Abstract
Background
Plant non-specific lipid transfer proteins (nsLtps) are small, basic proteins encoded by multigene families and have reported functions in many physiological processes such as mediating phospholipid transfer, defense reactions against phytopathogens, the adaptation of plants to various environmental conditions, and sexual reproduction. To date, no genome-wide overview of the Brassica rapa nsLtp (BrnsLtp) gene family has been performed. Therefore, as the first step and as a helpful strategy to elucidate the functions of BrnsLtps, a genome-wide study for this gene family is necessary.
Methodology/Principal Finding
In this study, a total of 63 putative BrnsLtp genes were identified through a comprehensive in silico analysis of the whole genome of B. rapa. Based on the sequence similarities, these BrnsLtps was grouped into nine types (I, II, III, IV, V, VI, VIII, IX, and XI). There is no type VII nsLtps in B. rapa, and a new type, XI nsLtps, was identified in B. rapa. Furthermore, nine type II AtLtps have no homologous genes in B. rapa. Gene duplication analysis demonstrated that the conserved collinear block of each BrnsLtp is highly identical to those in Arabidopsis and that both segmental duplications and tandem duplications seem to play equal roles in the diversification of this gene family. Expression analysis indicated that 29 out of the 63 BrnsLtps showed specific expression patterns. After careful comparison and analysis, we hypothesize that some of the type I BrnsLtps may function like Arabidopsis pathogenesis-related-14 (PR-14) proteins to protect the plant from phytopathogen attack. Eleven BrnsLtps with inflorescence-specific expression may play important roles in sexual reproduction. Additionally, BrnsLtpI.3 may have functions similar to Arabidopsis LTP1.
Conclusions/Significance
The genome-wide identification, bioinformatic analysis and expression analysis of BrnsLtp genes should facilitate research of this gene family and polyploidy evolution and provide new insight towards elucidating their biological functions in plants.
Introduction
Lipids are important chemicals involved in many aspects of development and growth in plants. The most common examples are the surface layers, cutin and suberin. These structures are made up of hydrophobic polyesters of fatty acid derivatives and are known to be associated with biotic and abiotic stress. Additionally, the lipids deposited in storage organs, such as seeds and fruits, are a crucial energy source for the growing populations of the world. Lipids and their derivatives also play roles in many important cell-signaling pathways [1]. More than 1000 chemically distinct lipid species are known to exist in eukaryotic cells [2], and these lipids can be classified into three major classes: glycerophospholipids, sphingolipids and sterols [3]. As the basic constituents of transport vesicles, most lipids are thought to be transported between organelles by vesicular transport. However, lipid transportation can also be detected when vesicular transport is impaired by ATP depletion and upon reduction in temperature or treatment with specific pharmacological drugs (e.g., colchicine and brefeldin A) [4], [5]. Additionally, lipid transport between organelles that does not occur via vesicular transportation has also been observed [6], [7].
Plant non-specific lipid transfer protein (nsLtp) was first isolated by gel filtration from potato tuber homogenates, and it was found that this protein stimulates the exchange of phospholipids between microsomal fractions and mitochondria [8]. The term plant “non-specific lipid transfer proteins” indicates that these Ltps can bind with various phospholipids with broad specificity [9]. Plant nsLtps are able to transfer phosphatidylcholine, phosphatidylinositol and phosphatidylglycerol from liposomes to mitochondria [10], and they also have the ability to transfer galactolipids. Additionally, nsLtps from some species, such as oilseed rape, spinach, and sunflower, are able to bind acyl-CoA [9], [11], [12]. The ability of plant nsLtps to bind fatty acids or acyl-CoA esters was determined by temperature-dependent ligand affinity or by separation of the acyl-Ltp complex by gel filtration [12]. Several isoforms of nsLtp in oilseed rape revealed the ability to both transfer phosphatidylcholine and bind oleoyl-CoA [9]. In the castor bean, the saturating binding capacities for oleic acid and oleoyl-CoA per mole of Ltp were 1:1 [12]. Taken together, this suggests that nsLtps have dual abilities to bind acyl chains and to transfer lipids.
It is generally accepted that amino acid sequences determine the spatial structures of proteins as well as their properties or functions. All known plant nsLtps are synthesized as precursors with an N-terminal signal peptide, and their mature proteins are small and basic characterized by an eight cysteine motif (ECM) as follows: C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C [13]. The cysteine (Cys) residues are arranged in four disulfide bonds to stabilize the tertiary structure of the hydrophobic cavity, the size plasticity of which allows for the binding of different lipid and hydrophobic compounds in vitro [14]. Plant nsLtps can be classified according to their molecular masses (MM) into two main types, nsLtp1 (9 kDa) and nsLtp2 (7 kDa) [14]. Computational and biochemical analyses have indicated that type 1 nsLtps are capable of accommodating lipids such as palmitic acid (C16:0) and the phospholipidacyl chains of 1, 2-dimyristoylphosphatidylglycerol [15], [16]. Nevertheless, less is currently known about the lipid binding abilities of type 2 nsLtps. Plant nsLtps from Arabidopsis, rice and Solanaceae can also be divided into several types (I, II, III, IV, V, VI, VII, VIII, IX, X and nsLTPY) based on their sequence similarity [17], [18].
Plant nsLtps were first proposed to be involved in membrane biogenesis as they were shown to transfer lipids between membranes in vitro [10]. However, an increasing number of studies have shown that almost all the nsLtps are extracellularly located and are secreted, and a possible role for these proteins in intracellular lipid transfer seems unlikely [19]. All known precursors of the nsLtps are tagged with N-terminal signal peptides, generally 21 to 27 amino acids in length for the type 1 family and 27 to 35 amino acids in length for the type 2 family [20], indicating they are secreted proteins. Thus, nsLtps may be involved in a range of other biological processes. Notwithstanding the amount of data available, the exact functions of nsLtps remain unclear. Multiple physiological functions of nsLtps have been suggested, including cutin synthesis [21]–[23], somatic embryogenesis [24]–[26], pollen development [27], [28], stigma and pollen adhesion [29]–[31], pollen tube growth and guidance [32], [33], cell wall extension [34], biotic stresses [23], [35]–[41], abiotic stresses [42]–[44], plant signaling [1], [45], [46], and seed maturation [47].
Plant nsLtp family is one of the most well-known protein families and is widely distributed in the plant kingdom. Boutrot et al. identified and classified 267 nsLtps sequences [17]. Recently, Wang et al. made a great contribution to the knowledge of plant nsLtps by constructing a systematic plant non-specific lipid transfer protein database (nsLTPDB), and they identified 595 nsLtps from 121 species [48]. However, the number of identified nsLtps in some species is still very low, such as in Brassica rapa, where only five members have been identified in nsLTPDB. Therefore, it is necessary to continue to identify nsLtps in these species. A subspecies of Chinese cabbage, Brassica rapa subsp. Pekinensis, originates from China and is one of the most economically significant vegetable crops in Asia. Additionally, its ‘A’ genome is an important resource for studying the evolutionary history from Arabidopsis thaliana to B. rapa, Brassica napus, or Brassica juncea, as well as underpinning the genetic improvement of Brassica-related crops. The recent release of the genome of accession Chiifu401–42 of B. rapa has enabled us to comprehensively identify and bioinformatically analyse the putative nsLtps in B. rapa. In the present study, we took advantage of the available B. rapa genome sequence to perform a genome-wide analysis of the putative nsLtps in this species. In total, we identified 63 genes encoding putative nsLtps in the B. rapa genome that could be classified into nine types based on the diversity of ECMs. We also conducted phylogenetic and gene duplication analyses of BrnsLtps. Furthermore, we employed publicly available UniGene data and quantitative RT-PCR data to analyse the expression patterns of these genes. The results of this study may guide research involving all members of the nsLtps family and facilitate our understanding of the influence of polyploidy on the evolution of plants.
