 | 0.370 |
| Yes | 5 | 2 | 3 | 0 | |
| PSA failure | |||||
| No | 80 | 35 | 26 | 19 | 0.024 |
| Yes | 18 | 2 | 11 | 5 | |
Note: Bold P-values are statistically significant.
Abbreviation: PSA, prostate-specific antigen.
Prognostic implications of HIVEP3 and SOX9 protein expression in PCa
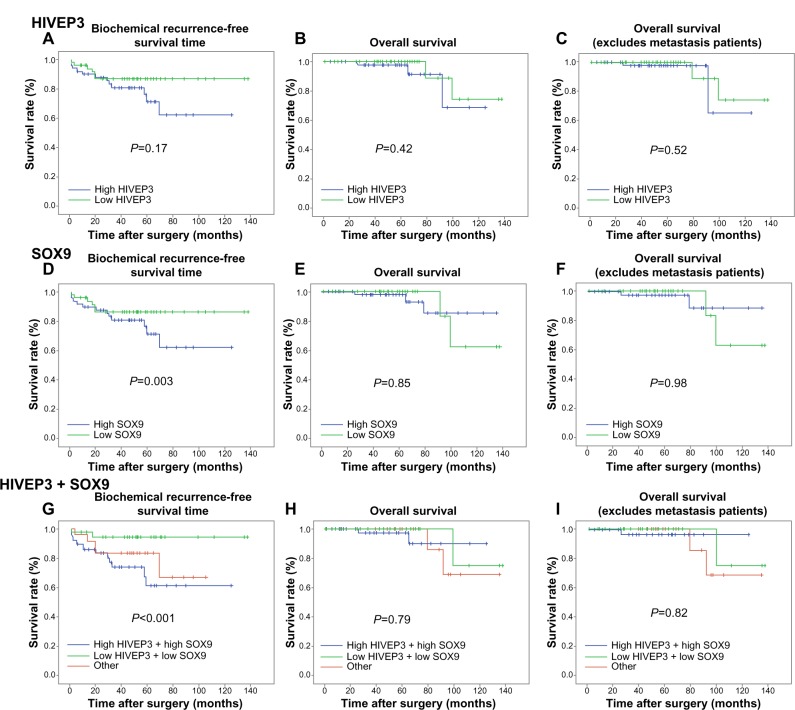
The association of the expression levels of HIVEP3 and SOX9 proteins with the biochemical recurrence-free survival, overall survival, and metastasis-free survival of PCa patients was analyzed using the Kaplan–Meier method (Figure 4). The data indicated that there were no significant differences in the biochemical recurrence-free survival, overall survival, and metastasis-free survival between the high expression group and the low expression group of HIVEP3 protein (all P>0.05 Figure 4A–C).
Figure 4.
Kaplan–Meier survival curves of the biochemical recurrence-free survival and overall survival.
Notes: Represents the Kaplan–Meier survival curves of the biochemical recurrence-free survival and overall survival for HIVEP3 (A–C, respectively), SOX9 (D–F, respectively), combined HIVEP3 and SOX9 (G–I, respectively) expression in PCa.
Abbreviation: PCa, prostate cancer.
In addition, the results by pairwise comparisons showed that there was a significant difference in the biochemical recurrence-free survival rates between patients with the high SOX9 expression and the low SOX9 expression (P=0.003, Figure 4D), but not in the overall survival and the metastasis-free survival (both P>0.05, Figure 4E and F). Interestingly, patients with HIVEP3-high/SOX9-high expression were likely to be with significantly shorter biochemical recurrence-free survival (P<0.001) than other groups (Figure 4G), but not in the overall survival and metastasis-free survival (both P>0.05, Figure 4H and I).
To further evaluate the prognosis value of HIVEP3 and/or SOX9 for biochemical recurrence-free survival of the PCa patients, a multivariate analysis was performed. Results, as shown in Table 5, suggested that: SOX9 (hazard ratio [HR] 0.276, 95% confidence interval [CI], 0.084 to approximately 0.906, P=0.034); HIVEP3/SOX9 (HR 1.86, 95% CI, 1.01 to approximately 3.62, P=0.008); Gleason score (HR 2.64, 95% CI, 1.61–4.32, P<0.001); and preoperative PSA (HR 1.02, 95% CI, 1.00–1.04, P=0.02) were also independent predictors for the biochemical recurrence-free survival of PCa patients.
Table 5.
