Abstract
The protozoan parasites Trypanosoma brucei spp. cause important human and livestock diseases in sub Saharan Africa. In the mammalian blood, two developmental forms of the parasite exist: proliferative ‘slender’ forms and arrested ‘stumpy’ forms that are responsible for transmission to tsetse flies. The slender to stumpy differentiation is a density-dependent response that resembles quorum sensing (QS) in microbial systems and is crucial for the parasite life cycle, ensuring both infection chronicity and disease transmission1. This response is triggered by an elusive ‘stumpy induction factor’ (SIF) whose intracellular signaling pathway is also uncharacterized. Laboratory-adapted (monomorphic) trypanosome strains respond inefficiently to SIF but can generate forms with stumpy characteristics when exposed to cell permeable cAMP and AMP analogues. Exploiting this, we have used a genome-wide RNAi library screen to identify the signaling components driving stumpy formation. In separate screens, monomorphic parasites were exposed to 8-(4-chlorophenylthio)-cAMP (pCPTcAMP) or 8-pCPT-2′-O-Me-5′-AMP to select cells that were unresponsive to these signals and hence remained proliferative. Genome-wide ion torrent-based RNA interference Target sequencing identified cohorts of genes implicated in each step of the signaling pathway, from purine metabolism, through signal transducers (kinases, phosphatases) to gene expression regulators. Genes at each step were independently validated in cells naturally capable of stumpy formation, confirming their role in density sensing in vivo, whilst the putative RNA-binding protein, RBP7, was required for normal QS and promoted cell-cycle arrest and transmission competence when overexpressed. This study reveals that QS signaling in trypanosomes shares similarities to fundamental quiescence pathways in eukaryotic cells, its components providing targets for QS-interference based therapeutics.
Protozoan parasites undergo developmental responses to adapt to the different environments encountered within their mammalian host, or during passage through their arthropod vectors2-4. As a preparation for transmission, specialized developmental forms are often generated to promote survival when ingested by a biting insect1,5. The abundance of these transmission stages can fluctuate during the course of a blood parasitaemia as can the abundance of the proliferative forms that sustain the infection. The balance of these different cell types determines the within-host dynamics of a parasite, ensuring that the population can maximize its longevity within a host, but also optimize its capacity for spread to new hosts6-8.
African trypanosomes, Trypanosoma brucei spp., are extracellular parasites responsible for Human African Trypanosomiasis (HAT) and the livestock disease ‘nagana’9. In the bloodstream, trypanosomes proliferate as morphologically ‘slender’ forms that evade host immunity by antigenic variation, generating characteristic waves of infection. As each wave of parasitaemia ascends, slender forms stop proliferating and undergo morphological and molecular transformation to stumpy forms, the parasite’s transmission stage10,11. This differentiation is parasite density-dependent12, resembling quorum-sensing systems common in microbial communities13. However, the differentiation-inducing factor SIF (‘stumpy induction factor’) is unidentified and, whilst some inhibitors of development have been identified14-16, the signal-response pathway that promotes stumpy formation is uncharacterised. Moreover, density-sensing is reduced in laboratory-adapted ‘monomorphic’ parasite strains16 although they can undergo cell-cycle arrest and the limited expression of some stumpy-specific genes when exposed to cell permeable analogues of cAMP or AMP16-18. This is distinct from cAMP-based signaling since only hydrolysable cAMP drives development, which is metabolized to AMP in the parasite17,19.
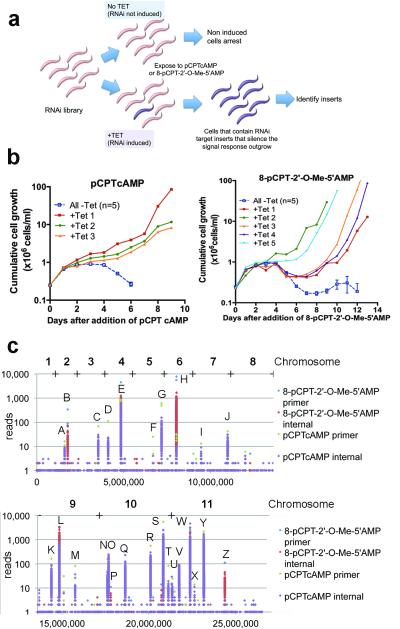
The availability of monomorphic parasite RNAi libraries capable of tetracycline-inducible gene silencing on a genome-wide scale20 and their ability to respond to hydrolysable-cAMP and AMP analogues allowed us to investigate genes that regulate stumpy formation. Thus, an RNAi library population of 2.5 ×107 cells (maintained with ~5 fold genome coverage) was selected with 100 μM pCPTcAMP or 10 μM 8-pCPT-2′-O-Me-5′-AMP17 in several replicate flasks, RNAi being induced, or not, with tetracycline (Figure 1a). Uninduced populations underwent division arrest and eventual death over 5 days (Figure 1b), whereas, three pCPTcAMP-selected and five 8-pCPT-2′-O-Me-5′-AMP-selected populations outgrew in the RNAi-induced populations, these being subject to DNA isolation and RNAi insert amplification (Figure 1b; Supp. Figure 1a). The resulting amplicon profiles varied in intensity but there was remarkable similarity between independently-selected populations under each regimen (Supp. Figure 1a). To analyse the amplicon complexity in depth, populations from each screen were subjected to Ion torrent™ sequencing21. Reads were aligned to the T. brucei TREU 927/4 reference genome (www.genedb.org), identifying 43 genes potentially targeted in either screen (Figure 1c; Supp. Dataset 1; Supp. Table 1). Twelve genes were common to both screens, 5 were 8-pCPT-2′-O-Me-5′-AMP-specific and 26 were pCPTcAMP-specific, likely reflecting the observed complexity in each amplicon population (Supp. Table 1). Analysing the reads for genome alignment and for the presence of the appropriate RNAi library primer flanks refined the list to 27-30 distinct gene targets (Supp. Table 2, Supp. Table 3, Supp. Figure 1b, Supp. Dataset 2).