Results
Identification of putative nsLTP genes in the B. rapa genome
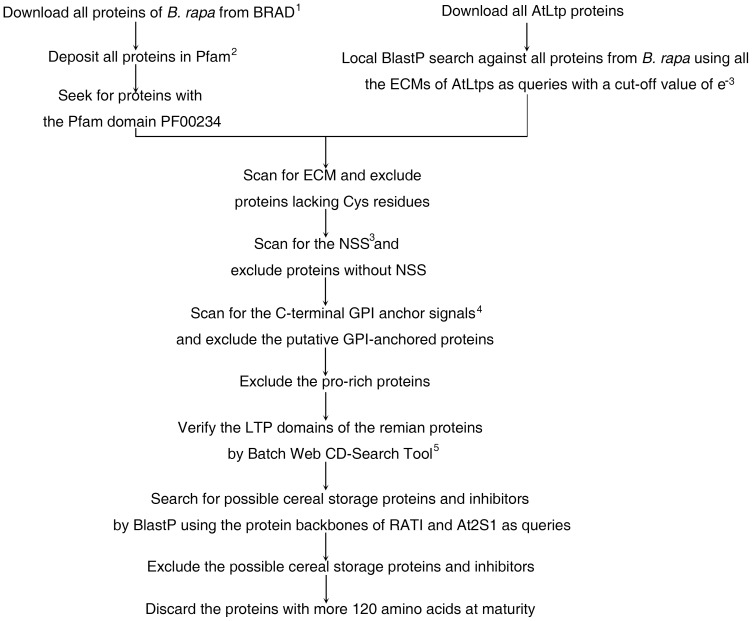
Previously, five nsLtp genes of B. rapa (GI numbers: 122939101, 48093506, 21591782, 3062791, and 1209260) were identified in nsLTPDB. Given that the whole genome of B. rapa is now available, we attempted to identify the entire collection of putative nsLtp genes in the B. rapa genome. Initially, a total of 152 protein sequence (PF00234: plant lipid transfer/seed storage/trypsin-alpha amylase inhibitor) were retrieved after all protein sequences from B. rapa (.pep file) were submitted to the Pfam database (Fig. 1). Then, 14 proteins lacking the Cys residues were omitted from the remaining set after manually scanning for the presence of the eight essential Cys residues (Fig. 1). After that, nine proteins lacking N-terminal signal sequences (NSS) and 22 proteins possessing C-terminal glycosylphosphatidylinositol (GPI) anchors were also excluded (Fig. 1). Subsequently, 32 proline-rich or hybrid proline-rich proteins, which are characterized by a high proportion of proline, histidine and glycine residues in the sequence located between the NSS and the ECM [49], [50], were also removed (Fig. 1). Next, the remaining 75 proteins were submitted to the Batch Web CD-Search Tool for verification of their LTP domains, and all but two proteins (Bra001685 and Bra035574) were found to possess LTP domains (Fig. 1). Subsequently, eight proteins (Bra006444, Bra010409, Bra019064, Bra019067, Bra026373, Bra026374, Bra026375, and Bra038483) similar to cereal storage proteins or trypsin-alpha amylase inhibitors were also discarded (Fig. 1). As the mature proteins of nsLtps have low molecular weight, six predicted mature proteins (Bra018484, Bra027016, Bra030873, Bra033087, Bra034559, and Bra037988) with more than 120 amino acids were not taken into consideration (Fig. 1). Finally, a search for misannotated putative nsLtp genes was conducted by local BlastP searches on the all proteins of B. rapa using the previously used 49 Arabidopsis nsLtps as query sequences [17] (Fig. 1). With this approach, four additional putative nsLtp genes (Bra008112, Bra015966, Bra021299, and Bra040156) were picked up. Ultimately, we identified a total of 63 BrnsLtps in the whole genome of B. rapa (Table 1, Table S1, Table S2). Furthermore, the MM and theoretical pI (isoelectric point) of each BrnsLtp were calculated and summarized in Table 1. The three-dimensional structures of all putative BrnsLtps were also predicted and showed in Figure S1.
Figure 1. The workflow of BrnsLtp identification and data mining.
1, BRAD, Brassica Database, http://brassicadb.org/brad/; 2, http://pfam.sanger.uk/; 3, NSS (N-terminal signal sequence) prediction, http://www.cbs.dtu.dk/services/SignalP; 4, GPI (glycosylphosphatidylinositol) anchor signal prediction, http://mendel.imp.ac.at/gpi/plant_server.html and http://psort.hgc.jp/form.html; 5, LTP domain prediction, http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi.
Table 1. Putative nsLtp genes identified in the genome of B. rapa.
| Name | BRAD-locus | Chromosome Location | Strand | Intron (bp) | ECMa | SPb(AAc) | MPd(AA) | MP(MMe) | MP(pIf) |
| Type I | |||||||||
| BrnsLtpI.1 | Bra000068 | ChrA03: 9142369, 9142755 | + | none | C-X9-C-X13-CC-X19-CXC-X21-C-X13-C | 25 | 103 | 10723.34 | 10.26 |
| BrnsLtpI.2 | Bra001345 | ChrA03: 15944546, 15944887 | + | none | C-X9-C-X14-CC-X19-CXC-X24-C-X13-C | 20 | 93 | 9651.88 | 10.28 |
| BrnsLtpI.3* | Bra005098 | ChrA05: 3409556, 3410188 | − | 228 | C-X9-C-X13-CC-X19-CXC-X23-C-X13-C | 18 | 116 | 11898.93 | 11.65 |
| BrnsLtpI.4* | Bra005099 | ChrA05: 3413125, 3413749 | − | 268 | C-X9-C-X14-CC-X19-CXC-X22-C-X13-C | 25 | 93 | 9414.92 | 11.08 |
| BrnsLtpI.5 | Bra006721 | ChrA03: 4792207, 4792704 | − | 150 | C-X9-C-X13-CC-X19-CXC-X22-C-X13-C | 23 | 92 | 9175.57 | 9.53 |
| BrnsLtpI.6 | Bra006736 | ChrA03: 4845130, 4845623 | + | 116 | C-X9-C-X13-CC-X19-CXC-X22-C-X13-C | 22 | 103 | 11229.85 | 7.31 |
| BrnsLtpI.7 | Bra012848 | ChrA03: 21927444, 21928521 | − | 718 | C-X9-C-X13-CC-X19-CXC-X22-C-X13-C | 24 | 95 | 9922.37 | 8.05 |
| BrnsLtpI.8* | Bra017112 | ChrA04: 16558054, 16558647 | + | 237 | C-X9-C-X13-CC-X19-CXC-X23-C-X13-C | 25 | 93 | 9253.68 | 11.59 |
| BrnsLtpI.9* | Bra017113 | ChrA04: 16555397, 16556028 | + | 278 | C-X9-C-X13-CC-X19-CXC-X22-C-X13-C | 25 | 92 | 9430.85 | 11.73 |
| BrnsLtpI.10* | Bra020322 | ChrA02: 6218834, 6219334 | − | 162 | C-X9-C-X13-CC-X19-CXC-X19-C-X13-C | 23 | 89 | 8936.34 | 9.78 |
| BrnsLtpI.11* | Bra020323 | ChrA02: 6223820, 6224155 | + | none | C-X9-C-X14-CC-X19-CXC-X19-C-X13-C | 23 | 88 | 8849.35 | 10.00 |
| BrnsLtpI.12 | Bra024983 | ChrA06: 24518522, 24518875 | + | none | C-X8-C-X16-CC-X19-CXC-X24-C-X9-C | 22 | 95 | 10052.28 | 10.07 |
| BrnsLtpI.13 | Bra029172 | ChrA03: 6690210, 6690542 | + | none | C-X9-C-X13-CC-X19-CXC-X19-C-X13-C | 23 | 87 | 8740.24 | 10.30 |
| BrnsLtpI.14 | Bra029719 | ChrA05: 22525095, 22525430 | + | none | C-X9-C-X13-CC-X19-CXC-X24-C-X13-C | 19 | 92 | 9535.75 | 9.82 |
| BrnsLtpI.15 | Bra036788 | ChrA09: 25812372, 25812722 | − | none | C-X9-C-X13-CC-X19-CXC-X23-C-X13-C | 25 | 91 | 9682.25 | 12.44 |
| BrnsLtpI.16 | Bra036789 | ChrA09: 25802085, 25802778 | − | 328 | C-X9-C-X14-CC-X19-CXC-X23-C-X13-C | 24 | 97 | 10473.18 | 8.28 |
| BrnsLtpI.17 | Bra037222 | ChrA09: 4927969, 4928711 | + | 392 | C-X9-C-X14-CC-X19-CXC-X21-C-X13-C | 22 | 94 | 9413.56 | 4.52 |
| BrnsLtpI.18* | Bra038907 | ChrA01: 12636983, 12637614 | + | 272 | C-X9-C-X13-CC-X19-CXC-X22-C-X13-C | 24 | 95 | 9913.47 | 8.34 |
| BrnsLtpI.19* | Bra038908 | ChrA01: 12629898, 12630245 | + | none | C-X9-C-X13-CC-X19-CXC-X23-C-X13-C | 24 | 91 | 9693.23 | 12.40 |
| Type II | |||||||||
| BrnsLtpII.1 | Bra008112 | ChrA02: 13195464, 13195760 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 30 | 68 | 7645.89 | 9.26 |
| BrnsLtpII.2 | Bra008375 | ChrA02: 15167775, 15168068 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 18 | 79 | 8467.68 | 4.89 |
| BrnsLtpII.3 | Bra014154 | ChrA08: 2994554, 2994847 | − | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 29 | 68 | 7103.21 | 10.12 |
| BrnsLtpII.4 | Bra015966 | ChrA07: 20026923, 20027216 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X5-C | 30 | 67 | 7626.94 | 9.58 |
| BrnsLtpII.5 | Bra018687 | ChrA06: 2603650, 2603949 | − | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 24 | 75 | 8036.32 | 8.22 |
| BrnsLtpII.6 | Bra021299 | ChrA01: 22700481, 22700774 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 29 | 68 | 7464.86 | 12.14 |
| BrnsLtpII.7 | Bra022308 | ChrA05: 18509672, 18509962 | − | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 28 | 68 | 7411.84 | 12.02 |