Univariate and multivariate analyses of different prognostic factors in 98 patients with prostate cancer
| Hazard ratio (95% CI) | P-value | ||
|---|---|---|---|
| Univariate | |||
| HIVEP3 | 1.34 (0.08–2.37) | 0.84 | |
| SOX9 | 0.15 (0.03–0.84) | 0.032 | |
| HIVEP3/SOX9 | 2.58 (1.36–4.96) | <0.001 | |
| Gleason score | 2.27 (1.22–4.24) | 0.010 | |
| Preoperative PSA | 1.02 (1.00–1.04) | 0.044 | |
| Clinical stage | 0.87 (0.27–2.79) | 0.82 | |
| Age | 0.95 (0.88–1.02) | 0.15 | |
| Multivariate | |||
| SOX9 | 0.28 (0.08–0.91) | 0.034 | |
| HIVEP3/SOX9 | 1.86 (1.01–3.62) | 0.008 | |
| Gleason score | 2.64 (1.61–4.32) | <0.001 | |
| Preoperative PSA | 1.02 (1.00–1.04) | 0.02 | |
Note: Bold P-values are statistically significant.
Abbreviations: CI, confidence interval; PSA, prostate-specific antigen.
Discussion
Since PCa is the most frequently occurring cancer in men and advanced metastatic PCa is currently incurable, it is of great significance to identify novel molecular markers that could improve the tumor classification and prognostic stratification of PCa. The four main findings of the present study are: 1) in an in vitro system, the downregulation of SOX9 by specific siRNA could significantly inhibit the expression of HIVEP3 in the PCa cell line; 2) the overexpression of SOX9 and HIVEP3 mRNA and proteins in the PCa tissues was further confirmed by qRT-PCR and immunohistochemistry analysis; 3) the expression levels of SOX9 and HIVEP3 in the PCa tissues were related to the severity of the tumor malignancy; and 4) the upregulation of SOX9 alone and the combined expression of HIVEP3/SOX9 were both independent predictors of shorter biochemical recurrence-free survival of patients with PCa.
Interestingly, the prognostic significance of the HIVEP3/SOX9 coexpression was more conspicuous than the SOX9 expression alone (P<0.001 versus P<0.003, respectively). These results suggest that the expression of HIVEP3 in the PCa cells may be closely related with that of SOX9. The two proteins may play important roles in the pathogenesis and aggressiveness of PCa, and the HIVEP3/SOX9 coexpression especially may be associated with the poor prognosis in PCa.
The four main functions of the SOX gene family are:19–22 1) SOX genes regulate specification and differentiation of many cell types, such as neurogenesis, neural crest development, chondrogenesis, male sex gonad or respiratory epithelium development, melanocyte differentiation, and the differentiation of Paneth cells in the gut; 2) SOX genes within the same subgroup often share functional roles; 3) SOX genes within the same subgroup can counteract the function of genes in another subgroup; and 4) the same SOX gene can mediate different stages of development in one cell type and/or developmental processes in more than one cell type. Based on the phylogenetic analysis of their high mobility group domains, the SOX genes can be separated into the subgroups A–J, A–H which are represented in mice and humans.23 Because they are expressed in many tissues, it is not surprising that the SOX genes are implicated in the etiology of many diseases and certain cancers. SOX9, together with SOX8 and SOX10, belongs to the SOX subgroup E.24,25 It is a downstream effector of SRY, which – in turn – is dependent on the activity of androgens and the androgen receptor. There have been several studies on the involvement of SOX9 in PCa. In 2004, Drivdahl et al26 found that the elevated expression of SOX9 in the PCa cell lines resulted in a decreased rate of cellular proliferation, cell cycle arrest in G0/G1, and an increased sensitivity to apoptosis. In 2007 and 2008, Wang et al27,28 also demonstrated that SOX9 was expressed in the PCa cells and was increased in relapsed hormone refractory PCa. In 2010, Thomsen et al29 identified SOX9 as part of a developmental pathway that is reactivated in prostate neoplasia where it promotes tumor cell proliferation. In 2012, Huang et al30 further confirmed that SOX9 may be required for prostate development and prostate cancer initiation. In line with the previous studies of our and other research groups, we here found that the upregulation of SOX9 was significantly associated with aggressive progression and poor prognosis of PCa patients.
