Abstract
Type 1 diabetes mellitus (T1DM) is a chronic disease resulting from destruction of insulin-producing pancreatic β cells. Genetic and environmental factors contribute to T1DM onset. Use of high-throughput DNA sequencing has allowed geneticists to perform genome-wide association studies (GWAS) to identify novel gene loci associated with T1DM. Interestingly, >50% of these genes encode products that are expressed in β cells. These studies, coupled with emerging molecular evidence that β cells are impaired by gain-of-function or loss-of-function of these loci, suggest an active role for the β cell in eliciting its own demise. Although immune dysregulation plays a vital role in T1DM pathogenesis, understanding the mechanisms contributing to β cell failure may lead to new strategies to preserve or improve β cell function in patients with T1DM.
Keywords: T1DM, islet, GWAS, insulin
Genetics and the development of type 1 diabetes
Diabetes mellitus is a disease of dysfunctional glucose homeostasis resulting from insufficient insulin secretion to meet the demand of peripheral insulin-responsive tissues. The two most common types of diabetes, type 1 diabetes (T1DM) and type 2 diabetes (T2DM), are generally regarded as distinct disease processes that share the common fate of insufficient pancreatic β cell function to maintain glucoseresponsive insulin release. T1DM has been long believed to be primarily a disease of immune dysregulation, leading to autoimmune destruction of the insulin-secreting pancreatic β cell and, ultimately, insulinopenia and hyperglycemia. Patients afflicted with T1DM require lifelong insulin therapy to maintain glycemic control. T1DM onset is on the rise and is a major cause of healthcare resource utilization worldwide arising from complications related to hyperglycemia-induced microvascular disease as well as increasing morbidity and mortality related to insulin therapy-induced hypoglycemia unawareness [1].
The etiology of T1DM has been a topic of much debate for several decades, with environmental risk factors and genetic susceptibility broadly suggested to contribute to the disease. Studies in monozygotic twins, who share an identical genome, demonstrate pairwise concordance of T1DM of 13–52% [2], suggesting that environmental and genetic causes may contribute similarly to disease. Over the past 35 years studies of the genetic contribution to T1DM have focused largely on loci implicated in the regulation of, and selection, against auto-reactive T lymphocytes, identified by candidate-gene analyses and linkage studies [3], although single-nucleotide polymorphisms (SNPs) within the human insulin (INS) gene, principally expressed in β cells, remain one of the highest risks for development of T1DM [4]. More recently, GWAS investigators have utilized high-throughput genomic sequencing to genotype SNPs in large cohorts of T1DM patients, their relatives, and controls, leading to the identification of a large number of previously unrecognized genomic loci that contribute to T1DM [5].
The identification of the relevant gene within a linkage disequilibrium (LD) block whose regulation is impacted by the disease-linked SNP is often complicated by the presence of multiple genes within the block. Many identified SNPs are not located within coding regions but instead in intergenic regions that may affect the function of transcriptional enhancers located far from the disease-relevant gene. Another relatively unexplored area is the impact of SNPs on the expression of noncoding RNAs that contribute to disease processes [6]. Nevertheless, lists of potential causative genes in loci associated with T1DM, and integration with other high-throughput approaches, are giving broad insight into novel pathways and systems associated with disease.
Notably, ~60% of apparent T1DM susceptibility genes are expressed in pancreatic β cells [7,8], and mounting evidence from gain and loss of function genetic animal models supports critical roles for these genes in the function and growth of β cells, including in the regulation of embryonic pancreas development, the endolysosomal pathway, signal transduction, and cytokine responses. This, along with evidence from the pancreatic pathology of patients with T1DM, supports the existence of compound defects of both the immune system and the β cell in the pathogenesis of T1DM (see Boxes 1, 2 and 3). Thus, we hypothesize that the β cell plays an active role in its own demise, rather than acting as an innocent bystander to autoimmune attack. In light of this hypothesis we will discuss recently identified T1D genetic loci and evidence from animal models and human pathology to suggest defects in the β cells of patients with T1DM beyond the previously suggested role of the β cell in eliciting inflammation and insulitis [9].
Box 1. Lessons from animal models of T1DM.
Investigating the early pathogenesis of T1DM in human subjects is highly challenging due to the inaccessibility of the target tissue for analysis and to the current inability to image pancreatic β cell mass in vivo. This has necessitated the use of animal models of T1DM to advance our understanding of pathogenesis and to test potential mechanism-based therapies. Two animal models are commonly used, the non-obese diabetic (NOD) mouse [79] and the Bio-Breeding/Worcester (BB/W) rat [80], both of which develop spontaneous diabetes and immune infiltration of the pancreatic islets, characteristic of human T1DM. The NOD model has risen in popularity due to extensive annotation of the mouse genome and to the ability to backcross mice with targeted deletion of specific gene loci onto this genetic background; however, the model remains limited. Compared to the pancreas of human T1DM subjects, the pancreas of diabetic NOD mice exhibits more aggressive insulitis and lacks β cell IFN-α and pan-islet MHC class I hyperexpression. Further, the residual β cells reported in human T1DM long after disease onset are not evident in the NOD model (detailed below) [81]. For reasons that are still not understood, the incidence of diabetes in NOD mice is gender-specific, and nearly ~80% of NOD females develop disease compared to ~20% of male mice by 30 weeks of age [79]. Perhaps related to these issues, therapeutic approaches which prevent or abrogate diabetes onset in NOD mice have met with limited translational success in human patients [82]. Despite these limitations, the NOD model has provided useful insights and generated hypotheses that can be tested in human tissue. Pre-diabetic NOD mice display a loss of first-phase insulin response following a glucose challenge, suggesting that β cell dysfunction pre-dates insulitis in NOD mice [83]. This defect may be due to the onset of β cell endoplasmic reticulum (ER) stress, observed in NOD mice and human patients with T1DM [84], and subsequent activation of cytokine signaling processes [85], which may serve as a trigger for autoimmunity in T1DM [86]. These studies are suggestive of β cell dysfunction appearing before, and possibly eliciting, the onset of autoimmune destruction.
Box 2. HLA and T1DM revisited.