Figure 1. Identification of trypanosome QS regulators.
a. Selection for genes whose RNAi silencing renders trypanosomes resistant to pCPTcAMP or 8-pCPT-2′-O-Me-5′-AMP, identifying molecules that promote stumpy formation.
b. RNAi libaries were exposed to pCPTcAMP or 8-pCPT-2′-O-Me-5′-AMP, RNAi being induced (1 μg/ml tetracycline), or not. The curves for uninduced samples are combined for clarity (mean ±s.e.m, n=5).
c. Ion Torrent™ read-density from the selected parasites aligned to the trypanosome genome. Since amplicons were fragmented prior to sequencing, reads with and without the flanking primers are shown.
As expected genes encoding enzymes involved in cAMP/AMP-analogue processing and cellular purine balance were identified, with six selected RNAi targets predicted to alter intracellular AMP levels (Supp. Figure 2a). For example, 8-pCPT-2′-O-Me-5′-AMP is converted to 8-pCPT-2′-O-Me-5′-adenosine in culture medium17, such that RNAi against adenosine kinase would prevent the conversion of the transported pCPT-adenosine analogue to its AMP equivalent22 (Supp. Figure 2a). Similarly depletion of adenylosuccinate synthetase (ADSS) and adenylosuccinate lyase (ADSL) reduce the conversion of IMP to AMP, potentially counteracting the effect of the membrane permeable cAMP or AMP analogs. The identified adenylate kinase, GMP synthase and IMP dehydrogenase targets are also predicted to rebalance purine levels within the parasites.
Signal transduction pathway genes were also unambiguously targeted by RNAi in the selected populations (Supp. Table 2; Supp. Table 3; Supp. Datasets 2 and 3). Potentially linking AMP balance to downstream cellular effects, an AMPK/SNF1/KIN11 homologue target (Tb927.3.4560) was identified, as was a MEK kinase (Tb927.2.2720), and predicted cell-cycle regulators of the NEK kinase (Tb927.10.5930/40/50; these three tandem genes being indistinguishable by RNAi phenotyping) and Dyrk/YAK kinase (Tb927.10.15020) families, the latter being required for cellular quiescence (G0) in yeast23 and Dictyostelium24. A dual-specificity phosphatase (Tb927.7.7160) and members of the protein phosphatase 1 gene family (PP1-4, PP1-5, PP1-625; Tb927.4.3620/30/40) were also selected, genes whose knock-down generates only limited cell growth defects in proliferative procyclic forms25. Finally regulatory and effector molecules were represented by the RBP7 RNA binding proteins (RBP7A and RBP7B, Tb927.10.12090/12100, indistinguishable by RNAi), whereas a number of hypothetical proteins with no detectable homologies to any gene were also identified. Overall, this suggested that representatives at many stages in the pCPTcAMP/8-pCPT-2′-O-Me-5′-AMP response pathway had been selected, from signal processing, through signal transduction to regulatory effector molecules.
To validate the identified genes, independent monomorphic and pleomorphic cell lines were initially generated targeting 12 discrete members (Supp. Table2). Nine genes were analysed in detail (Supp. Figures 2b, c), these representing different steps (‘signal processing’, ‘signal transduction’, ‘effector molecules’) in the predicted signal response pathway, and two hypothetical proteins (HYP1, Tb927.11.6600; HYP2, Tb927.9.4080) of unknown function, although HYP2 has a DksA zinc finger motif involved in prokaryotic rRNA transcriptional responses to nutritional status and quorum sensing. Several generated a growth inhibition when targeted by RNAi indicating roles in other important cellular processes (PP1, HYP1, ADSL, ADSS; Supp. Table 2; Supp. Figure 2b, 2c), although for ADSS this was alleviated in monomorphs by pCPTcAMP, likely due to the restoration of the purine balance by the analog (Supp. Figure 2c). Several targeted genes showed evidence of increased resistance to pCPTcAMP-mediated growth inhibition validating their selection in vitro (PP1, NEK, YAK, RBP7A/B, HYP 2, DS-PHOS; Supp. Figure 2b).