HIVEP3 belongs to the HIVEP gene family, which encodes unusually large proteins containing four to five zinc fingers.7 Each HIVEP protein contains two DNA-binding ZAS domains that are separated widely apart in the primary amino acid sequence. A ZAS domain is a composite structure containing two consecutive zinc fingers, followed by an acidic domain and a serine/threonine-rich region.8 Similar with SOX9, HIVEP3 also is a transcription factor and plays a role in and in the cell cycle. Allen et al31 have observed that anomalies and neoplasia growth in animals and cells deficient in HIVEP3, and other studies also found that the allelic loss, occurring at the chromosomal region of the human HIVEP3 counterpart in various tumors, suggested the tumor-suppressive function of HIVEP3. In contrast, our study showed that the HIVEP3 mRNA and protein were highly expressed in the PCa tissues and that both are associated with PSA failure status, indicating that the HIVEP3 may act as an oncogene in PCa. More interestingly, we found that the expression of HIVEP3 was significantly associated with that of SOX9. In vitro cell experiment, the loss of SOX9 also could inhibit the expression of HIVEP3, implying the relationship between them.
An accurate prediction of the probability of disease recurrence is essential for the proper therapy selection of PCa tissues. Thus, we investigated the prognostic significances of HIVEP3 and SOX9 in the present study. Our results revealed that the expression of SOX9 and/or HIVEP3 in PCa correlated with biochemical recurrence-free survival. However, the HIVEP3 overexpression alone was not associated with a prognosis in PCa patients, which is worthy of further validation using different cohorts.
In conclusion, our data offer the convincing evidence for the first time that the combined analysis of HIVEP3 and SOX9 may help to predict the tumor progression and prognosis of PCa patients. In particular, the overexpression of HIVEP3 in PCa might partly explain the poor prognosis of patients with an upregulation of SOX9. The exact roles of the two proteins in accelerating the development of PCa and promoting tumor progression need further clarification.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81170699, 81272813, 81200550), Science and Technology Research Project of Guangdong Province (2012B031800008), Guangdong Provincial Medical Research Fund (A2012489), Guangzhou Municipal Science and Technology Key Project (11C23150711), Key Projects of Bureau of Health in Guangzhou Municipality (201102A212015), and Projects of Guangdong Key Laboratory of Urology (2010A060801016).
Author contributions
Guo-qiang Qin, Hui-chan He, Zhao-dong Han, and Wei-de Zhong have made substantial contributions to the conception and design, acquisition of the data, analysis and interpretation of the data, and have been involved in drafting the manuscript. Yu-xiang Liang, Sheng-bang Yang, Ya-qiang Huang, Liang Zhou, Hao Fu, Jie-xian Li, and Fu-neng Jiang have made substantial contributions to the acquisition of the data, the analysis, and the interpretation of the data. All authors made significant contributions to the critical revision of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong AJ, Eisenberger MA, Halabi S, et al. Biomarkers in the management and treatment of men with metastatic castration-resistant prostate cancer. Eur Urol. 2012;61(3):549–559. doi: 10.1016/j.eururo.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lumen N, Fonteyne V, De Meerleer G, et al. Screening and early diagnosis of prostate cancer: an update. Acta Clin Belg. 2012;67(4):270–275. doi: 10.2143/ACB.67.4.2062671. [DOI] [PubMed] [Google Scholar]
- 4.Klotz L. Hormone therapy for patients with prostate carcinoma. Cancer. 2000;88(Suppl 12):3009–3014. doi: 10.1002/1097-0142(20000615)88:12+<3009::aid-cncr17>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Hu R, Denmeade SR, Luo J. Molecular processes leading to aberrant androgen receptor signaling and castration resistance in prostate cancer. Expert Rev Endocrinol Metab. 2010;5(5):753–764. doi: 10.1586/eem.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenberger MA, Blumenstein BA, Crawford ED, et al. Bilateral orchiectomy with or without flutamide for metastatic prostate cancer. N Engl J Med. 1998;339(15):1036–1042. doi: 10.1056/NEJM199810083391504. [DOI] [PubMed] [Google Scholar]
- 7.Hicar MD, Robinson ML, Wu LC. Embryonic expression and regulation of the large zinc finger protein KRC. Genesis. 2002;33(1):8–20. doi: 10.1002/gene.10084. [DOI] [PubMed] [Google Scholar]
