Abstract
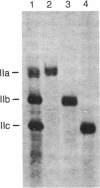
Purified eukaryotic nuclear RNA polymerase II consists of three subspecies that differ in the apparent molecular masses of their largest subunit, designated IIo, IIa, and IIb for polymerase species IIO, IIA, and IIB, respectively. Subunits IIo, IIa, and IIb are the products of a single gene. We present here the amino acid composition of calf thymus subunits IIa and IIb and the C-terminal amino acid sequence of subunit IIa (IIo) inferred from the nucleotide sequence of part of the mouse gene encoding this RNA polymerase subunit. The calculated amino acid composition of the peptide unique to subunit IIa indicates that subunit IIa contains a domain rich in serine, proline, threonine, and tyrosine. The sequence at the 3' end of the mouse RNA polymerase II largest subunit gene reveals that the C-terminal domain consists of 52 repeats of a seven amino acid block with the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser. This sequence is also unusual in that it contains a high percentage of potential phosphorylation sites.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bell G. I., Valenzuela P., Rutter W. J. Phosphorylation of yeast DNA-dependent RNA polymerases in vivo and in vitro. Isolation of enzymes and identification of phosphorylated subunits. J Biol Chem. 1977 May 10;252(9):3082–3091. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buhler J. M., Huet J., Davies K. E., Sentenac A., Fromageot P. Immunological studies of yeast nuclear RNA polymerases at the subunit level. J Biol Chem. 1980 Oct 25;255(20):9949–9954. [PubMed] [Google Scholar]
- Buhler J. M., Iborra F., Sentenac A., Fromageot P. The presence of phosphorylated subunits in yeast RNA polymerases A and B. FEBS Lett. 1976 Nov 15;72(1):37–41. doi: 10.1016/0014-5793(76)80893-9. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Stollar B. D. Conservation of a DNA-binding site in the largest subunit of eukaryotic RNA polymerase II. J Mol Biol. 1983 Nov 5;170(3):777–790. doi: 10.1016/s0022-2836(83)80131-4. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Christmann J. L., Dahmus M. E. Monoclonal antibody specific for calf thymus RNA polymerases IIO and IIA. J Biol Chem. 1981 Nov 25;256(22):11798–11803. [PubMed] [Google Scholar]
- Dahmus M. E., Kedinger C. Transcription of adenovirus-2 major late promoter inhibited by monoclonal antibody directed against RNA polymerases IIO and IIA. J Biol Chem. 1983 Feb 25;258(4):2303–2307. [PubMed] [Google Scholar]
- Dahmus M. E. Phosphorylation of eukaryotic DNA-dependent RNA polymerase. Identification of calf thymus RNA polymerase subunits phosphorylated by two purified protein kinases, correlation with in vivo sites of phosphorylation in HeLa cell RNA polymerase II. J Biol Chem. 1981 Apr 10;256(7):3332–3339. [PubMed] [Google Scholar]
- Dahmus M. E. Structural relationship between the large subunits of calf thymus RNA polymerase II. J Biol Chem. 1983 Mar 25;258(6):3956–3960. [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980 May 10;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf A. L. Amanitin-resistant RNA polymerase II mutations are in the enzyme's largest subunit. J Biol Chem. 1983 Nov 25;258(22):13403–13406. [PubMed] [Google Scholar]
- Greenleaf A. L., Borsett L. M., Jiamachello P. F., Coulter D. E. Alpha-amanitin-resistant D. melanogaster with an altered RNA polymerase II. Cell. 1979 Nov;18(3):613–622. doi: 10.1016/0092-8674(79)90116-8. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Haars R., Bautz E. K. In vitro proteolysis of a large subunit of Drosophila melanogaster RNA polymerase B. FEBS Lett. 1976 Dec 1;71(2):205–208. doi: 10.1016/0014-5793(76)80932-5. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Weeks J. R., Voelker R. A., Ohnishi S., Dickson B. Genetic and biochemical characterization of mutants at an RNA polymerase II locus in D. melanogaster. Cell. 1980 Oct;21(3):785–792. doi: 10.1016/0092-8674(80)90441-9. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J., Jendrisak J. J. Plant DNA-dependent RNA polymerases: subunit structures and enzymatic properties of the class II enzymes from quiescent and proliferating tissues. Biochemistry. 1978 May 16;17(10):1860–1866. doi: 10.1021/bi00603a009. [DOI] [PubMed] [Google Scholar]
- Higgins R. C., Dahmus M. E. Rapid visualization of protein bands in preparative SDS-polyacrylamide gels. Anal Biochem. 1979 Mar;93(2):257–260. doi: 10.1016/s0003-2697(79)80148-7. [DOI] [PubMed] [Google Scholar]
- Hodo H. G., 3rd, Blatti S. P. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1977 May 31;16(11):2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Ingles C. J., Biggs J., Wong J. K., Weeks J. R., Greenleaf A. L. Identification of a structural gene for a RNA polymerase II polypeptide in Drosophila melanogaster and mammalian species. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3396–3400. doi: 10.1073/pnas.80.11.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J., Himmelfarb H. J., Shales M., Greenleaf A. L., Friesen J. D. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2157–2161. doi: 10.1073/pnas.81.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedinger C., Gissinger F., Chambon P. Animal DNA-dependent RNA polymerases. Molecular structures and immunological properties of calf-thymus enzyme AI and of calf-thymus and rat-liver enzymes B. Eur J Biochem. 1974 May 15;44(2):421–436. doi: 10.1111/j.1432-1033.1974.tb03500.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Link G., Kidd G. H., Richter G., Bogorad L. Structural relationships among the multiple forms of DNA-dependent RNA polymerase II from cultured parsley cells. Eur J Biochem. 1978 Nov 15;91(2):363–368. doi: 10.1111/j.1432-1033.1978.tb12688.x. [DOI] [PubMed] [Google Scholar]
- Penefsky H. S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977 May 10;252(9):2891–2899. [PubMed] [Google Scholar]
- Ruet A., Sentenac A., Fromageot P., Winsor B., Lacroute F. A mutation of the B220 subunit gene affects the structural and functional properties of yeast RNA polymerase B in vitro. J Biol Chem. 1980 Jul 10;255(13):6450–6455. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles L. L., Jokerst R. S., Bingham P. M., Voelker R. A., Greenleaf A. L. Molecular cloning of sequences from a Drosophila RNA polymerase II locus by P element transposon tagging. Cell. 1982 Dec;31(3 Pt 2):585–592. doi: 10.1016/0092-8674(82)90314-2. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Eisenberg D. The structure of melittin. II. Interpretation of the structure. J Biol Chem. 1982 Jun 10;257(11):6016–6022. [PubMed] [Google Scholar]
- Weeks J. R., Coulter D. E., Greenleaf A. L. Immunological studies of RNA polymerase II using antibodies to subunits of Drosophila and wheat germ enzyme. J Biol Chem. 1982 May 25;257(10):5884–5892. [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]