The best characterized and highest risk gene locus associated with T1DM is the HLA locus which encodes MHC classes I and II [87], important in the recognition of self versus non-self antigens and in the activation of immune cascades. Specific genotypes of MHC class II, whose expression is generally restricted to immune cells, carry among the highest risk for development of T1DM [88]. MHC class I genotypes also carry T1DM risk independently of class II genotypes, and T1DM-associated alleles B*5701, B*3906 and A*0201 are among those carrying the highest risk [89]. Furthermore, global transgenic expression of the A*0201 diabetogenic class I allele in NOD mice accelerates diabetes onset [90]. It is not clear from these experiments whether class I expression on the β cell surface contributes to disease development.
The expression of class I on the surface of β cells has been extensively profiled in autopsy studies of patients with T1DM. In one histological study, the majority of the insulin-expressing islets in an autopsy series of 23 patients with recent-onset T1DM had marked overexpression of class I in all endocrine islet cells [91]. Class I-hyperexpressing β cells contain high IFN-α levels [92], which could lead to the induction of class I in other endocrine cell types. Class I hyperexpression is also observed in human islets cultured in the presence of IFN-α [93]. Transgenic overexpression of IFN-α in pancreatic β cells induced a T1DM-like phenotype in rodent models; however, class I expression was not assessed [94]. Recently, pancreatic specimens from donors with T1DM (as well as nondiabetic controls and non-diabetic patients with autoantibodies) were accessed from the Network for Pancreatic Organ donors with Diabetes (www.jdrfnpod.org). In these samples, the presence of class I hyperexpression throughout the islet was again observed, even in pseudoatrophic islets without β cells, but not in the islets of autoantibody-positive non-diabetic controls [95]. Of 72 T1DM samples studied, 11 patients with the high-risk A*0201 haplotype had coincident evidence of islet class I hyperexpression [95], perhaps connecting high-risk haplotypes with abnormalities in class I expression. Although class I hyperexpression in human islets cultured in high glucose concentrations has also been observed [96], there is no evidence to date to suggest that glucose can modulate IFN-α expression. These observations suggest that T1DM susceptibility conferred by variation in the HLA locus could be mediated in part by pancreatic β cells.
Box 3. Histopathology of the T1DM pancreas.
The natural course of β cell loss in the pancreas of patients with T1DM is distinct from that of the autoimmune-susceptible NOD mouse model, which often displays severe islet infiltrates and rapid progression to C-peptide-negative diabetes [79]. The study of non-diabetic patients with a high risk for development of T1DM with elevated serum autoantibodies has been limited [97]. The largest of these studies assessed 62 such patients for the existence of insulitis and found insulitis to be extremely rare, only occurring in two patients [98]. Similarly, the severity of immune infiltration in T1DM pancreata is remarkably mild [97], but does consist of cytokine-producing T lymphocytes and macrophages, as well as other immune cell types. T1DM pancreatic specimens also possess a small number of insulin-positive pancreatic β cells even as long as 56 years after diagnosis [95,99]. The number of remaining β cells may be further underestimated, as degranulated insulin+ β cells could be missed by immunohistochemistry techniques unless other β cell-specific markers are also utilized, a phenomenon previously reported in NOD mice [100]. The long-term survival of β cells and a relatively mild lymphocytic infiltrate suggests that functional defects within the β cell of patients of T1DM should be considered as a cause of early glucose intolerance and hyperglycemia. Taken together, the possibility of a dysfunctional β cell (possibly related to inherited defects in T1DM loci), in addition to the MHC class I hyperexpression (Box 2) necessary to recruit autoreactive T lymphocytes, could lead in tandem to a slow decline in β cell mass and function coincident with disease onset.
Exploration of novel T1DM genes and pathways to dysfunction in the β cell
Regulation of pancreatic development
Several loci associated with T1DM encode proteins that are expressed in the pancreas during embryonic development, including the Krüppel-like zinc finger transcription factor Gli-similar 3 (GLIS3) [10], the transmembrane protein delta-like homolog 1 (DLK1) [11], and the proenzyme chymotrypsinogen B1/B2 (CTRB1/2) [10,12] (see Glossary). The identification of GLIS3 mutations in patients with syndromic neonatal diabetes and hypothyroidism (NDH) [13] and of GLIS3 polymorphisms associated with T1DM and T2DM [14] suggests a role for GLIS3 in β cells. Loss of Glis3 function, studied in mutant mice lacking the fifth zinc finger region responsible for DNA binding, leads to a decrease in neurogenin-3+ (Ngn3) endocrine progenitors, and markedly diminished pancreatic endocrine mass at birth, with severe postnatal hyperglycemia and hypoinsulinemia [15]. Acute loss of function of Glis3 in adult β cells leads to downregulation of insulin gene expression and, ultimately, hyperglycemia and enhanced β cell apoptosis [16]. These studies suggest that the association of Glis3 with T1DM relates to its critical role in the development and adult function of the β cell.
The transmembrane protein DLK1, also known as preadipocyte factor 1 (PREF-1), is subject to genomic imprinting with expression only from the paternally inherited allele [17], and this is particularly interesting in light of the observation that the risk of transmission of T1DM to offspring from diabetic fathers is 1.7-fold higher than from diabetic mothers [18]. In rodent models, Dlk1 is broadly expressed, with high levels found in the developing pancreas at E12.5 [17,19] and later restriction to adult pancreatic β cells, pituitary somatotrophs, bone marrow, adrenal gland, and skeletal muscle [19,20]. DLK1 is believed to play a role in the transition of immature proliferative cells to mature differentiated cell types. However, the functional role of Dlk1 in the pancreas remains controversial, with initial studies suggesting a role in mediating the effects of growth hormone and prolactin on the induction of b cell replication [20,21]. Conditional loss of function of Dlk1 in pancreatic β cells did not recapitulate the neonatal lethality and weight-loss reported in whole-body Dlk1 knockout mice with normal islet architecture up to 6 weeks after birth. Glycemic control, β cell replication, and β cell mass were not examined [22].
The observation of pancreatic proenzyme chymotrypssinogen B1/B2 (CTRsB1/2) as a T1DM susceptibility locus is intriguing but confusing. Chymotrypsinogen is primarily expressed in pancreatic acinar cells where it functions as an endopeptidase [23]. It is expressed as early as embryonic (E) day E14.5 primarily in developing pancreatic tip cells which give rise to acinar cells in adult rodents [24]. It is unclear whether chymotrypsinogen plays a developmental role in the pancreas; its expression may serve simply as a marker for the developing acinar cell. In T1DM patients, mild pancreatic exocrine deficiency not requiring pancreatic enzyme replacement is noted in up to 80% of patients [25]; however, a decade later the mild exocrine insufficiency resolved, suggesting that exocrine deficiency represents an early defect following T1DM onset [26]. Chymotrypsinogen is also an autoantigen in patients with T1DM [27]. It is intriguing to speculate that these observations relate to the anatomic and functional relationship between islets and the surrounding acinar parenchyma [28].