| BrnsLtpII.8 | Bra025378 | ChrA06: 21876530, 21876814 | + | none | C-X7-C-X14-CC-X8-CXC-X25-C-X6-C | 23 | 71 | 7607.87 | 5.43 |
| BrnsLtpII.9* | Bra027111 | ChrA09: 8544965, 8545258 | − | none | C-X7-C-X13-CC-X8-CXC-X21-C-X6-C | 24 | 73 | 7856.14 | 5.06 |
| BrnsLtpII.10* | Bra027114 | ChrA09: 8562465, 8562758 | − | none | C-X7-C-X13-CC-X8-CXC-X21-C-X6-C | 24 | 73 | 7972.22 | 4.86 |
| BrnsLtpII.11 | Bra028162 | ChrA04: 6295941, 6296249 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 24 | 78 | 8399.71 | 4.81 |
| BrnsLtpII.12 | Bra030699 | ChrA08: 20754782, 20755075 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 24 | 73 | 7913.12 | 4.64 |
| BrnsLtpII.13 | Bra032265 | ChrA05: 12176382, 12176675 | − | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 29 | 68 | 7233.42 | 10.38 |
| BrnsLtpII.14 | Bra033084 | ChrA02: 21491213, 21491515 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 26 | 74 | 7845.08 | 6.75 |
| BrnsLtpII.15 | Bra040627 | ChrA02: 9924592, 9924879 | + | none | C-X7-C-X13-CC-X8-CXC-X23-C-X6-C | 21 | 74 | 7663.76 | 8.34 |
| Type III | |||||||||
| BrnsLtpIII.1 | Bra009282 | ChrA10: 15963803, 15964270 | − | 177 | C-X9-C-X16-CC-X9-CXC-X12-C-X6-C | 29 | 67 | 6969.07 | 4.43 |
| BrnsLtpIII.2 | Bra028294 | ChrA01: 18866657, 18866944 | − | none | C-X9-C-X16-CC-X9-CXC-X12-C-X6-C | 31 | 64 | 6730.60 | 4.90 |
| BrnsLtpIII.3 | Bra029135 | ChrA03: 6509041, 6509295 | − | none | C-X9-C-X16-CC-X9-CXC-X12-C-X6-C | 20 | 64 | 6729.72 | 6.78 |
| Type IV | |||||||||
| BrnsLtpIV.1* | Bra002906 | ChrA10: 6849704, 6850036 | + | none | C-X9-C-X15-CC-X9-CXC-X22-C-X8-C | 23 | 87 | 9362.02 | 9.46 |
| BrnsLtpIV.2* | Bra002907 | ChrA10: 6843872, 6844180 | + | none | C-X9-C-X15-CC-X9-CXC-X19-C-X7-C | 28 | 74 | 7850.28 | 10.71 |
| BrnsLtpIV.3 | Bra002914 | ChrA10: 6777869, 6778177 | − | none | C-X9-C-X15-CC-X9-CXC-X19-C-X7-C | 28 | 74 | 7774.18 | 10.10 |
| BrnsLtpIV.4 | Bra020696 | ChrA02: 23700424, 23700738 | − | none | C-X9-C-X15-CC-X9-CXC-X24-C-X7-C | 28 | 76 | 7992.20 | 4.70 |
| BrnsLtpIV.5 | Bra020839 | ChrA08: 11767442, 11767795 | + | none | C-X10-C-X11-CC-X12-CXC-X23-C-X7-C | 25 | 92 | 9845.14 | 4.52 |
| BrnsLtpIV.6 | Bra022364 | ChrA05: 18159874, 18160179 | + | none | C-X9-C-X19-CC-X9-CXC-X24-C-X6-C | 23 | 78 | 8313.64 | 9.02 |
| BrnsLtpIV.7 | Bra028980 | ChrA03: 5697175, 5697474 | − | none | C-X9-C-X15-CC-X9-CXC-X19-C-X6-C | 26 | 73 | 7472.52 | 4.46 |
| BrnsLtpIV.8 | Bra035573 | ChrA02: 7465564, 7465998 | − | 72 | C-X9-C-X15-CC-X9-CXC-X22-C-X8-C | 30 | 90 | 9604.48 | 9.78 |
| Type V | |||||||||
| BrnsLtpV.1 | Bra005153 | ChrA05: 3743608, 3744053 | − | 98 | C-X14-C-X14-CC-X12-CXC-X24-C-X10-C | 23 | 92 | 9521.52 | 12.31 |
| BrnsLtpV.2 | Bra014853 | ChrA04: 3607346, 3607959 | + | 224 | C-X14-C-X14-CC-X11-CXC-X24-C-X10-C | 23 | 106 | 11085.73 | 9.16 |
| Type VI | |||||||||
| BrnsLtpVI.1 | Bra011229 | ChrA01: 3189969, 3190301 | − | none | C-X10-C-X16-CC-X9-CXC-X22-C-X9-C | 19 | 91 | 9647.42 | 10.20 |
| BrnsLtpVI.2* | Bra034567 | ChrA08: 12794464, 12794909 | − | 101 | C-X10-C-X17-CC-X9-CXC-X22-C-X9-C | 28 | 86 | 9277.77 | 9.89 |
| BrnsLtpVI.3* | Bra034568 | ChrA08: 12792882, 12793311 | − | 85 | C-X10-C-X17-CC-X9-CXC-X22-C-X9-C | 28 | 86 | 9285.70 | 9.82 |
| BrnsLtpVI.4* | Bra034570 | ChrA08: 12788663, 12789092 | − | 85 | C-X10-C-X17-CC-X9-CXC-X22-C-X9-C | 28 | 86 | 9285.70 | 9.82 |
| Type VIII | |||||||||
| BrnsLtpVIII.1 | Bra015984 | ChrA07: 19946287, 19946673 | − | none | C-X6-C-X14-CC-X12-CXC-X25-C-X8-C | 22 | 106 | 11657.72 | 8.11 |
| Type IX | |||||||||
| BrnsLtpIX.1 | Bra001252 | ChrA03: 15530671, 15530955 | − | none | C-X13-C-X15-CC-X9-CXC-X22-C-X6-C | 17 | 77 | 7975.40 | 6.93 |
| BrnsLtpIX.2 | Bra006901 | ChrA09: 26056900, 26057265 | − | none | C-X13-C-X15-CC-X9-CXC-X22-C-X6-C | 24 | 97 | 10290.02 | 4.72 |
| BrnsLtpIX.3 | Bra012819 | ChrA03: 22081654, 22082025 | + | none | C-X13-C-X15-CC-X9-CXC-X22-C-X6-C | 26 | 97 | 10396.21 | 4.72 |
| Type XI | |||||||||
| BrnsLtpXI.1 | Bra000287 | ChrA03: 10350000, 10350350 | + | none | C-X9-C-X18-CC-X13-CXC-X24-C-X9-C | 29 | 87 | 9167.88 | 6.86 |
| BrnsLtpXI.2 | Bra018483 | ChrA05: 8507982, 8508317 | + | none | C-X9-C-X20-CC-X13-CXC-X24-C-X9-C | 23 | 88 | 9021.55 | 4.35 |
| BrnsLtpXI.3 | Bra018544 | ChrA05: 9107060, 9107431 | − | none | C-X9-C-X18-CC-X13-CXC-X25-C-X9-C | 26 | 97 | 10165.23 | 9.81 |
| BrnsLtpXI.4* | Bra024980 | ChrA06: 24509174, 24509509 | + | none | C-X8-C-X16-CC-X13-CXC-X24-C-X9-C | 22 | 89 | 9465.57 | 10.35 |
| BrnsLtpXI.5* | Bra024981 | ChrA06: 24511564, 24511899 | + | none | C-X8-C-X16-CC-X13-CXC-X24-C-X9-C | 22 | 89 | 9465.57 | 10.35 |
| BrnsLtpXI.6* | Bra024982 | ChrA06: 24516109, 24516444 | + | none | C-X8-C-X16-CC-X13-CXC-X24-C-X9-C | 22 | 89 | 9465.57 | 10.35 |
| nsLTPY | |||||||||
| BrnsLtpY.1 | Bra024207 | ChrA03: 26879106, 26879879 | + | 242, 106 | C-X9-C-X14-CC-X30-CXC-X23-C-X13-C | 21 | 120 | 13391.43 | 5.33 |
| BrnsLtpY.2 | Bra040156 | ChrA01: 4374192, 4374566 | + | none | C-X10-C-X14-CC-X16-CXC-X21-C-X13-C | 20 | 104 | 11873.64 | 7.04 |
ECM, eight-cysteine motif; bSP, signal peptide; cAA, number of amino acids; dMP, mature protein; eMM, molecular mass in Dalton; fpI, isoelectric point (cysteine residues were not taken into account in the pI caculation). A cluster of tandem duplication repeats was indicated by an asterisk after the gene names. The values in ECM allowing direct identification of the nsLtp type are indicated in bold italic.
Sequence analysis and classification of putative BrnsLtps
Boutrot et al. [17] pointed out that the relationship between MM and nsLtp type was more complicated than previously thought and, as a consequence, was no longer considered to be a good criterion for classifying nsLtps. They also employed an alternative method to cluster 49 out of the 52 rice nsLtps and 45 out of the 49 Arabidopsis nsLtps into nine types based on sequence similarity [17]. Liu et al. [18] also used this method to classify 135 Solanaceae nsLtps into five types. Recently, Edstam et al. [51] even found that sequence similarity was not sufficient tool for a classification system if the sequences include genes from non-flowering plants. In this study, as for B. rapa is a flowering plant, we still applied the sequence similarity method to sort the BrnsLtps, and our results indicated that 61 out of the 63 BrnsLtps also could be divided into nine types (I, II, III, IV, V, VI, VIII, IX, and XI) mainly based on the identity matrix (data not shown) calculated from the multiple sequence alignments (Table 1, Fig. 2). Similarly, the majority (34 out of 63) of BrnsLtps belong to type I or type II nsLtps (Table 1, Fig. 2).