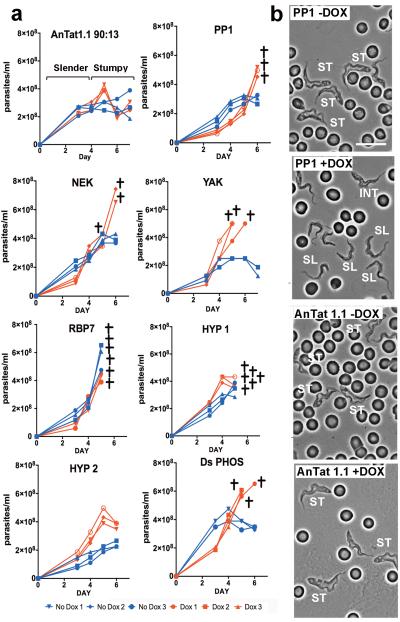
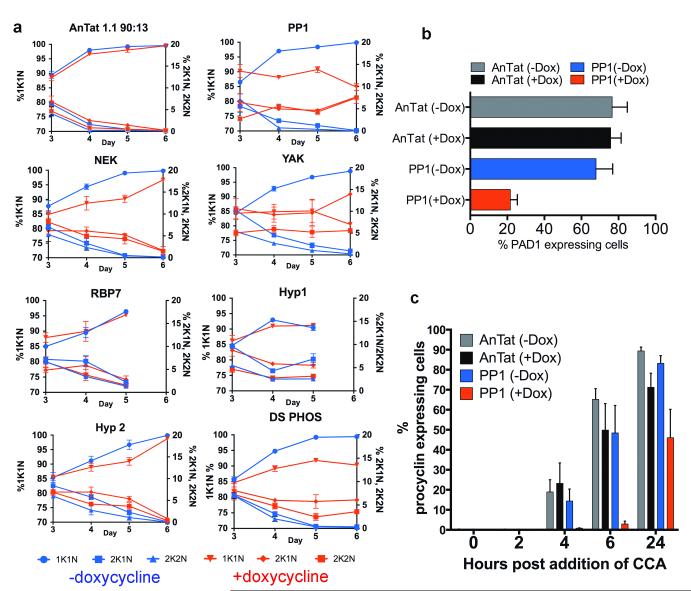
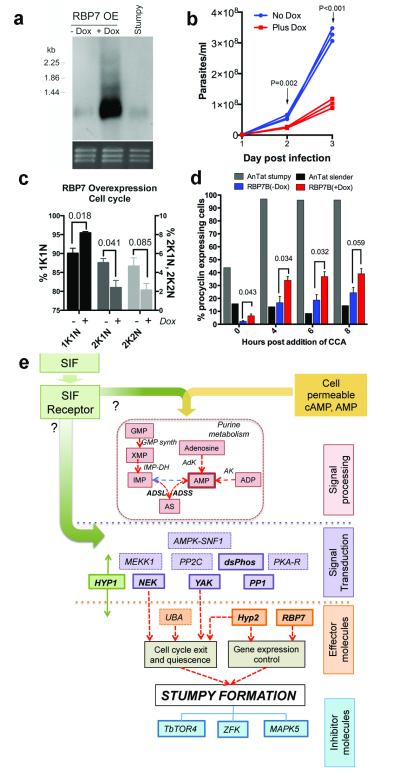
To analyse the physiological relevance of the identified genes in developmental quorum-sensing, nine pleomorphic RNAi cell lines were investigated for their response to the SIF signal in vivo. Figure 2 shows that the parental AnTat 1.1 90:13 line generated a highly-enriched population of arrested stumpy forms, these accumulating from day 4 onwards (Figure 2a). For ADSL and ADSS parasitaemias were strongly suppressed for at least 5 days, matching their growth characteristics in vitro (Supp. Figure 2c, 3). In contrast, all of the other target RNAi lines exhibited abrogated or delayed stumpy formation over 4-6 days, with mice requiring sacrifice at the exceptionally high parasitaemias generated in the case of PP1, NEK, YAK and DS-PHOS (Figure 2a). The induced parasites also retained a slender morphology or showed delayed progression to stumpy morphology compared to the control or uninduced infections (Figure 2b and Supp. Figure 4a). In two cases (‘RBP7′, ‘HYP1′), the cell lines generated elevated growth when uninduced, reflecting leaky RNAi for these lines (Supp. Figure 4b). Such leaky RNAi would be positively selected for genes involved in density-dependent cell-cycle arrest. Confirming the reduction of stumpy formation in each cell line in vivo, cell-cycle analysis during the course of the parasitaemias revealed reduced accumulation of cells with a 1 kinetoplast and 1 nucleus (1K1N) configuration, indicating loss or a delay of the cell-cycle arrest in G1/G0 characteristic of stumpy forms (Figure 3a). Detailed analysis of the molecular characteristics of the populations confirmed that the parasites showed reduced or delayed expression of the stumpy-specific surface protein PAD126 (PP1 depleted cells are shown in Figure 3b and Supp. Figure 5a) as well as reduced mitochondrial elaboration (Supp. Figure 5b). As expected for a slender-enriched population, the PP1-depleted cells were less able to differentiate to procyclic forms after exposure to cis-aconitate (CCA), indicated by reduced procyclin expression (Figure 3c; Supp. Fig. 5c; Supp. Figure 6a) and kinetoplast repositioning (Supp. Figure 6b). Hence, by the key parameters, RNAi targeting of genes predicted to operate at different steps in a signaling pathway prevented or delayed stumpy formation in vivo, confirming their involvement in physiological quorum sensing. With respect to an effector function, the overexpression of RBP7B (Tb927.10.12100) (Figure 4a) promoted premature cell-cycle arrest (Figure 4b, c; Supp. Figure 7) and increased capacity for differentiation to procyclic forms in pleomorphic lines (Figure 4d) albeit incompletely in the population, similar to the RBP6-mediated regulation of development in tsetse forms27. Transcriptome analysis revealed few widespread changes in gene expression upon perturbed RBP7 expression (Supp. Figure 8 and 9; Supp. Dataset 4), although RNA regulators and procyclin transcripts were elevated in RBP7 over-expressing cells whilst histones were downregulated compared to RBP7 depleted cells (Supp. Figure 9; Supp. Data set 4), consistent with their cell cycle arrest and differentiation competence.