Regulation of the endolysosomal pathway
Endosomes have been classically connected with T1DM via roles in processing and presentation of pancreatic islet antigens in MHC class II complexes on immune cells [29]. MHC class I cell-surface presentation can also be regulated by the endolysosomal pathway [30], and this could potentially serve as a mechanism for the maintenance of hyperexpression on the β cell surface. In pancreatic β cells, endosomes play an essential role within the secretory pathway [31]. Endosomes and lysosomes comprise a network of intracellular organelles whose functions range from cellular defense against viral attack to processing of ligand-bound receptor complexes and internalization of macromolecules from the extracellular matrix [32]. In the classical endocytic pathway (reviewed in [33]), the unidirectional portion of this system is composed of the early endosome (EE), late endosome (LE), and lysosome. Maturation of EEs to LEs and ultimate fusion with lysosomes is regulated by vacuolar H+ ATPase-mediated endosomal acidification, endosomal membrane small-GTPase association, and membrane tethering by the class C Vps protein complexes [33].
Two recently identified T1DM loci have been linked to the endolysosomal pathway in recent GWAS. Several groups identified SNPs associated with T1DM at chromosome 16p13 [34,35]. These SNPs reside within the CLEC16A gene locus (originally known as KIAA0350) which was initially believed to encode a C-type lectin protein [35]. Recent studies of the Drosophila melanogaster ortholog of CLEC16A, known as Ema, have shown that Ema does not contain a C-type lectin signature [36]. Ema was shown to localize specifically to late endosomes and to regulate maturation of late endosomes and fusion with lysosomes via direct interaction with the class C vacuole/ endosome tethering (Vps) and homotypic fusion and protein sorting (HOPS) protein complex, which is essential for fusion and degradation of internalized cargo [36]. These studies also demonstrate dysfunctional processing of both ligand-bound epidermal growth factor receptors (EGFR) and components of the bone morphogenetic protein (BMP)– SMAD signaling cascade, leading to inappropriate prolongation of these signals [36]. These findings are of particular interest given the critical importance of EGFR signaling for maintenance of β cell development, mass, and responses to incretins [37]. Further, BMP–SMAD signaling has been shown to be essential for glucose-stimulated insulin secretion (GSIS) via an autocrine/paracrine effect within the islet [38]. Recent preliminary studies suggest that Clec16a regulates GSIS in β cell lines, and this appears to be related to maintenance of mitochondrial oxygen consumption and ATP generation [39]. The endosomal dysfunction observed in β cell lines following Clec16a RNA interference provides proof-of-concept of the broad relevance of endosomal function in β cells beyond antigen processing.
Endosomal proteins often share homologous roles with components of autophagy. Drosophila Ema null mutants possess dramatic reductions in the size of autophagosomes, which are unable to appropriately degrade their cargo [40]. Whether dysfunctional autophagy leads to β cell failure remains controversial [41]. Future study of the role of CLEC16A in β cell survival related to induction of autophagy could provide insights into how β cells respond to a hostile milieu of cytokine release, immune cell infiltration, and increased insulin demand as surrounding β cells die.
Genetic studies identifying T1DM-associated SNPs within chromosome 12q13 identify a rich locus with multiple potential disease-relevant targets [42], including the small GTPase RAB5B which encodes a critical component for EE function. GTP-bound Rab5 is recruited to the surface of EEs where it assists in the initial sorting and processing of internalized cargo. Rab5 is a key component of EEs together with its effector Vps34/p150, a phosphoinositide-3-kinase (PI3K) complex that generates phosphatidylinositol 3-phosphate (PI-3-P), an essential component of EE identity [43]. Whether the T1DM-associated SNPs in this region affect RAB5B expression in the β cell remains to be determined.
Extracellular signaling receptors and intracellular signaling pathways
Cell-surface receptors and intracellular signaling components comprise more than 50% of the genes associated with T1DM. The likely interplay between environmental factors, including viruses, and local cytokine release in the islet suggests that T1DM risk could be engendered by how these external effects are processed and managed. Some of these signaling components are ubiquitously expressed and specific roles in the β cell have not been explored. For instance, the T1DM gene TYK2 [11] encodes a broadly expressed tyrosine kinase important in IFN-α signaling [44], but we do not know if the deleterious effect of islet IFN-α expression (discussed above) is dependent on Tyk2. Here, we will focus on T1DM-associated receptors and intracellular signaling components that have been demonstrated to play functional roles in the pancreatic β cell. These loci include the heregulin receptor ERBB3 [10], the interferon-induced helicase IFIH1 [45], and the tyrosine phosphatase PTPN2 [10,42].
Located in the gene-rich linkage disequilibrium block on chromosome 12q13, SNPs within an exon of ERBB3 gene are associated with T1DM [46]. ErbB3 is expressed primarily in the developing pancreatic ducts and mesenchyme and ErbB3 null embryos (which die at E13.5 due to cardiac defects) exhibit pancreatic hypoplasia at E13.5 [47,48]. ErbB3 expression in the adult pancreas is sensitive to cytokines, with an increase in ductal expression of ErbB3 in transgenic NOD mice overexpressing IFN-γ selectively in the pancreas. In NOD mice, pan-islet cell ErbB2 and α-cell ErbB4 expression increase following immune infiltration [48]. It is unclear whether and how ErbB3 influences β cell development and function and what role(s) pancreatic ErbB receptors play following immune infiltration and cytokine release.