Figure 2. Multiple sequence alignment of the putative mature BrnsLtp proteins.
The conserved cysteine residues are marked against a dark blue background. The names of different types of BrnsLtps are indicated with different color backgrounds. And the accession number of each gene was showed in the parentheses. Consensus residues Thr/Ser-X1-X2-Asp-Arg/Lys and Pro-Tyr-X-Ile-Ser are marked by rectangles. Tryptophan residues (W) are indicated with yellow circles.
To clearly understand the sequence characteristics of BrnsLtps, we conducted a multiple sequence alignment using the deduced mature proteins from the 63 BrnsLtps (Fig. 2). The results showed that all the predicted mature BrnsLtps had a total number of amino acids that varied from 64 to 120 (Table 1 and Fig. 2). It was found that the eight Cys residues were highly conserved in all of the 63 BrnsLtps and that these residues may form four disulfide bonds to stabilize the tertiary structure of the hydrophobic cavity (Fig. 2; Figure S1). The proteins nsLtp1 (9 kDa) and nsLtp2 (7 kDa), however, have different disulfide bond patterns. The disulfide bond linkage of nsLtp1 at Cys1-Cys6 and Cys5-Cys8 differs from that of nsLtp2 at Cys1-Cys5 and Cys6-Cys8 [52]. Additionally, X is a hydrophilic residue in the CXC motif of nsLtp1; however, a hydrophobic residue was found at the X position in nsLtp2 [52]. These conserved hydrophobic or hydrophilic residues may play significant roles in the biological functions of nsLtps [53]. Our results showed that there were 11 different residues (Tyr, Leu, Phe, Val, Ile, Ala, Gly, Arg, Lys, Glu, and Gln) at the X position of the CXC motif in the 63 BrnsLtps (Fig. 2). Among of them, seven (Tyr, Leu, Phe, Val, Ile, Ala, and Gly) and four (Arg, Lys, Glu, and Gln) amino acids belong to the hydrophobic and hydrophilic residue groups, respectively. Leu is the most frequent residue that appears in the CXC motif of BrnsLtps, while Tyr and Gln appeared only once (Fig. 2). It is worth mentioning that the Leu residue is also the most frequent residue in the CXC of nsLTPs in Arabidopsis and rice, identified previously [17]. Douliez et al. [14] found that all plant nsLtp1 proteins had two highly conserved residues located in Thr/Ser-X1-X2-Asp-Arg/Lys and Pro-Tyr-X-Ile-Ser. Here, we also found that all type I nsLtps, save BrnsLtpI.12, possessed a Thr/Ser-X1-X2-Asp-Arg/Lys; however, only eight type I nsLtps had Pro-Tyr-X-Ile-Ser (Fig. 2 and Table S2). In other words, the Thr/Ser-X1-X2-Asp-Arg/Lys is more conserved and Pro-Tyr-X-Ile-Ser is more variable (Fig. 2). It was reported that no tryptophan residues were found in the sequence of nsLTP1 [14] and that the hydrophobic tunnel of Ace-AMP1 was probably interrupted by bulky, aromatic tryptophan and phenylalanine residues such that it could not bind and transfer lipids [54]. Our results indicated that seven BrnsLtps (BrnsLtpI.3, BrnsLtpII.8, BrnsLtpIV.2, BrnsLtpIV.3, BrnsLtpIV.7, BrnsLtpXI.3, and BrnsLtpY.2) possessed tryptophan residues (Fig. 2 and Table S2). Additionally, multiple alignments revealed a variable number of inter-cysteine amino acid residues (summarized in Table 2). Therefore, these BrnsLtp types can be identified according to the typical spacings for this motif. Our results showed that in the B. rapa genome, there are also no type VII nsLtps sequences, which contain 27 residues between the conserved Cys6 and Cys7, just as in Arabidopsis (Table 2). Additionally, we found a new type of BrnsLtps that contains 13 residues between Cys4 and Cys5, which we designated type “XI” to follow the Greek numeral “X”, which had already been used to identify another new type nsLtps found in Solanaceae [18]. Edstam et al. pointed out that novel nsLtps types may also have evolved during land plant evolution [51]. Although BrnsLtpI.12 was more similar to type XI than to type I BrnsLtps based on the sequence identity (Fig. 2), we consider this protein as type I nsLtps as for it contains 19 residues between Cys4 and Cys5. Furthermore, the mature proteins of BrnsLtpIV.5 and BrnsLtpIV.6 also shared low identities (less than 30%) with other type IV BrnsLtps (data not shown), we classified these two proteins into type IV nsLtps according to the typical spacings for ECM motifs and the following results of Bayesian estimation (Figure S2).
Table 2. Diversity of eight cysteine motifs in different types of BrnsLtps.
| nsLTP type | ECM and number of flanking amino acid residues | ||||||
| 1 | 2 | 3, 4 | 5 6 | 7 | 8 | ||
| Type I | X3,5,9,10 | C-X9 | C-X13,14,16 | C-C-X19 | C-X-C-X19,21-24 | C-X9,13 | C-X1,2,4,7,15,16,20 |
| Type II | X0,2,7-9,12,13 | C-X7 | C-X13,14 | C-C-X8 | C-X-C-X21,23,25 | C-X5,6 | C-X0,2 |
| Type III | X2 | C-X9 | C-X16 | C-C-X9 | C-X-C-X12 | C-X6 | C-X1,4 |
| Type IV | X2,3,8,13 | C-X9,10 | C-X11,15,19 | C-C-X9,12 | C-X-C-X19,22-24 | C-X6-8 | C-X0,3,7,15 |
| Type V | X3 | C-X14 | C-X14 | C-C-X11,12 | C-X-C-X24 | C-X10 | C-X6,21 |
| Type VI | X2,9 | C-X10 | C-X16,17 | C-C-X9 | C-X-C-X22 | C-X9 | C-X7,8 |
| Type VIII | X11 | C-X6 | C-X14 | C-C-X12 | C-X-C-X25 | C-X8 | C-X21 |
| Type IX | X2,19 | C-X13 | C-X15 | C-C-X9 | C-X-C-X22 | C-X6 | C-X1,4 |
| Type XI | X3,9,14 | C-X8,9 | C-X16,18,20 | C-C-X13 | C-X-C-X24,25 | C-X9 | C-X0–2 |
The consensus motif of each nsLtp type was deduced from the analysis of the mature sequences of 63 putative BrnsLtps. The values allowing direct identification of the nsLtp type are underlined. Cysteine residues are indicated in bold italic. Character “X” represents any amino acid, and the Arabic numeral following “X” stands for the numbers of amino acid residues.
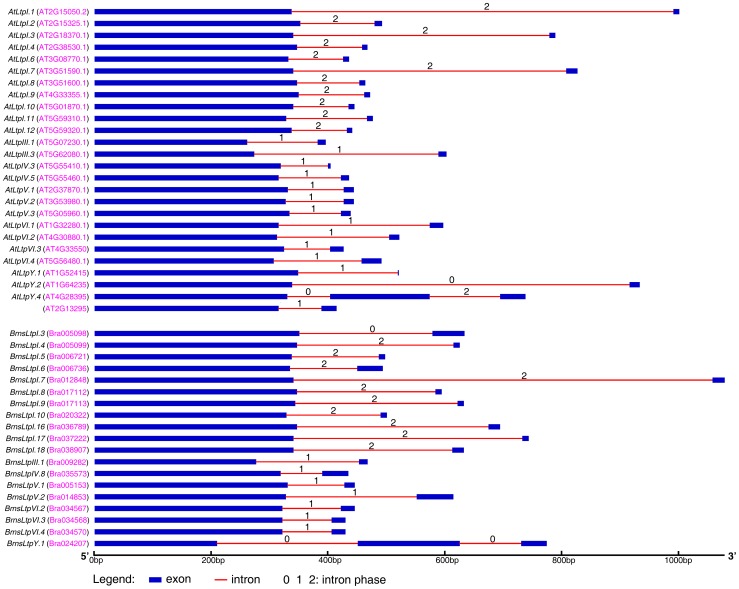
Additionally, we examined the exon-intron organization of BrnsLtp family members. Our result indicated that only 19 BrnsLtps (six types) had introns, including 11 type I, one type III, one type IV, two type V, three type VI, and one nsLTPY nsLtps (Table 1 and Fig. 3). And all these 19 BrnsLtps save BrnsLtpY.1 were predicted to be interrupted by a single intron positioned 7 to 59bp upstream of the stop codon (Table 1 and Fig. 3). Except for BrnsLtpIV.8, no introns were identified in the coding regions of type II, IV, VIII, IX, XI, and nsLTPY BrnsLtp genes (Table 1). A previous study showed that 25 Arabidopsis Ltps (11 type I, two type III, two type IV, three type V, four type VI, and three nsLTPY) possess introns [17]. Actually, Wang et al. [52] discovered several additional putative Ltp coding genes in Arabidopsis. Therefore, we also analysed the gene structure of these additional AtLtps, and our results demonstrated that only the protein encoded by AT2G13295 was predicted to have a single intron (Fig. 3). Exon shuffling may play an important role in the origin of both ancient and modern genes [55]. So the statistical analysis of intron phases (the position of introns within or between codons) is a good choice to evaluate the evolution between orthologous genes and paralogous genes. After comparison, we found that except for BrnsLtpI.3 and nsLTPY nsLtps, the same type nsLtps had identical intron phase distribution (Fig. 3).