Figure 2. RNAi to the identified genes prevents growth control and morphological transformation.
a. In vivo growth of pleomorphic RNAi lines targeting distinct genes identified from the genome-wide screen for QS-signal resistance. RNAi was induced by provision of doxycyline to the infected animals (n=3, red lines), with parallel infections remaining uninduced (n=3, blue lines). Infections were terminated when the ascending parasitaemias were predicted to become lethal within 12 hours (crosses).
b.Morphology of PP1 RNAi cells and parental T. brucei AnTat1.1 90:13 cells (each grown in mice ±doxycycline) at 6 days post-infection. The induced PP1 cells remained predominently slender in morphology. Cells with slender (SL), intermediate (INT) or stumpy (ST) morphology are labelled. Bar=15μm.
Figure 3. Silencing the identified genes reduces G1 arrest and differentiation competence.
a. Cell-cycle status of pleomorphic RNAi lines. n=6; mean ±s.e.m. percentage 1 kinetoplast (K), 1 nucleus (N) (G0/G1, plus S-phase cells; left Y-axis), 2K1N (G2-phase cells) or 2K2N (post-mitotic cells) (both right Y-axis) are shown. Test genes showed a signficant difference (GLMM, p<0.001) in comparison to AnTat1.1 90:13 +doxycycline on at least one day of infection.
b. PAD1 expression on day 6 post-infection (n=3/group, mean ±s.e.m.). PP1 RNAi cells show reduced PAD1 expression (GLM, F1,4=22.35, p=0.009).
c. PP1-depleted cells show signficantly reduced Procyclin expression during differentiation (GLM, F1,4=10.87, p=0.030) Bars represents mean ±s.e.m; n=3.
Figure 4. RBP7 drives cell cycle arrest and differentiation competence.
a. Inducible overexpression of RBP7B mRNA on day 3 post-infection. Stumpy RNA is also shown; Ethidium bromide stained rRNA indicates loading.
b. AnTat1.1 90:13 induced with doxycycline (red lines) to overexpress RBP7B show reduced parasitaemia in mice (n=3 per group). GLM, F1,4=55.25, p=0.002 and F1,4=233.1, p<0.001 on day 2 and 3, respectively.
c. Cells accumulate in G1 upon RBP7B ectopic expression. Values shown are mean ±s.e.m at day 3 post-infection; n=3 per group. GLM, F1,4=15.1, p=0.018 (1K1N); F1,4=8.9, p=0.041 (2K1N); F1,4=5.17, p=0.085 (2K2N).
d. Parasites isolated on day 3 post-infection were exposed to 6mM cis aconitate (CCA) and EP-procyclin expression monitored by flow cytometry. At 0h enhanced cold induction of EP procyclin expression is seen in the induced population. n=3; GLM; F1,4=8.54, p=0.043 (0h); F1,4=9.99, p=0.034 (4h); F1,4=10.36, p=0.032 (6h) F1,4=6.84 , p=0.059 (8h).
e. Schematic of the proposed SIF-signaling pathway in T. brucei. Major identified components (Supp. Table 2) are shown; those in bold italics are experimentally confirmed. The order and potential branching in the pathway is unknown as is the position of pathway inhibitors, TbTOR4 (Tb927.1.1930) ZFK (Tb927.11.9270) and MAPK5 (Tb927.6.4220).