The importance of environmental factors in T1DM pathogenesis is well established, but there is no consensus as to which factors are most important. Viral infections remain one of the most likely candidates, although the evidence for viral infection and, more importantly, the specific viral types responsible for T1DM are still subject to much debate [49]. Detection of viral dsRNAs by RIG-I and IFIH1 leads to activation of the innate immune system and of transcription factors, such as IFN-regulatory factors (IRFs) and nuclear factor κB (NF-κB), ultimately leading to the production of type 1 IFN and other proinflammatory cytokines [50]. In the pancreatic β cell, exposure to the synthetic dsRNA polyinosinic–polycytidylic acid (PIC) leads to increased NF-κB and IRF-3 activity, and release of type 1 IFN, which is suggestive of activation of the RIG-I-like helicase (RLH) pathway. Treatment with dsRNA leads to β cell apoptosis that is dependent on autocrine IFN signaling because islets of IRF-3 or IFN-α receptor knockout mice are resistant to PIC-induced cell death [51]. Similarly, PIC-treated islets demonstrate an increase in RIG-I and IFIH1 expression, whereas treatment with a 5′-triphosphate ssRNA analog induces RIG-I action and β cell death [52]. 5′-Triphosphate ssRNA analogs also induce NF-κB and IFN-β promoter activity that appears to be dependent on RIG-I [52]. Similarly, loss of function studies in PIC-treated isolated β cells demonstrates that IFIH1 regulates the expression of cytokines and chemokines [53]. Taken together, these studies suggest that RIG-I and IFIH1, although important for function of host innate immunity to viral infections, play a deleterious role in the pancreatic β cell, a concept supported by human genetic studies of rare protective IFIH1 variants that reduce IFIH1 expression [54,55].
Intracellular signals transmitted from cell-surface receptors rely heavily on the phosphorylation of tyrosine residues. The balance of tyrosine phosphorylation is dependent on protein tyrosine kinases, which phosphorylate tyrosine residues, and the opposing effects of protein tyrosine phosphatases (PTPs). In pancreatic β cells, cytokine exposure leads to PTPN2 cytoplasmic localization, which reduces STAT1 and STAT3 phosphorylation [56]. PTPN2 loss of function in primary β cells serves to increase cytokine- induced STAT1 phosphorylation and β cell apoptosis [57], and promotes PIC-induced cell death, suggesting a protective role in β cells following cytokine and viral insults [53].
T1DM, NF-κB, and sensitivity to cytokine-induced β cell apoptosis
Immune-cell secreted cytokines, including IL-1β, TNF-α, and IFN-γ, act synergistically to promote β cell apoptosis. Following cytokine binding to cell-surface receptors, signaling cascades lead to activation of multiple transcription factors, including NF-κB, a central player in cellular stress responses, cell growth, and cell survival [58]. In resting cells, NF-κB resides within the cytoplasm bound to the trimeric inhibitor of NF-κB (IκB) kinase protein complex [59]. Cytokine signals lead to phosphorylation and, ultimately, ubiquitination and proteosomal degradation of the IκB kinase complex [60], allowing NF-κB to translocate to the nucleus to activate its target genes. Genetic models indicate positive roles for NF-κB during pancreas development and negative roles in the mature β cell, such that disruption of NF-κB signaling in the mature β cell protects from both insulitis and cytokine-induced cell death [61–63].
NF-κB-regulated anti-apoptotic genes are induced following cytokine exposure of human islets [64]. Genetic studies have implicated one such target, TNFAIP3, as being associated with T1DM [10]. In pancreatic β cells, TNFAIP3 serves as an early inhibitor of cytokine-induced nitric oxide (NO) production and inducible NO signaling (iNOS), mediators of cytokine-related toxicity. Indeed, TNFAIP3 is the most highly induced anti-apoptotic gene following cytokine exposure of islets, and this effect is directly mediated by NF-κB [65]. TNFAIP3 overexpression also mediates cytoprotective effects in islets, not only preventing cytokine-induced β cell apoptosis, but also allowing for enhanced islet allograft survival in transplantation models [66,67]. It remains to be determined how the disease-associated SNP influences TNFAIP3 expression and whether the beneficial effects of TNFAIP3 are lost in patients harboring the risk allele.
Systems-based approaches to identify novel β cell related T1DM genes
Recently, the integration of high-throughput gene expression profiling with GWAS data sets and protein-interaction prediction algorithms is allowing greater insight into the relevant networks underlying the pathogenesis of β cell failure in T1DM [68]. Several studies have utilized global expression profiling following exposure of human islets or β cell lines to cytokines. Of course, reliance on these datasets will illuminate our understanding of cytokine-induced β cell failure in T1DM while minimizing other pathways important for β cell compromise in T1DM.
Huntington-interacting protein 14 (HIP14) is one such gene identified by overlapping in silico phenome–interactome network analysis with a T1DM genome-wide linkage dataset [69]. Within the pancreatic islet, HIP14 is exclusively expressed in pancreatic β cells and expression is downregulated in the context of cytokine exposure. Knockdown of HIP14 expression leads to β cell apoptosis and decreases glucose-stimulated insulin release, whereas overexpression reduces NF-κB activity and protects against cytokine-induced β cell apoptosis [69].
Combined proteome–transcriptome–genome approaches identified galectin-3 as a candidate gene/protein in T1DM susceptibility. Galectin-3 expression is induced by IL-1β exposure in rat and human islets, with overexpression of galectin-3 protecting β cells against IL-1β, in part through a blockade of JNK phosphorylation [70].
Studies within a T1DM-associated linkage region on chromosome 21 have also identified several genes with altered expression following cytokine exposure [71], including carbonyl transferase (CBR1), which functions as a nitrosylated glutathione reductase important for β cell survival during oxidative stress [72], and the E3 ligase protein tetratricopeptide repeat domain 3 (TTC3), which facilitates the ubiquitination and degradation of the survival factor Akt/PKB [73].
Expression profiling in cytokine-treated human islets has been further applied to non-translated RNAs. MicroRNA (miRNA) expression profiling in human islets identified induction of miR-21, miR-34a, and miR-146a following cytokine treatment, which were also induced during the progression of diabetes in NOD mice [74]. MiR-146a expression was also directly regulated by NF-κB activity, and the induction of miR-146a may exert negative effects on β cell function related to binding to the 3′ untranslated region (3′UTR) of HIP14, which carries an miR-146a seed sequence [69,74]. Further, the miR-21 target, tumor suppressor programmed cell death 4 (PDCD4), plays a role in β cell survival in NOD and STZ-treated mice [75]. MiR-29a/b/c is another miRNA induced in prediabetic NOD mice and cytokine-treated islets, and leads to insulin secretory dysfunction as well as apoptosis by reducing expression of the anti-apoptotic protein Mcl1 [76]. Thus, systems-based approaches identify both coding and non-coding RNAs as novel factors involved in β cell survival and contributing to T1DM risk.