Figure 3. Gene structure of the BrnsLtps and AtLtps.
Only those genes with introns (26 AtLtps and 19 BrnsLtps) are showed. The accession number of each gene is displayed in red font inside the parentheses. Intron phases are analysed based on the exon information. Phase 0 is designated introns between exons, phase 1 is designated introns between the first and the second nucleotide in a codon, and phase 2 is designated introns between the second and the third nucleotide in a codon [55].
Phylogenetic analysis of the putative AtnsLtps and BrnsLtps
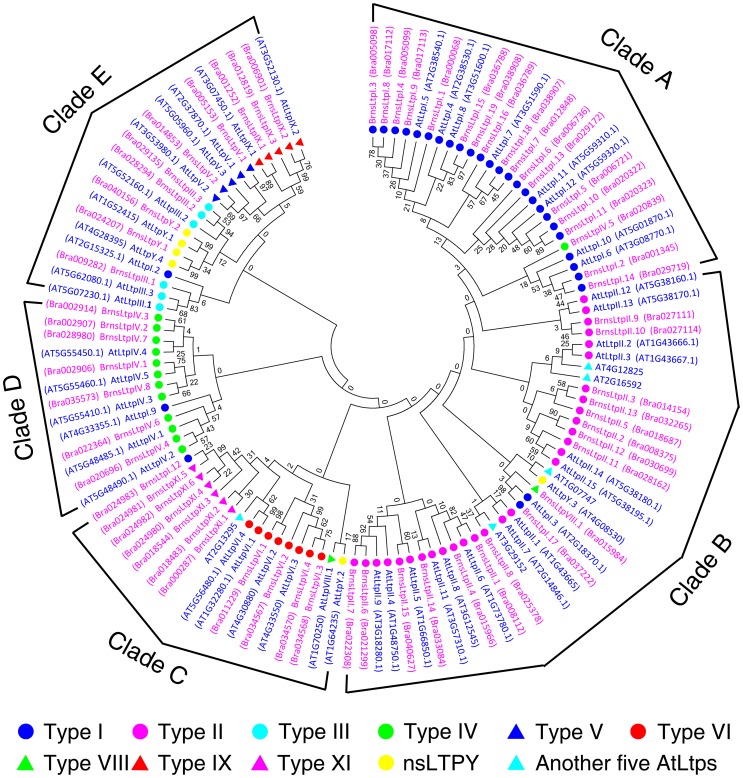
In order to analyse the phylogenetic organization of the nsLtp families in Arabidopsis and B. rapa, we constructed a phylogenetic tree using Neighbor-Joining from the alignment of the amino acids of the ECMs. Boutrot et al. [17] previously pointed out that AtLtpI.1 and AtLtpII.10 were lacking Cys residues, and therefore, we discarded these two proteins from the 49 Arabidopsis nsLtps for the phylogenetic tree construction. Recently, Wang et al. [52] found additional AtLtp genes such as AT1G07747, AT1G52415, AT2G16592, AT2G13295, AT3G29152, and AT4G12825. Only four other AtLtps were discovered by Wang et al. [52] on account of the fact that AT1G52415 is the same as AtLtpY.1, which was identified previously by Boutrot et al. [17]. Thus, a total of 52 AtnsLtps and 63 BrnsLtps were used to conduct a protein-based phylogenetic analysis (Fig. 4). Our results showed that these 115 nsLtps can be divided into five clades (Fig. 4). There were 18, 17, 11, seven, and ten BrnsLtps in each clade from A–E, respectively (Fig. 4). Almost all type I BrnsLtps were grouped in clade A, all type II were grouped in clade B, and all type IV BrnsLtps were grouped in clade D. The C and E clades were each composed of proteins from two or four main types, respectively (Fig. 4). The phylogeny of BrnsLtps and AtLtps was also determined with Bayesian estimation using the MrBayes program (Figure S2). Similar to the Neighbor-Joining phylogenetic results, the Bayesian estimation also indicated that almost all the sequences belonging to the same type are grouped and constitute monophyletic groups, except for the type II nsLtps (Figure S2). Type II nsLtps are close in the tree but are not grouped in a single clade. This may be because several AtLtps (AtLtpII.1, AtLtpII.2, AtLtpII.3, AtLtpII.7, AtLtpII.11, AtLtpII.12, AtLtpII.13, AT1G07747, and AT4G12825) were more distantly related to other type II nsLtps (Figure S2). Furthermore, all type II BrnsLtps except for BrnsLtpII.5 appeared to be monophyletic (Figure S2). Previously, Boutrot et al. [17] pointed out that the general organization of the tree is consistent with the classification of nsLtps types except for type II nsLtps via phylogenetic analysis from the alignments of 45, 49, and 122 sequences of Arabidopsis, rice, and wheat nsLtps using maximum-likelihood inference. Amazingly, we also found that the nine type II AtLtps not grouped in a clade had no homologous genes in the three subgenomes of B. rapa (Table 3). It is an interesting question as to why these genes were missed during the triplication evolution process from A. thaliana to B. rapa. It was also worth noting that when the tree is built with only rice and wheat sequences, type II nsLtps seems to be monophyletic [17]. Does this mean that the monocotyledon plants discarded these genes during the evolutionary divergence between monocots and dicots?
Figure 4. Phylogenetic tree of BrnsLtps.
The amino acids of the ECMs were used for Neighbor-Joining phylogenetic tree construction using the MEGA 5.05 software. The ten types of nsLtps and another five AtnsLtps are indicated with circles or triangles of different colors. And the accession number of each gene is showed in the parentheses nearby the corresponding gene name.
Table 3. Identification of homologous nsLtp genes between A. thaliana and three subgenomes in B. rapa.
| A. thaliana | CCBa | LFb | MF1c | MF2d | A. thaliana | CCB | LF | MF1 | MF2 |
| AtLtpI.1 | / | / | / | / | AtLtpV.1 | J | BrnsLtpV.1 | / | / |
| AtLtpI.2 | / | / | / | / | AtLtpV.2 | N | / | BrnsLtpV.2 | / |
| AtLtpI.3 | H | / | / | BrnsLtpI.17 | AtLtpV.3 | R | Bra009154 | / | / |
| AtLtpI.4 | J – Tandem | BrnsLtpI.3 BrnsLtpI.4 – Tandem | BrnsLtpI.8 BrnsLtpI.9 – Tandem | BrnsLtpI.1 | AtLtpVI.1 | B | / | / | / |
| AtLtpI.5 | J – Tandem | BrnsLtpI.3 BrnsLtpI.4 –Tandem | BrnsLtpI.8 BrnsLtpI.9 – Tandem | BrnsLtpI.1 | AtLtpVI.2 | U | BrnsLtpVI.1 | / | Bra010269 |
| AtLtpI.6 | F | BrnsLtpI.14 | / | BrnsLtpI.2 | AtLtpVI.3 | U | / | / | BrnsLtpVI.2 BrnsLtpVI.3 BrnsLtpVI.4 – Tandem |
| AtLtpI.7 | N – Tandem | BrnsLtpI.15 | BrnsLtpI.7 | BrnsLtpI.18 BrnsLtpI.19 – Tandem | AtLtpVI.4 | W | Bra002813 | / | / |
| AtLtpI.8 | N – Tandem | BrnsLtpI.15 | BrnsLtpI.7 | BrnsLtpI.18 BrnsLtpI.19 – Tandem | AtLtpVIII.1 | E | / | Bra007907 Bra007908 Bra007909 – Tandem | / |
| AtLtpI.9 | U | / | / | / | AtLtpIX.1 | F | Bra029649 | / | BrnsLtpIX.1 |
| AtLtpI.10 | R | / | / | / | AtLtpIX.2 | N | BrnsLtpIX.2 | BrnsLtpIX.3 | / |
| AtLtpI.11 | W – Tandem | / | BrnsLtpI.10 BrnsLtpI.11 – Tandem | BrnsLtpI.6 | AtLtpY.1 | C | / | / | / |
| AtLtpI.12 | W – Tandem | / | BrnsLtpI.10 BrnsLtpI.11 – Tandem | BrnsLtpI.6 | AtLtpY.2 | / | / | / | / |
| AtLtpII.1 | C | / | / | / | AtLtpY.3 | / | / | / | / |
| AtLtpII.2 | C | / | // | / | AtLtpY.4 | U | / | BrnsLtpY.1 | / |
| AtLtpII.3 | C | / | / | / | AT1G07747 | A | / | / | / |
| AtLtpII.4 | C | / | BrnsLtpII.3 | BrnsLtpII.13 | AT2G16592 | H – Tandem | / | / | / |
| AtLtpII.5 | E | / | BrnsLtpII.15 | / | AT2G13295 | / | / | / | / |
| AtLtpII.6 | E | BrnsLtpII.4 | BrnsLtpII.1 | / | AT3G29152 | L | BrnsLtpII.8 | / | / |
| AtLtpII.7 | / | / | / | / | AT4G12825 | T | / | / | / |
| AtLtpII.8 | F | / | / | / | / | / | / | / | BrnsLtpI.5 |
| AtLtpII.9 | F | BrnsLtpII.7 | BrnsLtpII.6 | Bra001701 | AT5G46890 | V – Tandem | BrnsLtpXI.4 BrnsLtpXI.5 BrnsLtpXI.6 BrnsLtpI.12 – Tandem | / | Bra017517 Bra017518 – Tandem |
| AtLtpII.10 | / | / | / | / | / | / | / | / | BrnsLtpI.13 |
| AtLtpII.11 | N | / | / | / | / | / | BrnsLtpI.16 | / | / |
| AtLtpII.12 | S – Tandem | / | / | / | / | / | / | BrnsLtpII.2 | / |
| AtLtpII.13 | S – Tandem | / | / | / | / | / | BrnsLtpII.5 | / | / |