By employing a stringent genome-wide in vitro selection, these experiments have provided a first identification of the molecules required to promote the development of trypanosome transmission stages in the mammalian bloodstream (Supp. Table 2 and Figure 4e). Although inhibitors of stumpy formation14-16 would not be identified in the screens, the reproducibility of the enriched amplicons in independent selections and the use of Ion Torrent™-based deep-sequencing indicates that many genes involved in the promotion of stumpy formation have been identified. Moreover, this set is significantly enriched for genes whose RNAi reduces differentiation to procyclic forms in monomorphic lines (P=0.0008 X2 test; Supp. Figure 10), indicating that monomorphs need to progress through a stumpy-like (quiescent) form to differentiate to procyclic forms. Although the use of membrane-permeable analogues prevents identification of the SIF receptor at the parasite surface, our results reveal that selecting resistance to AMP analogues identifies purine salvage enzymes, but also genes important in the physiological SIF signaling pathway (Figure 4e). Interestingly, the AMPK/SNF1/KIN11 homologue (Tb927.3.4560) is a potential AMP/ATP energy sensor that could inhibit TbTORC4, whose activity is proposed to prevent stumpy formation16. Combined with the confirmation of downstream transduction components such as PP1, NEK, YAK kinase and a dual-specificity phosphatase our analyses reveal that QS-signaled production of stumpy forms in trypanosomes shares components with quiescence regulation in mammalian stem cells28 and the starvation responses and developmental transitions of unicellular eukaryotes29.
This assembly of the molecular regulators of quorum-sensing provides the first detailed catalogue of an environmental signaling pathway in trypanosomes, providing molecular insight into microbial sociality relevant to both virulence and transmission in a major eukaryotic pathogen. As drivers of the irreversible arrest of stumpy forms in the mammalian bloodstream these molecules also represent novel therapeutic targets via quorum sensing interference30, whose pharmacological activation would generate a stringent anti-virulence effect.
Methods
RNAi screens
The RNAi library was removed from liquid Nitrogen and added to 10% HMI-9 containing 2 μg/ml blasticidin and 1 μg/ml phleomycin to maintain transfectant selection. The library was divided into three cultures of 2.5×107cells at 2.5×105cells/ml and 1μg/ml tetracycline added to one to induce dsRNA. 24hours after induction two cultures were maintained at 5×107cells at 2.5×105cells/ml, to ensure full genome coverage, in medium containing 10μM 8pCPT-2-O’-Me-5′AMP or 100μM 8-(4-chlorophenylthio)-cAMP. DNA was harvested from the third culture without drug addition (“before experiment”; BE). Both treated cultures were divided into five smaller volumes (1×107cells in each) for optimum growth conditions. Cell counts were made every 24hours using the Beckman Z2 coulter particle count & size analyser or haemocytometer as appropriate. Proliferating cultures were split to 2.5×105cells/ml when necessary.
Tetracycline and 10 μM 8pCPT-2-O’-Me-5′AMP or 100 μM 8-(4-chlorophenylthio)-cAMP were washed from resistant cultures. 1.25×107cells were aliquoted to a 50ml falcon tube and centrifuged at 2000rpm for 10minutes. The supernatant was removed and pellet re-suspended in 50ml fresh 20%HMI-9. Washed cells were incubated at 37°C for a few days, passaging back to 2.5×105cells/ml when necessary; before freezing as above at −80°C.
Genomic DNA extraction
Genomic DNA was harvested from resistant cultures. After cells were aliquoted for freezing and washing the remaining culture was divided between two falcon tubes, centrifuged at 2000rpm for 10minutes and the supernatant poured off. The pellets were re-suspended, mixed together and the resulting solution divided between six 1.5ml eppendorfs. Genomic DNA was extracted using a Qiagen DNeasy blood and tissue kit into six columns, eluting into 150 μl and stored at −20 °C.
PCR was carried out on genomic DNA from resistant cultures and Escherichia coli transformed with pGEM-T-Easy plasmids or the amplicons subject to Ion Torrent™ sequencing. All PCRs were carried out in 0.2ml PCR tubes using the Thermo Electron Corporation SPRINT thermal cycler. Typical 50 μl PCR reaction:10 μl 5xFlexibuffer, 4 μl 2.5 μM dNTPs, 3 μl 25 mM MgCl2, 2 μl forward and reverse primers, 0.5 μl Taq polymerase, 0.5 μl DNA sample and 27.5 μl dH2O. Typical PCR conditions:95°c for 5minutes, 30 cycles of (95°C for 30 seconds, 55°C for 45seconds and 72°C for 2minutes), followed by 72°C for 4minutes. The primers used are shown below.
| Lib2 Forward |
TAGCCCCTCGAGGGCCAGT | RNAi insert specific: for amplifying RNAi inserts from genomic DNA |
| Lib2 Reverse |
GGAATTCGATATCAAGCTTGGC | |
| M13 Forward |
GTAAAACGACGGCCAGTG | pGEM-T-Easy vector specific: for amplifying or sequencing RNAi inserts ligated into pGEM-T-Easy |
| M13 Reverse |
GGAAACAGCTATGACCATG |
Primer sequences used in PCR reactions.