Concluding remarks
T1DM is a genetically complex and heterogeneous disease that we are still struggling to comprehend (Box 4). Here we discuss recent GWAS-identified T1DM loci and suggest a role for the β cell in mediating its own demise with defects in discrete pathways working both independently and together to lead to decreased functional β cell mass and insulinopenia over time (Figure 1). Establishing a direct role for these loci in β cell survival and function still requires additional evidentiary support, such as conditional deletion and/or overexpression in the developing or adult β cell, and assessment of these models on autoimmune disease-susceptible genetic backgrounds, to delineate a direct role for the β cell in crosstalk with the immune system. The development of humanized mouse models, in which a functional human immune system is reconstituted and human islets or β cells derived from induced pluripotent stem-cell (iPS) precursors derived from patients with T1DM are transplanted, could bypass the limitations of current animal models of T1DM [77], although the current poor efficiency and functionality of iPS-generated β cells still requires optimization [78]. It is imperative that we understand the β cell specific contribution to T1DM disease pathogenesis because the need to target β cell functional and growth defects will impact upon therapeutic efforts to stimulate endogenous β cell regeneration in the context of immune modulation.
Box 4. Outstanding questions.
Correlation versus causation. Do alterations in expression in T1DM-associated genes by SNPs lead to defects in β cell function or survival in pre-diabetic patients?
Molecular validation of SNPs. How do GWAS-identified T1DM SNPs regulate the expression of candidate T1DM genes in the pancreatic β cell?
Future therapies. Can β cell therapies targeting associated genes, in conjunction with immune therapies, be utilized to improve β cell function in patients with T1DM?
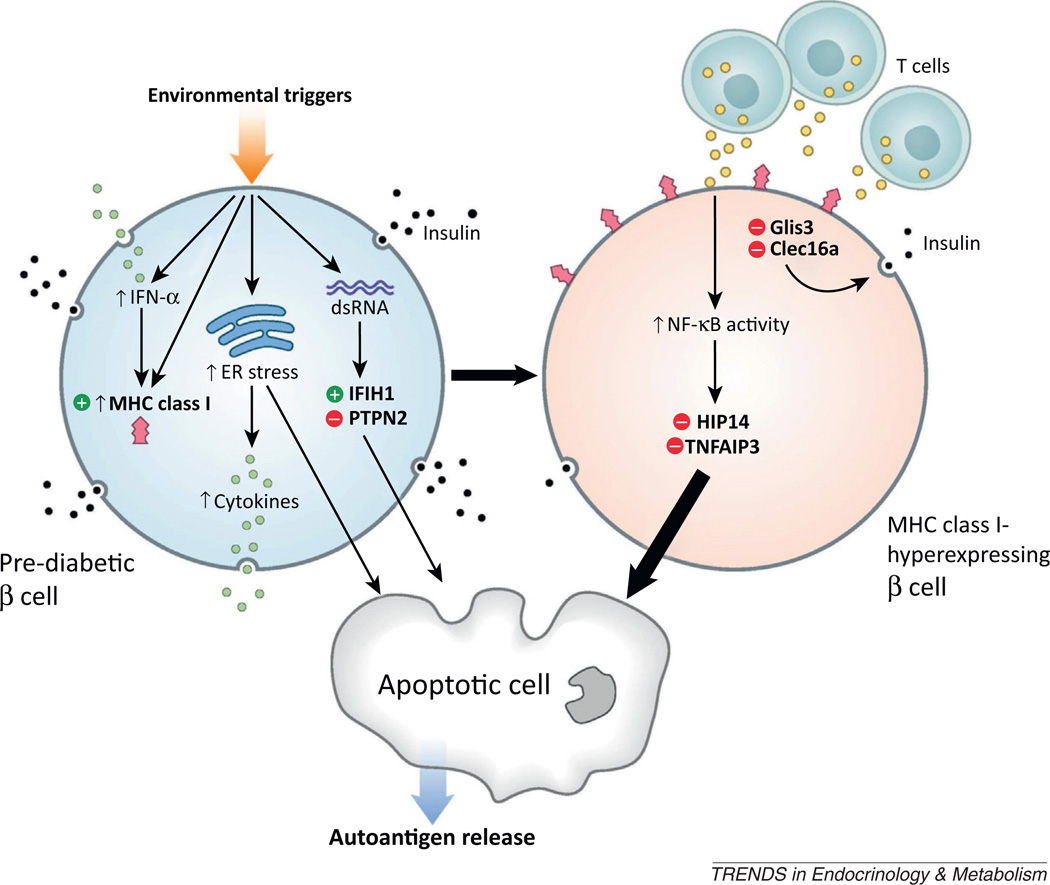
Figure 1.
The genetics of T1DM illuminates potential pathways to β cell failure and diabetes. A model of the events resulting in β cell failure, with a pre-diabetic β cell (upper left) subjected to various environmental stressors (including viruses) that activate IFN-α expression and MHC class I hyperexpression on the b cell surface (upper right). Induction of ER stress and β cell cytokine release (green) as well as dsRNA responses (in the case of viral attack) and downstream genetic events (activation of IFIH1 and loss of protective PTPN2) can lead to β cell apoptosis (bottom). MHC class I-hyperexpressing b cells that survive initial environmental insults (upper right) exhibit reduced insulin secretion (black granules), mediated by loss of Clec16a and Glis3 function, and are subject to immune attack by infiltrating T lymphocytes, which release high concentrations of cytokines (yellow), subsequently leading to activation of NF-κB signaling and significant β cell apoptosis (bottom) in the absence of protective HIP14 and TNFAIP3 function. Circulating autoantigens released following β cell apoptosis lead to autoantibody formation and further mobilization of the immune system against the remaining β cells.
Acknowledgments
We acknowledge the many authors whose work could not be cited due to reference limitations. We acknowledge Mary Leonard of University of Pennsylvania Biomedical Art and Design for the artwork shown in the figure and funding support from the Margaret Q. Landenberger Foundation, the Charles H. Humpton, Jr. Endowment, and the National Institutes of Health (K08-DK089117 and 5-P01-DK-049210-15).
Glossary
- Autophagy
a catabolic process of cell self-preservation that compensates for starvation by degradation of intracellular organelles.
- Chymotrypsinogen
a proteolytic enzyme synthesized in the acinar cells of the pancreas and stored inside membrane-bounded granules at the apex of the acinar cell. It functions as an endopeptidase responsible for cleavage of ingested peptides at aliphatic amino acid residues to yield oligopeptides that are further hydrolyzed into amino acids that can be absorbed in the digestive tract.