| AtLtpII.14 | S | / | / | / | / | / | BrnsLtpII.9 BrnsLtpII.10 – Tandem | / | / |
| AtLtpII.15 | S – Tandem | BrnsLtpII.11 | / | / | / | / | / | / | BrnsLtpII.12 |
| AtLtpIII.1 | R | BrnsLtpIII.1 | / | / | / | / | / | BrnsLtpII.14 | / |
| AtLtpIII.2 | W | BrnsLtpIII.2 | / | BrnsLtpIII.3 | AT4G22666 | U | / | / | BrnsL tpIV.5 Bra020840 – Tandem |
| AtLtpIII.3 | X | / | / | / | / | / | BrnsLtpIV.6 | / | / |
| AtLtpIV.1 | V – Tandem | / | BrnsLtpIV.4 | / | / | / | BrnsLtpVIII.1 | / | / |
| AtLtpIV.2 | V – Tandem | / | BrnsLtpIV.4 | / | / | / | / | / | BrnsLtpXI.1 |
| AtLtpIV.3 | W | BrnsLtpIV.3 | / | / | AT4G12470 | Tandem | Bra029456 Bra029458 – Tandem | Bra000775 Bra000777 Bra000778 – Tandem | Bra018481 BrnsLtpXI.2 Bra018484– Tandem |
| AtLtpIV.4 | W | BrnsLtpIV.2 | BrnsLtpIV.8 Bra035574 – Tandem | BrnsLtpIV.7 Bra028981 – Tandem | / | / | / | / | |
| AtLtpIV.5 | W | BrnsLtpIV.1 | / | / | / | / | BrnsLtpY.2 | / | / |
CCB, conserved collinear block; bLF, the least fractionated blocks of B. rapa ; cMF1, the medium fractionated blocks of B. rapa ; dMF2, the most fractionated blocks of B. rapa .
Chromosomal localization and gene duplication
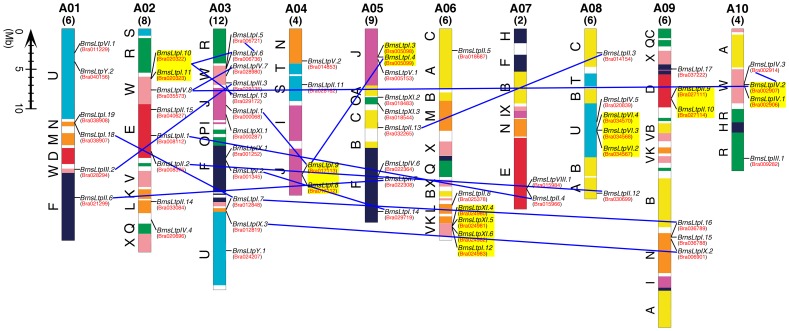
The approximate position of each BrnsLtp on the physical map of the B. rapa genome was marked based on information obtained from the Brassica Database (BRAD). The 63 BrnsLtps were randomly distributed across ten chromosomes of B. rapa, from A01 to A10 (Fig. 5). Twelve genes were located on chromosome A03, nine genes on chromosome A05, eight genes on chromosome A02, six genes each on chromosomes A01, A06, A08, and A09, four genes each on chromosome A04 and A10, and two genes on chromosome A07 (Fig. 5). Additionally, comparison of the nsLtp genes those are homologous between Arabidopsis and the three subgenomes of B. rapa revealed that 27 BrnsLtp genes are located on the least fractionated blocks (LF), 16 BrnsLtps are on the medium fractionated blocks (MF1), and 20 BrnsLtps are on the most fractionated blocks (MF2) (Table 3).
Figure 5. Genomic localization of the BrnsLtp genes on the chromosomes of B. rapa.
Chromosome numbers are indicated above each chromosome. The number of BrnsLtp genes distributed on each B. rapa chromosome is indicated by an Arabic numeral in the bracket, which is under the relative chromosome number. And the accession number of each gene was showed in the parentheses underneath the corresponding gene name. The BrnsLtp genes present on duplicated chromosomal segments are connected by blue lines between the two relevant chromosomes. Tandem duplicated genes are marked on a yellow background. The conserved collinear blocks on each chromosome are labeled A to X and are color-coded according to inferred ancestral chromosomes following an established convention.
Wang et al. [56] confirmed that the genome of B. rapa is almost a complete triplication of the genome of the small cruciferous model plant A. thaliana. Cannon et al. [57] pointed out both segmental and tandem gene duplications play important roles in the expansion and evolution of gene families in plant genomes. In this study, we investigated the influence of duplications on the expansion of the BrnsLtp gene family during evolution. Our results also confirmed that segmental duplication, as well as tandem duplication, contributed to the expansion of this gene family in B. rapa (Fig. 5 and Table 3). Except for chromosome A06, the other nine chromosomes exhibited segmental nsLtp gene duplications (Fig. 5). Commonly, genes separated by ≤5 genes are considered to be tandem duplicates. According to this principle, we found that seven chromosomes, except for chromosomes A01, A03, and A07, had tandem nsLtp gene duplications (Fig. 5 and Table 3). These entire 17 tandem duplicated genes belong to five nsLtp types (I, II, IV, VI, and XI) (Fig. 5 and Table 3). A previous study showed that 18 out of the 49 Arabidopsis nsLtp genes belonging to three types (I, II, and IV) are tandem duplication repeats [17]. Although the results in Table 3 showed that BrnsLtpIV.1 and BrnsLtpIV.2 were not tandem duplicated, we inferred that there was an existing duplication between these two genes based on three reasons: 1. the genes homologous to BrnsLtpIV.1 and BrnsLtpIV.2 are AtLtpIV.5 (AT5G55460.1) and AtLtpIV.4 (AT5G55450.1), respectively, and these two Arabidopsis nsLtp genes are duplication repeats [17]; 2. The BRAD-loci of BrnsLtpIV.1 and BrnsLtpIV.2 are Bra002906 and Bra002907 (Table 1) respectively, which is adjacent; and 3. BrnsLtpIV.1, BrnsLtpIV.2, and BrnsLtpIV.3 share 80.34% similarity in their coding sequences (data not shown) (Figure S3). As for BrnsLtpIV.3 (Bra002914) and BrnsLtpIV.2 (Bra002907), which are separated by six other gene loci, we consider BrnsLtpIV.3 to be a segmental duplication of BrnsLtpIV.1 and BrnsLtpIV.2 rather than a tandem duplication (Fig. 5). In spite of the homologous nsLtp genes determined from publicly available data that provided useful information (Table 3), we found some minor defects in BRAD after careful analysis. For example, BrnsLtpII.2 may be homologous to BrnsLtpII.12 because their coding sequences and their deduced protein sequences are up to 92.18% and 83.51% identical, respectively (Fig. 6). With the exception of BrnsLtpI.6/10/11, BrnsLtpI.5 and BrnsLtpI.13 may also be homologous to AtLtpI.11/12 (Fig. 6). Though the results of Table 3 show that BrnsLtpI.18 and BrnsLtpI.19 share a tandem duplication between them, the identity of their coding sequences was low, only 50.96%. This low identity (51.91%) was also observed between BrnsLtpI.15 and BrnsLtpI.7 (data not shown). However, BrnsLtpI.16 has high sequence similarity with BrnsLtpI.7/18 not only in the coding sequences (89.71%) but also in the protein sequences (82.92%) (Fig. 6). Therefore, BrnsLtpI.7/18 might be homologous to BrnsLtp16 rather than to BrnsLtpI.15/19 (Fig. 6). Furthermore, comparison of the homologous nsLtp genes in Arabidopsis and the three B. rapa subgenomes revealed that the conserved collinear block of each BrnsLtp is highly identical to that in Arabidopsis (Fig. 5 and Table 3).
Figure 6. Alignments of the coding sequences and the deduced protein sequences of selected BrnsLtp genes in B. rapa.