- 8.Allen CE, Mak CH, Wu LC. The kappa B transcriptional enhancer motif and signal sequences of V(D)J recombination are targets for the zinc finger protein HIVEP3/KRC: a site selection amplification binding study. BMC Immunol. 2002;3:10. doi: 10.1186/1471-2172-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oukka M, Wein MN, Glimcher LH. Schnurri-3 (KRC) interacts with c-Jun to regulate the IL-2 gene in T cells. J Exp Med. 2004;199(1):15–24. doi: 10.1084/jem.20030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wein MN, Jones DC, Shim JH, et al. Control of bone resorption in mice by Schnurri-3. Proc Natl Acad Sci U S A. 2012;109(21):8173–8178. doi: 10.1073/pnas.1205848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen CE, Wu LC. Downregulation of KRC induces proliferation, anchorage independence, and mitotic cell death in HeLa cells. Exp Cell Res. 2000;260(2):346–356. doi: 10.1006/excr.2000.5029. [DOI] [PubMed] [Google Scholar]
- 12.Oukka M, Kim ST, Lugo G, Sun J, Wu LC, Glimcher LH. A mammalian homolog of Drosophila schnurri, KRC, regulates TNF receptor-driven responses and interacts with TRAF2. Mol Cell. 2002;9(1):121–131. doi: 10.1016/s1097-2765(01)00434-8. [DOI] [PubMed] [Google Scholar]
- 13.Endo Y, Deonauth K, Prahalad P, Hoxter B, Zhu Y, Byers SW. Role of Sox-9, ER81 and VE-cadherin in retinoic acid-mediated trans-differentiation of breast cancer cells. PLoS One. 2008;3(7):e2714. doi: 10.1371/journal.pone.0002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer EM, Marchionni L, Huang Z, et al. Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene. 2008;27(57):7180–7191. doi: 10.1038/onc.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Z, Hurley PJ, Simons BW, et al. Schaeffer EM. Sox9 is required for prostate development and prostate cancer initiation. Oncotarget. 2012;3(6):651–663. doi: 10.18632/oncotarget.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Leav I, Ibaragi S, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68(6):1625–1630. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67(2):528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 18.Zhong WD, Qin GQ, Dai QS, et al. SOXs in human prostate cancer: implication as progression and prognosis factors. BMC Cancer. 2012;12:248. doi: 10.1186/1471-2407-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samant GV, Schupp MO, François M, et al. Sox factors transcriptionally regulate ROBO4 gene expression in developing vasculature in zebrafish. J Biol Chem. 2011;286(35):30740–30747. doi: 10.1074/jbc.M111.220665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui J, Shen X, Zhao H, Nagahama Y. Genome-wide analysis of Sox genes in Medaka (Oryzias latipes) and their expression pattern in embryonic development. Cytogenet Genome Res. 2011;134(4):283–294. doi: 10.1159/000329480. [DOI] [PubMed] [Google Scholar]
- 21.Gracz AD, Magness ST. Sry-box (Sox) transcription factors in gastrointestinal physiology and disease. Am J Physiol Gastrointest Liver Physiol. 2011;300(4):G503–G515. doi: 10.1152/ajpgi.00489.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei L, Cheng D, Li D, et al. Identification and characterization of Sox genes in the silkworm, Bombyx mori. Mol Biol Rep. 2011;38(5):3573–3584. doi: 10.1007/s11033-010-0468-5. [DOI] [PubMed] [Google Scholar]
- 23.Jakob S, Lovell-Badge R. Sex determination and the control of Sox9 expression in mammals. FEBS J. 2011;278(7):1002–1009. doi: 10.1111/j.1742-4658.2011.08029.x. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchiya M, Ogawa H, Suzuki T, Sugiyama N, Haraguchi T, Hiraoka Y. Exportin 4 interacts with Sox9 through the HMG Box and inhibits the DNA binding of Sox9. PLoS One. 2011;6(10):e25694. doi: 10.1371/journal.pone.0025694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sashikawa Kimura M, Mutoh H, Sugano K. SOX9 is expressed in normal stomach, intestinal metaplasia, and gastric carcinoma in humans. J Gastroenterol. 2011;46(11):1292–1299. doi: 10.1007/s00535-011-0443-5. [DOI] [PubMed] [Google Scholar]
- 26.Drivdahl R, Haugk KH, Sprenger CC, Nelson PS, Tennant MK, Plymate SR. Suppression of growth and tumorigenicity in the prostate tumor cell line M12 by overexpression of the transcription factor SOX9. Oncogene. 2004;23(26):4584–4593. doi: 10.1038/sj.onc.1207603. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67(2):528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Leav I, Ibaragi S, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68(6):1625–1630. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- 29.Thomsen MK, Ambroisine L, Wynn S, et al. Transatlantic Prostate Group SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010;70(3):979–987. doi: 10.1158/0008-5472.CAN-09-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z, Hurley PJ, Simons BW, et al. Sox9 is required for prostate development and prostate cancer initiation. Oncotarget. 2012;3(6):651–663. doi: 10.18632/oncotarget.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen CE, Muthusamy N, Weisbrode SE, Hong JW, Wu LC. Developmental anomalies and neoplasia in animals and cells deficient in the large zinc finger protein KRC. Genes Chromosomes Cancer. 2002;35(4):287–298. doi: 10.1002/gcc.10128. [DOI] [PubMed] [Google Scholar]