- Delta-like homolog 1 (DLK1)
a transmembrane protein, also known as preadipocyte factor 1 (PREF-1), that contains EGF-like repeats homologous to Notch ligands but lacks a Notch interaction domain. DLK1 is cleaved by tumor necrosis factor α activating enzyme to generate a soluble protein called fetal antigen 1 (FA1).
- Gli-similar 3 (GLIS3)
a Krüppel-like zinc finger transcription factor that is a member of the Gli-similar family of transcriptional regulators. These are structurally similar to the Gli subfamily of transcription factors that contain with five zinc fingers in tandem, that are essential for DNA binding, and both activator and repressor domains.
- Heregulin receptor ErbB3
an EGFR-like transmembrane receptor that exhibits broad roles in development. Following ligand binding, ErbB receptors form dimers, resulting in tyrosine phosphorylation and recruitment of downstream effectors. Although ErbB3 is structurally similar to other ErbB receptors, it is unique in that it binds ligands neuregulin-1 and -2 and requires heterodimers with EGFR or ErbB4 to transmit downstream growth-factor signals.
- Human leukocyte antigen (HLA) locus
a superlocus that encodes cell-surface antigen-presenting proteins. HLAs corresponding to MHC class I present peptides produced from proteins digested in the proteasome. HLAs corresponding to MHC class II present antigens to T lymphocytes. HLAs corresponding to MHC class III encode components of the complement system.
- Huntington-interacting protein 14 (HIP14)
a well-characterized neuronal palmitoyl acyltransferase enzyme that has been implicated in the pathogenesis of Huntington disease through palmitoylation of huntingtin (HTT). HIP14 is important for intracellular trafficking and exocytosis in neurons through its substrate specificity for several vesicle-associated proteins.
- Interferon-induced helicase (IFIH1)
also known as MDA5 or helicard, a member of a RIG-I-like helicase (RLH) in concert with RIG-I, which functions as a cytoplasmic viral RNA detector.
- Major histocompatibility complex (MHC)
also termed human leukocyte antigen (HLA) in humans, a family of cell-surface molecules that are divided into three subgroups – class I, class II, and class III – and which determine compatibility of donors for organ transplant as well as susceptibility to autoimmune disease via crossreacting immunization.
- Retinoic acid-inducible gene 1 (RIG-I)
a RIG-I-like receptor dsRNA helicase and part of the RIG-I-like receptor (RLR) family, which functions as a pattern recognition receptor and a sensor for viruses.
- Tumor necrosis factor α-induced protein 3 (TNFAIP3)
also called A20, a zinc finger protein that is strongly induced by cytokines and protective following TNFα exposure in endothelial cells.
- Tyrosine-protein phosphatase non-receptor type (PTPN2)
also known as the T cell protein tyrosine phosphatase TC-PTP, a ubiquitously expressed protein tyrosine phosphatase initially identified in a peripheral T cell cDNA library. Although it contains a nuclear localization signal, PTPN2 will redistribute to the cytosol under particular stresses (hyperosmolarity, oxidative stress, and cold shock) to modulate EGFR, MAPK/JNK, and Jak/ STAT signaling.
References
- 1.Centers for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in The United States, 2011. United States: Department of Health and Human Services; 2011. [Google Scholar]
- 2.Hyttinen V, et al. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52:1052–1055. doi: 10.2337/diabetes.52.4.1052. [DOI] [PubMed] [Google Scholar]
- 3.Bluestone JA. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell GI, et al. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 5.Cooper JD, et al. Confirmation of novel type 1 diabetes risk loci in families. Diabetologia. 2012;55:996–1000. doi: 10.1007/s00125-012-2450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran I, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eizirik DL, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutlu B, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med. Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eizirik DL, et al. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace C, et al. The imprinted DLK1–MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy MV, et al. Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun. 2011;12:208–212. doi: 10.1038/gene.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senee V, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 14.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang HS, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol. Cell. Biol. 2009;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Sustained expression of the transcription factor GLIS3 is required for normal beta cell function in adults. EMBO Mol. Med. 2013;5:92–104. doi: 10.1002/emmm.201201398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt JV, et al. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 18.Harjutsalo V, et al. Differential transmission of type 1 diabetes from diabetic fathers and mothers to their offspring. Diabetes. 2006;55:1517–1524. doi: 10.2337/db05-1296. [DOI] [PubMed] [Google Scholar]
- 19.Yevtodiyenko A, Schmidt JV. Dlk1 expression marks developing endothelium and sites of branching morphogenesis in the mouse embryo and placenta. Dev. Dyn. 2006;235:1115–1123. doi: 10.1002/dvdy.20705. [DOI] [PubMed] [Google Scholar]
- 20.Carlsson C, et al. Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/delta-like protein in pancreatic islets: molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology. 1997;138:3940–3948. doi: 10.1210/endo.138.9.5408. [DOI] [PubMed] [Google Scholar]
- 21.Friedrichsen BN, et al. Expression, biosynthesis and release of preadipocyte factor-1/delta-like protein/fetal antigen-1 in pancreatic beta-cells: possible physiological implications. J. Endocrinol. 2003;176:257–266. doi: 10.1677/joe.0.1760257. [DOI] [PubMed] [Google Scholar]
- 22.Appelbe OK, et al. Conditional deletions refine the embryonic requirement for Dlk1. Mech. Dev. 2013;130:143–159. doi: 10.1016/j.mod.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitcomb DC, Lowe ME. Human pancreatic digestive enzymes. Dig. Dis. Sci. 2007;52:1–17. doi: 10.1007/s10620-006-9589-z. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman BG, et al. Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol. 2008;9:R99. doi: 10.1186/gb-2008-9-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frier BM, et al. Exocrine pancreatic function in juvenile-onset diabetes mellitus. Gut. 1976;17:685–691. doi: 10.1136/gut.17.9.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creutzfeldt W, et al. Follow-up of exocrine pancreatic function in type-1 diabetes mellitus. Digestion. 2005;72:71–75. doi: 10.1159/000087660. [DOI] [PubMed] [Google Scholar]