(A) Alignment of the BrnsLtpII.2 and BrnsLtpII.12 coding sequences. (B) Alignment of the BrnsLtpII.2 and BrnsLtpII.12 protein sequences. (C) Alignment of the BrnsLtpI.5/6/10/11/13 coding sequences. (D) Alignment of the BrnsLtpI.5/6/10/11/13 protein sequences. Nucleic acid bases or amino acid residues in positions conserved in 100, 75, and 50% of all sequences are shaded in dark blue, purple, and light blue, respectively. The asterisks in (B) and (D) indicate the cysteine residues of the deduced protein backbones.
Expression analysis of BrnsLtp genes
The B. rapa expressed sequence tags (ESTs) in GenBank are a valuable resource for gene discovery, genome annotation, and analysis of gene expression in this species. To investigate the expression patterns of BrnsLtps, we first took full advantage of the B. rapa ESTs data to analyse EST counts from six different organs (buds, flowers, leaves, roots, and siliques) (Table 4, Table S3). Our results showed that there were no EST data for eight BrnsLtps, EST data for 12 BrnsLtps were available only for unspecified tissue, and the most abundant nsLtp expression was attributed to type I BrnsLtps (Table 4, Table S3). Wang et al. [52] also found that rice type I nsLtps had the most ESTs counts after analysis of the public rice microarray data. Furthermore, the ESTs of 15 BrnsLtps seems to be exclusively detected in a single organ (Table 4). These genes included three genes present in the bud with UniGene numbers Bra.140 (BrnsLtpIII.2/3) and Bra.20831 (BrnsLtpIX.2); one flower gene with UniGene number Bra.25295 (BransLtpIX.1); seven leaf genes with UniGene numbers Bra.18420 (BrnsLtpI.12 and BrnsLtpXI.4/5/6), Bra.2602 (BrnsLtpIV.2/3), and Bra.30903 (BrnsLtpXI.2); three root genes with UniGene numbers Bra.17303 (BrnsLtpI.17), Bra.5092 (BrnsLtpIV.6), and Bra.26893 (BrnsLtpV.1); and one silique gene with the UniGene number Bra.12938 (BrnsLtpIV.7) (Table 4). If we consider the bud and flower as a whole (inflorescence), then the ESTs of six other genes appeared to be specifically detected in this reproductive tissue, including Bra.1371 (BrnsLtpI.16/18), Bra.503 (BrnsLtpII.15), Bra.21209 (BrnsLtpIII.1), Bra.21558 (BrnsLtpIX.3), and Bra.389 (BrnsLtpY.1) (Table 4). In total, 21 out of 63 BrnsLtps were likely to show specific expression patterns. Moreover, no ESTs were found specifically in the stem, and only four genes (BrnsLtpI.1/3/4/8) had ESTs in the stem in the current databases (Table 4).
Table 4. Expression analysis of BrnsLtp genes by UniGene (Transcripts Per Million, TPM).
| Name | UNa | Bb | Fc | Ld | Re | Sf | Sig | WPh | UTi | Other | Name | UN | B | F | L | R | S | Si | WP | UT | Other |
| BrnsLtpI.1 | Bra.119 | 2180 | 856 | 1724 | 349 | 949 | 4151 | √ | √ | √ | BrnsLtpII.14 | Bra.11101 | – | – | – | – | – | – | – | √ | – |
| BrnsLtpI.2 | Bra.1471 | 363 | 0 | 172 | 279 | 0 | 125 | √ | √ | √ | BrnsLtpII.15 | Bra.503 | 1635 | 2336 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpI.3 | Bra.71 | 2180 | 1012 | 57 | 0 | 474 | 4403 | – | – | √ | BrnsLtpIII.1 | Bra.21209 | 1453 | 233 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpI.4 | Bra.119 | 2180 | 856 | 1724 | 349 | 949 | 4151 | √ | √ | √ | BrnsLtpIII.2 | Bra.140 | 302 | 0 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpI.5 | Bra.13066 | – | – | – | – | – | – | – | √ | – | BrnsLtpIII.3 | Bra.140 | 302 | 0 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpI.6 | Bra.13066 | – | – | – | – | – | – | – | √ | – | BrnsLtpIV.1 | – | – | – | – | – | – | – | – | – | – |
| BrnsLtpI.7 | Bra.8616 | 2604 | 1869 | 0 | 0 | 0 | 0 | √ | √ | – | BrnsLtpIV.2 | Bra.2602 | 0 | 0 | 114 | 0 | 0 | 0 | √ | – | √ |
| BrnsLtpI.8 | Bra.71 | 2180 | 1012 | 57 | 0 | 474 | 4403 | – | – | √ | BrnsLtpIV.3 | Bra.2602 | 0 | 0 | 114 | 0 | 0 | 0 | √ | – | √ |
| BrnsLtpI.9 | Bra.1589 | 0 | 233 | 574 | 139 | 0 | 880 | √ | √ | √ | BrnsLtpIV.4 | Bra.3442 | 0 | 311 | 229 | 0 | 0 | 251 | – | – | – |
| BrnsLtpI.10 | Bra.9345 | 0 | 0 | 57 | 0 | 0 | 3270 | √ | – | – | BrnsLtpIV.5 | – | – | – | – | – | – | – | – | – | – |
| BrnsLtpI.11 | Bra.293 | 121 | 311 | 57 | 0 | 0 | 2390 | – | – | √ | BrnsLtpIV.6 | Bra.5092 | 0 | 0 | 0 | 209 | 0 | 0 | – | – | – |
| BrnsLtpI.12 | Bra.18420 | 0 | 0 | 57 | 0 | 0 | 0 | – | – | √ | BrnsLtpIV.7 | Bra.12938 | 0 | 0 | 0 | 0 | 0 | 125 | – | √ | – |
| BrnsLtpI.13 | Bra.10478 | 0 | 0 | 57 | 0 | 0 | 1509 | – | √ | √ | BrnsLtpIV.8 | Bra.10653 | – | – | – | – | – | – | – | √ | – |
| BrnsLtpI.14 | Bra.148 | 121 | 4985 | 57 | 0 | 0 | 1635 | – | √ | – | BrnsLtpV.1 | Bra.26893 | 0 | 0 | 0 | 139 | 0 | 0 | – | – | – |
| BrnsLtpI.15 | Bra.3744 | 0 | 0 | 114 | 69 | 0 | 0 | √ | √ | √ | BrnsLtpV.2 | Bra.3434 | 0 | 0 | 114 | 139 | 0 | 125 | – | – | – |
| BrnsLtpI.16 | Bra.1371 | 5390 | 4907 | 0 | 0 | 0 | 0 | – | √ | – | BrnsLtpVI.1 | Bra.7258 | – | – | – | – | – | – | – | √ | – |
| BrnsLtpI.17 | Bra.17303 | 0 | 0 | 0 | 698 | 0 | 0 | – | – | √ | BrnsLtpVI.2 | – | – | – | – | – | – | – | – | – | – |
| BrnsLtpI.18 | Bra.1371 | 5390 | 4907 | 0 | 0 | 0 | 0 | – | √ | – | BrnsLtpVI.3 | – | – | – | – | – | – | – | – | – | – |
| BrnsLtpI.19 | Bra.9008 | 0 | 389 | 57 | 0 | 0 | 0 | – | – | – | BrnsLtpVI.4 | – | – | – | – | – | – | – | – | – | – |
| BrnsLtpII.1 | – | – | – | – | – | – | – | – | – | – | BrnsLtpVIII.1 | – | – | – | – | – | – | – | – | – | – |
| BrnsLtpII.2 | Bra.11434 | – | – | – | – | – | – | – | √ | – | BrnsLtpIX.1 | Bra.25295 | 0 | 233 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpII.3 | Bra.17924 | 0 | 311 | 0 | 69 | 0 | 0 | √ | – | √ | BrnsLtpIX.2 | Bra.20831 | 60 | 0 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpII.4 | – | – | – | – | – | – | – | – | – | – | BrnsLtpIX.3 | Bra.21558 | 121 | 77 | 0 | 0 | 0 | 0 | √ | – | – |
| BrnsLtpII.5 | Bra.11200 | – | – | – | – | – | – | – | √ | – | BrnsLtpXI.1 | Bra.31801 | – | – | – | – | – | – | – | – | – |
| BrnsLtpII.6 | Bra.4920 | 0 | 311 | 57 | 628 | 0 | 251 | √ | – | √ | BrnsLtpXI.2 | Bra.30903 | 0 | 0 | 57 | 0 | 0 | 0 | – | – | – |
| BrnsLtpII.7 | Bra.4920 | 0 | 311 | 57 | 628 | 0 | 251 | √ | – | √ | BrnsLtpXI.3 | Bra.10370 | – | – | √ | – | – | – | – | – | √ |
| BrnsLtpII.8 | – | – | – | – | – | – | – | – | – | – | BrnsLtpXI.4 | Bra.18420 | 0 | 0 | 57 | 0 | 0 | 0 | – | – | √ |
| BrnsLtpII.9 | Bra.11200 | – | – | – | – | – | – | – | √ | – | BrnsLtpXI.5 | Bra.18420 | 0 | 0 | 57 | 0 | 0 | 0 | – | – | √ |
| BrnsLtpII.10 | Bra.11200 | – | – | – | – | – | – | – | √ | – | BrnsLtpXI.6 | Bra.18420 | 0 | 0 | 57 | 0 | 0 | 0 | – | – | √ |
| BrnsLtpII.11 | Bra.6760 | – | – | – | – | – | – | – | √ | – | BrnsLtpY.1 | Bra.389 | 181 | 77 | 0 | 0 | 0 | 0 | – | – | – |
| BrnsLtpII.12 | Bra.11434 | – | – | – | – | – | – | – | √ | – | BrnsLtpY.2 | Bra.7621 | – | – | – | – | – | – | – | √ | – |
| BrnsLtpII.13 | Bra.17924 | 0 | 311 | 0 | 69 | 0 | 0 | √ | – | √ |
UN, unigene number; bB, bud; cF, flower; dL, leaf; eR, root; fS, seed; gSi, silique; hWP, whole plant; iUT, unspecified tissue; “√” and “–” represent “exist” and “not exist”, respectively. Underlined indicated specific expression.