Validation through independent RNAi constructs
RNAi target gene fragments were selected based on default settings of the RNAit software31. They were amplified from AnTat 1.1 90:1332 genomic DNA (for pleomorph RNAi lines) using a forward primer carrying flanking BamHI and HindIII or Nde sites and a reverse primer carrying flanking XhoI and XbaI sites for cloning into the stem-loop pALC14 plasmid33. The constructs were linearized with NotI and transfected into AnTat 1.1 90:13 cells by electroporation. Stable transfectants were selected in the presence of 1 μg/ml puromycin using the methods described in MacGregor et al.(2013)34
For the monomorph RNAi lines, the target fragments were amplified using forward primer carrying flanking XbaI and BamHI and reverse primer carrying XhoI and SmaI sites for cloning into the stem loop pRPaiSL (MCS 1/2) plasmid35. The constructs were linearised with AscI and transfected into 2T1 cells by electroporation. Stable transfectants were selected in the presence of 2.5 μg/ml hygromycin.
For overexpression of RBP7B, the coding region was amplified and inserted into the expression vector pDEX-577-Y 36. Several independent cell lines were isolated and their growth analysed in vitro or in vivo in the presence or absence of tetracycline, or doxycycline, respectively.
Ion Torrent™ sequencing and bioinformatics analysis
Ion Torrent™ sequencing was carried out at the University of Edinburgh Wellcome Trust Clinical Research Facility Genetics Core. For library preparation, each sample was assigned an Ion Xpress barcode, Xpress Barcode 1 or Xpress barcode 2 and samples were run on a 316D chip, generating 2,801,839 total reads, with each sample (pCPT-cAMP, 8pCPT-2-O’-Me-5′AMP screen outputs) generating >1.3 million reads.
The quality of raw sequence data was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cutadapt software was used to identify and remove primer sequences (parameters: -O 10 -m 25 -n 3 -q 20; version: 1.0); this in effect partitioned the reads into those that had a given primer (e.g. “Lib2F, trimmed”) and those that did not (e.g. Lib2F, not trimmed”). Partitioning was done for each primer for each sample. Sequences were aligned to the Trypanosoma brucei brucei genome (obtained from ftp.sanger.ac.uk/pub4/pathogens/Trypanosoma/brucei/Latest_Whole_Genome_Sequence/Tb927_WGS_24_08_2012/chromosomes/) using bowtie2 (parameters: --very-sensitive; version: 2.0.2), piped through samtools (parameters: -bhS; version: 0.1.18), and stored in indexed BAM format. Read depths were calculated using simple scripts.
The annotated T. b. brucei genome was viewed using Artemis software (http://www.sanger.ac.uk/resources/software/artemis/ ; http://ukpmc.ac.uk/abstract/MED/11120685), and coding segment (CDS) region coordinates extracted prior to conversion to bedtools bedfile format using simple scripts. Overlap analyses, to determine reads mapping to CDS regions (“gene-based” tallies), were performed using the “GenomicFeatures” Bioconductor package (www.bioconductor.org).
Transcriptome analysis
Quality of sequence data provided by BGI Hong Kong was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). No additional processing of the primary data was required.
Sequence alignment to the T. brucei brucei genome
Paired end sequences were aligned to the Trypanosoma brucei genome (obtained from ftp.sanger.ac.uk/pub4/pathogens/Trypanosoma/brucei/Latest_Whole_Genome_Sequence/Tb927_WGS_24_08_2012/chromosomes/ ) using bowtie2 (parameters: --very-sensitive-local; version: 2.0.2), piped through samtools (parameters: -bhS; version: 0.1.18), and stored in indexed BAM format.
Alignment to protein-coding regions
The annotated T. b. brucei genome was viewed using Artemis software (http://www.sanger.ac.uk/resources/software/artemis/ ; http://ukpmc.ac.uk/abstract/MED/11120685), and coding segment (CDS) region coordinates extracted prior to conversion to bedfile file format. Bedtools (parameters: multicov -bams ; version: bedtools 2.15.0) was used to generate coverages for each CDS for each sample replicate. RPKM-like values were calculated by dividing coverage by CDS size.
Pair-wise comparisons of the samples
Statistical analyses of the sample groups were undertaken in the R environment using Bioconductor packages. Differential expression was explored using linear models and empirical Bayes methods, using the limma Bioconductor package37. RPKM-like values were offset by 1, and logged. Prior to quantile normalisation and groupwise comparisons, the data were filtered to remove loci whose mean values were below the 10% quantile for all samples.
Northern blotting
RNA preparation and analysis was carried out as described38
Antibodies
Antibodies used in this study were: Anti-EP Procyclin, sourced from Cedar Lane, Canada (cat. no. CLP001A) Anti-PAD1, as detailed in Dean et al, (2009)26
In vitro pCPTcAMP resistance validation
Pleomorphic cells were seeded at 1 ×105 cells/ml. RNAi was induced in one flask (1 μg/ml doxycycline) while the other was left uninduced. After 24 hours, each flask was split, one being exposed to 100 μM of pCPTcAMP (Sigma). Assays were performed in triplicate, growth being monitored every 24 hours. Cells were maintained at ≤ 1 ×106 cells/ml using HMI-9, supplementing with doxycycline and pCPTcAMP, where needed.