- 27.Kim YJ, et al. IDDM patients’ sera recognize a novel 30-kD pancreatic autoantigen related to chymotrypsinogen. Immunol. Invest. 1993;22:219–227. doi: 10.3109/08820139309063404. [DOI] [PubMed] [Google Scholar]
- 28.Malaisse-Lagae F, et al. Exocrine pancreas: evidence for topographic partition of secretory function. Science. 1975;190:795–797. doi: 10.1126/science.1105788. [DOI] [PubMed] [Google Scholar]
- 29.Moustakas AK, Papadopoulos GK. Molecular properties of HLA-DQ alleles conferring susceptibility to or protection from insulin-dependent diabetes mellitus: keys to the fate of islet beta-cells. Am. J. Med. Genet. 2002;115:37–47. doi: 10.1002/ajmg.10342. [DOI] [PubMed] [Google Scholar]
- 30.Reusch U, et al. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 1999;18:1081–1091. doi: 10.1093/emboj/18.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner MD, Arvan P. Protein traffic from the secretory pathway to the endosomal system in pancreatic beta-cells. J. Biol. Chem. 2000;275:14025–14030. doi: 10.1074/jbc.275.19.14025. [DOI] [PubMed] [Google Scholar]
- 32.Platta HW, Stenmark H. Endocytosis and signaling. Curr. Opin. Cell Biol. 2011;23:393–403. doi: 10.1016/j.ceb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awata T, et al. Association of type 1 diabetes with two Loci on 12q13 and 16p13 and the influence coexisting thyroid autoimmunity in Japanese. J. Clin. Endocrinol. Metab. 2009;94:231–235. doi: 10.1210/jc.2008-0718. [DOI] [PubMed] [Google Scholar]
- 35.Hakonarson H, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, et al. The novel endosomal membrane protein Ema interacts with the class C Vps-HOPS complex to promote endosomal maturation. J. Cell Biol. 2010;188:717–734. doi: 10.1083/jcb.200911126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miettinen P, et al. EGF receptor in pancreatic beta-cell mass regulation. Biochem. Soc. Trans. 2008;36:280–285. doi: 10.1042/BST0360280. [DOI] [PubMed] [Google Scholar]
- 38.Goulley J, et al. BMP4-BMPR1A signaling in beta cells is required for and augments glucose-stimulated insulin secretion. Cell Metab. 2007;5:207–219. doi: 10.1016/j.cmet.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Faculty Synopses. Monogenic disorders of insulin secretion: congenital hyperinsulinism and neonatal diabetes, March 15–16, 2012. Pediatr. Diabetes. 2012;13:337–361. [Google Scholar]
- 40.Kim S, et al. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1072–E1081. doi: 10.1073/pnas.1120320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen ZF, et al. The double-edged effect of autophagy in pancreatic beta cells and diabetes. Autophagy. 2011;7:12–16. doi: 10.4161/auto.7.1.13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 44.Velazquez L, et al. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. [Google Scholar]
- 45.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 46.Keene KL, et al. Evidence for two independent associations with type 1 diabetes at the 12q13 locus. Genes Immun. 2012;13:66–70. doi: 10.1038/gene.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erickson SL, et al. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 48.Kritzik MR, et al. Expression of ErbB receptors during pancreatic islet development and regrowth. J. Endocrinol. 2000;165:67–77. doi: 10.1677/joe.0.1650067. [DOI] [PubMed] [Google Scholar]
- 49.Coppieters KT, et al. Immunology in the clinic review series: focus on type 1 diabetes and viruses: the role of viruses in type 1 diabetes: a difficult dilemma. Clin. Exp. Immunol. 2012;168:5–11. doi: 10.1111/j.1365-2249.2011.04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Dogusan Z, et al. Double-stranded RNA induces pancreatic beta-cell apoptosis by activation of the toll-like receptor 3 and interferon regulatory factor 3 pathways. Diabetes. 2008;57:1236–1245. doi: 10.2337/db07-0844. [DOI] [PubMed] [Google Scholar]
- 52.Garcia M, et al. Regulation and function of the cytosolic viral RNA sensor RIG-I in pancreatic beta cells. Biochim. Biophys. Acta. 2009;1793:1768–1775. doi: 10.1016/j.bbamcr.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Colli ML, et al. MDA5 and PTPN2, two candidate genes for type 1 diabetes, modify pancreatic beta-cell responses to the viral by-product double-stranded RNA. Hum. Mol. Genet. 2010;19:135–146. doi: 10.1093/hmg/ddp474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nejentsev S, et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Downes K, et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS ONE. 2010;5:e12646. doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore F, et al. PTPN2, a candidate gene for type 1 diabetes, modulates interferon-gamma-induced pancreatic beta-cell apoptosis. Diabetes. 2009;58:1283–1291. doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santin I, et al. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic beta-cell apoptosis via regulation of the BH3-only protein Bim. Diabetes. 2011;60:3279–3288. doi: 10.2337/db11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melloul D. Role of NF-kappaB in beta-cell death. Biochem. Soc. Trans. 2008;36:334–339. doi: 10.1042/BST0360334. [DOI] [PubMed] [Google Scholar]
- 59.Mercurio F, et al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 60.Palombella VJ, et al. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 61.Eldor R, et al. Conditional and specific NF-kappaB blockade protects pancreatic beta cells from diabetogenic agents. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5072–5077. doi: 10.1073/pnas.0508166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S, et al. NF-kappa B prevents beta cell death and autoimmune diabetes in NOD mice. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1913–1918. doi: 10.1073/pnas.0610690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Norlin S, et al. Nuclear factor-κB activity in β-cells is required for glucose-stimulated insulin secretion. Diabetes. 2005;54:125–132. doi: 10.2337/diabetes.54.1.125. [DOI] [PubMed] [Google Scholar]
- 64.Sarkar SA, et al. Cytokine-mediated induction of anti-apoptotic genes that are linked to nuclear factor kappa-B (NF-kappaB) signalling in human islets and in a mouse beta cell line. Diabetologia. 2009;52:1092–1101. doi: 10.1007/s00125-009-1331-x. [DOI] [PubMed] [Google Scholar]
- 65.Liuwantara D, et al. Nuclear factor-kappaB regulates beta-cell death: a critical role for A20 in beta-cell protection. Diabetes. 2006;55:2491–2501. doi: 10.2337/db06-0142. [DOI] [PubMed] [Google Scholar]
- 66.Grey ST, et al. A20 inhibits cytokine-induced apoptosis and nuclear factor kappaB-dependent gene activation in islets. J. Exp. Med. 1999;190:1135–1146. doi: 10.1084/jem.190.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grey ST, et al. Genetic engineering of a suboptimal islet graft with A20 preserves beta cell mass and function. J. Immunol. 2003;170:6250–6256. doi: 10.4049/jimmunol.170.12.6250. [DOI] [PubMed] [Google Scholar]