Cell cycle analysis
Methanol-fixed blood smears were re-hydrated in PBS for 10 min and stained with DAPI (100 ng/ml) for 5 minutes. 250 cells were analysed per slide.
In vitro differentiation to procyclic forms
Parasites were incubated at 3 ×106 cells/ml in SDM79 with 6mM cis-aconitate (CCA), 27°C. Samples were collected for flow cytometry at 0-24 hours.
Flow cytometry
Approximately 3 ×106 cells were 2% formaldehyde/0.05% Glutaraldehyde-fixed for >1 h at 4 °C. Subsequently, the cell suspension was pelleted, washed twice with PBS and re-suspended in 200μl mouse α-EP-procyclin (Cedar Lane, cat. no. CLP001A; 1:500) or rabbit α-PAD126 (1:100). After washing and staining with secondary antibody (1:1000 α mouse-FITC and α-rabbit-Cy5) cells were analysed using a Becton Dickenson LSRII Flow cytometer and analysed using FlowJO software (Tree Star Inc.). Unstained cells and secondary antibody-only stained cells provided negative controls.
Mitotracker assays
Bloodstream-form trypanosomes (2 ×106 /ml) were incubated in HMI-medium containing 100 nM Mitotracker Red CMXROS (Molecular Probes) for 30 min at 37°C. Then the cells were washed with HMI-9 and incubated for a further 20 min in the absence of Mitotracker, after which the parasites were fixed for 2 min at 4°C with 0.4% paraformaldehyde (prepared fresh in PBS). The cells were then washed once with PBS and air-dried smears were prepared. The slides were fixed for 10 min in methanol at 20°C, before rehydration for 10 min in PBS, followed by DAPI staining and mounting in MOWIOL.
Statistical analyses and animal studies
Animal experiments were carried out according to the UK Animals (Scientific) Procedures act under a licence (60/4373) issued by the UK Home Office and approved by the University of Edinburgh Local ethical committee.
Animal groups were selected from age and sex matched MF1 mice, with comparisons plus or minus doxycycline carried out on mice from the same batch randomly allocated.
Six female age-matched cyclophosphamide-treated MF1 mice were inoculated i.p. One group (n=3) was provided with doxycycline (200 μg/ml in 5% sucrose) in their drinking water immediately pre-inoculation, the other group (n=3) received 5% sucrose only. Parasitaemias were scored over 5-7 days with humane end-points conforming to UK Home Office requirements.
Three mice per group were used to generate data for statistical analysis. Preliminary analyses suggested that this sample size would be sufficient to detect differences between cell lines and treatment groups (i.e. plus/minus doxycycline). To analyse significance, data was collected on %1K1N values from the test genes and one control (Antat 1.1 90:13) with and without the presence of doxycycline. Blinding was not carried out. Data was analysed with Proc Mixed SAS version 9.3.1, SAS Institute Inc., Cary, NC or Minitab 16 Statistical Software, using using a general linear model (GLM), general linear mixed model (GLMM), or a X2 test. Data was examined prior to analysis to ensure normality and no transformations were required. P values of less than 0.05 were considered statistically significant. A Bonferroni correction was used for analysis of RNAi cell lines over multiple days (Figure 3), to account for multiple comparisons. No animals were excluded from the analysis, although humane end points were applied at peak parasitaemias for some groups where quorum sensing was lost, causing enhanced virulence.
Supplementary Material
Acknowledgements
This work was funded by a Wellcome Trust Programme grant (088293MA) to KM and by a Wellcome Trust strategic award (095831MA) to the Centre for Immunity, Infection and Evolution. We thank Dr Margo Chase–Topping for statistical support.
Footnotes
Data deposition Data in this submission has been submitted to the NCBI GEO database with accession code GSE46501
Competing financial Interests The authors declare no competing financial interests in relation to this work
Reprints and permissions information is available at www.nature.com/reprints
References
- 1.MacGregor P, Szoor B, Savill NJ, Matthews KR. Trypanosomal immune evasion, chronicity and transmission: an elegant balancing act. Nature reviews. 2012;10:431–438. doi: 10.1038/nrmicro2779. doi:10.1038/nrmicro2779 nrmicro2779 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldenberg S, Avila AR. Aspects of Trypanosoma cruzi stage differentiation. Advances in parasitology. 2011;75:285–305. doi: 10.1016/B978-0-12-385863-4.00013-7. doi:10.1016/B978-0-12-385863-4.00013-7B978-0-12-385863-4.00013-7 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Aly AS, Vaughan AM, Kappe SH. Malaria parasite development in the mosquito and infection of the mammalian host. Annual review of microbiology. 2009;63:195–221. doi: 10.1146/annurev.micro.091208.073403. doi:10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthews KR. Controlling and coordinating development in vector-transmitted parasites. Science. 2011;331:1149–1153. doi: 10.1126/science.1198077. doi:331/6021/1149 [pii] 10.1126/science.1198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker DA. Malaria gametocytogenesis. Mol Biochem Parasitol. 172:57–65. doi: 10.1016/j.molbiopara.2010.03.019. doi:S0166-6851(10)00070-8 [pii]10.1016/j.molbiopara.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank SA. Models of parasite virulence. The Quarterly review of biology. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 7.Gjini E, Haydon DT, Barry JD, Cobbold CA. Critical interplay between parasite differentiation, host immunity, and antigenic variation in trypanosome infections. The American naturalist. 2010;176:424–439. doi: 10.1086/656276. doi:10.1086/656276. [DOI] [PubMed] [Google Scholar]
- 8.Mackinnon MJ, Read AF. Virulence in malaria: an evolutionary viewpoint. Philosophical transactions of the Royal Society of London. 2004;359:965–986. doi: 10.1098/rstb.2003.1414. doi:10.1098/rstb.2003.1414 VKADVXQANEWR38DD [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett MP, Croft SL. Management of trypanosomiasis and leishmaniasis. British medical bulletin. 2012;104:175–196. doi: 10.1093/bmb/lds031. doi:10.1093/bmb/lds031 lds031 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson M. Notes on the polymorphism of Trypanosoma gambiense in the blood and its relation to the exogenous cycvle in Glossina palpalis. Proceedings of the Royal society B. 1912;85:241–539. [Google Scholar]
- 11.Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. British medical bulletin. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 12.Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. Journal of cell science. 1997;110(Pt 21):2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- 13.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annual review of cell and developmental biology. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. doi:10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 14.Vassella E, et al. Deletion of a novel protein kinase with PX and FYVE-related domains increases the rate of differentiation of Trypanosoma brucei. Mol Microbiol. 2001;41:33–46. doi: 10.1046/j.1365-2958.2001.02471.x. [DOI] [PubMed] [Google Scholar]
- 15.Domenicali Pfister D, et al. A Mitogen-activated protein kinase controls differentiation of bloodstream forms of Trypanosoma brucei. Eukaryot Cell. 2006;5:1126–1135. doi: 10.1128/EC.00094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barquilla A, et al. Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14399–14404. doi: 10.1073/pnas.1210465109. doi:10.1073/pnas.1210465109 1210465109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laxman S, Riechers A, Sadilek M, Schwede F, Beavo JA. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19194–19199. doi: 10.1073/pnas.0608971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacGregor P, Matthews KR. Identification of the regulatory elements controlling the transmission stage-specific gene expression of PAD1 in Trypanosoma brucei. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks533. doi:gks533 [pii]10.1093/nar/gks533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould MK, et al. Cyclic AMP Effectors in African Trypanosomes Revealed by Genome-Scale RNA Interference Library Screening for Resistance to the Phosphodiesterase Inhibitor CpdA. Antimicrobial agents and chemotherapy. 2013;57:4882–4893. doi: 10.1128/AAC.00508-13. doi:10.1128/AAC.00508-13 AAC.00508-13 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsford S, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome research. 2011 doi: 10.1101/gr.115089.110. doi:gr.115089.110 [pii] 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. doi:10.1038/nature10242 nature10242 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Vodnala M, et al. Adenosine kinase mediates high affinity adenosine salvage in Trypanosoma brucei. J Biol Chem. 2008;283:5380–5388. doi: 10.1074/jbc.M705603200. doi:10.1074/jbc.M705603200 M705603200 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Molecular and cellular biology. 1991;11:4045–4052. doi: 10.1128/mcb.11.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souza GM, Lu S, Kuspa A. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development. 1998;125:2291–2302. doi: 10.1242/dev.125.12.2291. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Tu X, Wang CC. Okadaic acid overcomes the blocked cell cycle caused by depleting Cdc2-related kinases in Trypanosoma brucei. Experimental cell research. 2006;312:3504–3516. doi: 10.1016/j.yexcr.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Dean SD, Marchetti R, Kirk K, Matthews K. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459:213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. doi:10.1126/science.1229641 338/6112/1352 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietras EM, Warr MR, Passegue E. Cell cycle regulation in hematopoietic stem cells. The Journal of cell biology. 2011;195:709–720. doi: 10.1083/jcb.201102131. doi:10.1083/jcb.201102131 jcb.201102131 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annual review of genetics. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. doi:10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 30.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. doi:10.1038/nrd3013 nrd3013 [pii] [DOI] [PubMed] [Google Scholar]
Methods References
- 31.Redmond S, Vadivelu J, Field MC. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol. 2003;128:115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 32.Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes & development. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Molecular and cellular biology. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacGregor P, Rojas F, Dean S, Matthews KR. Stable transformation of pleomorphic bloodstream form Trypanosoma brucei. Mol Biochem Parasitol. 2013 doi: 10.1016/j.molbiopara.2013.06.007. doi:S0166-6851(13)00092-3 [pii] 10.1016/j.molbiopara.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol Biochem Parasitol. 2005;144:142–148. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly S, et al. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. doi:S0166-6851(07)00099-0 [pii] 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 38.Tasker M, Wilson J, Sarkar M, Hendriks E, Matthews K. A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol Biol Cell. 2000;11:1905–1917. doi: 10.1091/mbc.11.5.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.