- 68.Bergholdt R, et al. Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression. Diabetes. 2012;61:954–962. doi: 10.2337/db11-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berchtold LA, et al. Huntingtin-interacting protein 14 is a type 1 diabetes candidate protein regulating insulin secretion and beta-cell apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E681–E688. doi: 10.1073/pnas.1104384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karlsen AE, et al. Immune-mediated beta-cell destruction in vitro and in vivo-A pivotal role for galectin-3. Biochem. Biophys. Res. Commun. 2006;344:406–415. doi: 10.1016/j.bbrc.2006.03.105. [DOI] [PubMed] [Google Scholar]
- 71.Bergholdt R, et al. Transcriptional profiling of type 1 diabetes genes on chromosome 21 in a rat beta-cell line and human pancreatic islets. Genes Immun. 2007;8:232–238. doi: 10.1038/sj.gene.6364379. [DOI] [PubMed] [Google Scholar]
- 72.Rashid MA, et al. Carbonyl reductase 1 protects pancreatic beta-cells against oxidative stress-induced apoptosis in glucotoxicity and glucolipotoxicity. Free Radic. Biol. Med. 2010;49:1522–1533. doi: 10.1016/j.freeradbiomed.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 73.Suizu F, et al. The E3 ligase TTC3 facilitates ubiquitination and degradation of phosphorylated Akt. Dev. Cell. 2009;17:800–810. doi: 10.1016/j.devcel.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 74.Roggli E, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruan Q, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12030–12035. doi: 10.1073/pnas.1101450108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roggli E, et al. Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61:1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brehm MA, et al. Advancing animal models of human type 1 diabetes by engraftment of functional human tissues in immunodeficient mice. Cold Spring Harb. Perspect. Med. 2012;2:a007757. doi: 10.1101/cshperspect.a007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nostro MCG, Keller G. Generation of beta cells from human pluripotent stem cells: potential for regenerative medicine. Semin. Cell Dev. Biol. 2012;23:701–710. doi: 10.1016/j.semcdb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Makino S, et al. Breeding of a non-obese, diabetic strain of mice. Jikken Dobutsu. 1980;29:1–13. doi: 10.1538/expanim1978.29.1_1. [DOI] [PubMed] [Google Scholar]
- 80.Rossini AA, et al. Spontaneous diabetes in the gnotobiotic BB/W rat. Diabetes. 1979;28:1031–1032. doi: 10.2337/diab.28.11.1031. [DOI] [PubMed] [Google Scholar]
- 81.van Belle TL, et al. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol. Rev. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 82.Shoda LK, et al. A comprehensive review of interventions in the NOD mouse and implications for translation. Immunity. 2005;23:115–126. doi: 10.1016/j.immuni.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 83.Ize-Ludlow D, et al. Progressive erosion of beta-cell function precedes the onset of hyperglycemia in the NOD mouse model of type 1diabetes. Diabetes. 2011;60:2086–2091. doi: 10.2337/db11-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marhfour I, et al. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55:2417–2420. doi: 10.1007/s00125-012-2604-3. [DOI] [PubMed] [Google Scholar]
- 85.Tersey SA, et al. Islet beta-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes. 2012;61:818–827. doi: 10.2337/db11-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Sullivan-Murphy B, Urano F. ER stress as a trigger for beta-cell dysfunction and autoimmunity in type 1 diabetes. Diabetes. 2012;61:780–781. doi: 10.2337/db12-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nerup J, et al. HL-A antigens and diabetes mellitus. Lancet. 1974;2:864–866. doi: 10.1016/s0140-6736(74)91201-x. [DOI] [PubMed] [Google Scholar]
- 88.Jahromi MM, Eisenbarth GS. Cellular and molecular pathogenesis of type 1A diabetes. Cell. Mol. Life Sci. 2007;64:865–872. doi: 10.1007/s00018-007-6469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Noble JA, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59:2972–2979. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marron MP, et al. Functional evidence for the mediation of diabetogenic T cell responses by HLA-A2.1 MHC class I molecules through transgenic expression in NOD mice. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13753–13758. doi: 10.1073/pnas.212221199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Foulis AK, et al. Aberrant expression of class II major histocompatibility complex molecules by B cells and hyperexpression of class I major histocompatibility complex molecules by insulin containing islets in type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 1987;30:333–343. doi: 10.1007/BF00299027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Foulis AK, et al. Immunoreactive alpha-interferon in insulin-secreting beta cells in type 1 diabetes mellitus. Lancet. 1987;2:1423–1427. doi: 10.1016/s0140-6736(87)91128-7. [DOI] [PubMed] [Google Scholar]
- 93.Pujol-Borrell R, et al. Differential expression and regulation of MHC products in the endocrine and exocrine cells of the human pancreas. Clin. Exp. Immunol. 1986;65:128–139. [PMC free article] [PubMed] [Google Scholar]
- 94.Stewart TA, et al. Induction of type I diabetes by interferon-alpha in transgenic mice. Science. 1993;260:1942–1946. doi: 10.1126/science.8100367. [DOI] [PubMed] [Google Scholar]
- 95.Coppieters KT, et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med. 2012;209:51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pavlovic D, et al. Effect of interferon-gamma and glucose on major histocompatibility complex class I and class II expression by pancreatic beta- and non-beta-cells. J. Clin. Endocrinol. Metab. 1997;82:2329–2336. doi: 10.1210/jcem.82.7.4055. [DOI] [PubMed] [Google Scholar]
- 97.In’t Veld P. Insulitis in human type 1 diabetes: the quest for an elusive lesion. Islets. 2011;3:131–138. doi: 10.4161/isl.3.4.15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.In’t Veld P, et al. Screening for insulitis in adult autoantibody-positive organ donors. Diabetes. 2007;56:2400–2404. doi: 10.2337/db07-0416. [DOI] [PubMed] [Google Scholar]
- 99.Keenan HA, et al. Residual insulin production and pancreatic ss-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59:2846–2853. doi: 10.2337/db10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sherry NA, et al. Effects of autoimmunity and immune therapy on beta-cell turnover in type 1 diabetes. Diabetes. 2006;55:3238–3245. doi: 10.2337/db05-1034. [DOI] [PubMed] [Google Scholar]