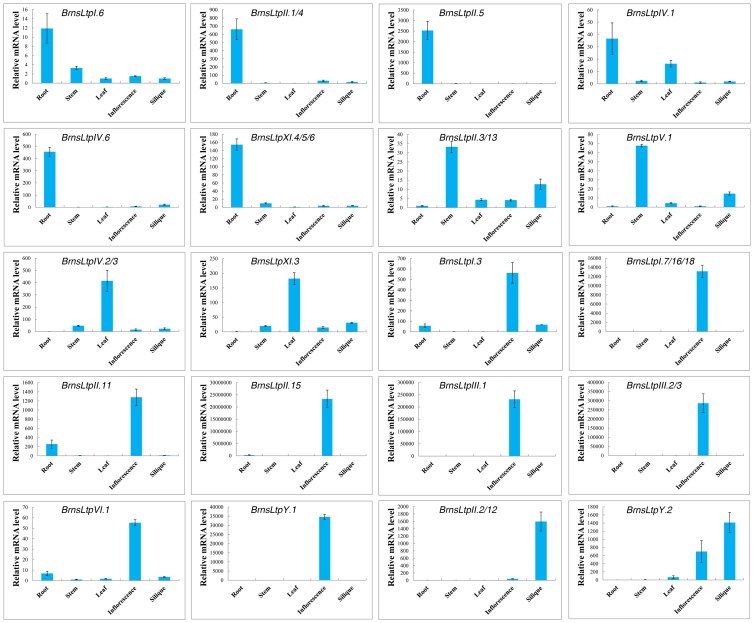
To validate the expression patterns of BrnsLtp genes indicated by the UniGene data, we employed quantitative RT-PCR analysis for five different tissues of B. rapa, including the roots, stems, leaves, inflorescences, and siliques (Fig. 7, Figure S4, Table S4). Our results showed that the expression profiles of the ten genes were in agreement with the EST data. For example, seven genes (BrnsLtpI.16/18, BrnsLtpIII.1, BrnsLtpIII.2/3, BrnsLtpII.15, and BrnsLtpY.1) were highly expressed in the inflorescence, while BrnsLtpIV.2/3 and BrnsLtpIV.6 were strongly expressed in the leaf and root, respectively (Fig. 7). As for the coding sequence of BrnsLtpI.7, it was highly identical to that of BrnsLtpI.16/18 (Figure S3); this similarity meant that a common primer was shared by these three genes for quantitative RT-PCR analysis. Although the relevant UniGene number of BrnsLtpI.7 was different from that of BrnsLtpI.16/18, their ESTs seems to be only detected in the inflorescence (Table 4). Therefore, we considered that BrnsLtpI.7 and BrnsLtpI.16/18 were specifically expressed in the inflorescence, which consistent with the results shown in Figure 7. The expression patterns of the other five BrnsLtp genes (including BrnsLtpI.12, BrnsLtpV.1, and BrnsLtpXI.4/5/6) were not in accordance with the results obtained from digital expression analysis (Table 4, Fig. 7, Figure S4). For example, BrnsLtpV.1 was highly expressed in the stem rather than in the root, while BrnsLtpXI.4/5/6 was highly expressed in the root rather than in the leaf (Table 4, Fig. 7). Additionally, another 14 BrnsLtp genes were found with specific expression patterns, such as five genes (BrnsLtpI.6, BrnsLtpII.1/4, BrnsLtpII.5, and BrnsLtpIV.1) that were expressed in the root, two genes (BrnsLtpII.3/13) in the stem, one gene (BrnsLtpXI.3) in the leaf, three genes (BrnsLtpI.3, BrnsLtpII.11, and BrnsLtpVI.1) in the inflorescence, and three genes (BrnsLtpII.2/12 and BrnsLtpY.2) in the silique (Fig. 7). In sum, 29 out of 63 BrnsLtps were found with specific expression patterns, and 11 BrnsLtps were specifically expressed in the inflorescence (Fig. 7). The cis-acting regulatory elements (CREs) located in the promoter sequence are considered to regulate the gene expression level. Therefore, we investigated the CREs of the 11 BrnsLtps with inflorescence-specific expression patterns. And our results indicated that a certain number of POLLEN1LELAT52 [58] and GTGANTG10 [59] CREs that belong to the late pollen genes are present in the promoters of all these 11 BrnsLtps (Table 5). Additionally, we found that some BrnsLtps might be homologous to certain nsLtps of other species or varieties with known functions, such as BrnsLtpI.8 and AtLTP1 (AtLtpI.5) (72.88% identity), BrnsLtpI.5 and AtLTP3/4 (AtLtpI.12/11) (82.61% and 80.87% identities, respectively), BrnsLtpI.19 and AtLTP5 (AtLtpI.8) (86.44% identity), and BrnsLtpIV.4 and AtDIR1 (AtLtpIV.1) (66.35% identity); BrnsLtpI.4 was identical to BcLTP (Fig. 8, Table 6). However, due to difficulties in designing specific primers (Figure S3), we did not determine the expression profiles of BrnsLtpI.4/5/8/19 by quantitative RT-PCR. A previous study indicated that AtDIR1 is expressed in seedlings, flowers and leaves [60]. A recent study also showed that long distance movement of DIR1 and the role of DIR1-like during systemic acquired resistance in Arabidopsis [61]. Here, our results showed that BrnsLtpIV.4 was highly expressed in the root, inflorescence, and silique and less so in the stem and leaf (Figure S4). Though BrnsLtpIV.4 was homologous to AtDIR1 (AtLtpIV.1) (Table 3), their expression patterns were different. This may be due to the differences in regulatory elements in promoter regions via deletion and/or acquisition of regulatory sequences during evolution. In addition, the expression profile of BrnsLtpII.14 was not presented here because the result was not ideal.
Figure 7. Quantitative RT-PCR analysis for selected BrnsLtp genes in tissues and organs of B. rapa with specific expression patterns.
Table 5. Analysis of the number of plant CREs belonging to pollen genes in the promoters of selected BrnsLtps.
| Gene Name | BRAD-Locus | Promoter Length (bp) | Plant Cis-acting Regulatory Elements | |
| POLLEN1LELAT52 | GTGANTG10 | |||
| BrnsLtpI.3 | Bra005098 | 1927a | 14 | 7 |
| BrnsLtpI.7 | Bra012848 | 896a | 4 | 3 |
| BrnsLtpI.16 | Bra036789 | 2000b | 21 | 18 |
| BrnsLtpI.18 | Bra038907 | 2000b | 10 | 5 |
| BrnsLtpII.11 | Bra028162 | 2000b | 12 | 11 |
| BrnsLtpII.15 | Bra040627 | 2000b | 14 | 10 |
| BrnsLtpIII.1 | Bra009282 | 1348c | 8 | 14 |
| BrnsLtpIII.2 | Bra028294 | 482a | 2 | 5 |
| BrnsLtpIII.3 | Bra029135 | 1456c | 4 | 13 |
| BrnsLtpVI.1 | Bra011229 | 2000b | 9 | 11 |
| BrnsLtpY.1 | Bra024207 | 2000b | 4 | 9 |
a: the promoter sequence is interrupted by many uncertain nucleotides marked with “N”; b: the length of intergenic region is more than 2000bp; c: the length of intergenic region is less than 2000bp.
Figure 8. Alignments of the precursor protein sequences of nsLtps with known functions and their homologous in B. rapa.
Table 6. The summary of the nsLtp genes with known functions.
| Gene name (species or varieties) | Accession number | Known functions | Original reference |
| LTP1 (Arabidopsis thaliana) | At2g38540 | Calmodulin-binding protein and cell differentiation | [95], [26] |
| LTP3 (Arabidopsis thaliana) | At5g59320 | Freezing and drought stress | [43] |
| LTP4 (Arabidopsis thaliana) | At5g59310 | Pathogen defence | [96] |
| LTP5 (Arabidopsis thaliana) | At3g51600 | Pollen tube tip growth and fertilization | [32] |
| DIR1 (Arabidopsis thaliana) | At5g48485 | Systemic resistance signalling | [45] |
| LTP2 (Hordeum vulgare cv. Bomi) | AAA03283.1 | Antibacterial activity | [97] |
| LTP4 (Hordeum vulgare cv. Bomi) | Q43875.1 (Swiss-Prot) Q42842.1 (Swiss-Prot) | Response to bacterial pathogens | [98] |
| BcMF15 (Brassica campestris ssp. chinensis) | EF600901 | Microspore development | [99] |
| BcLTP (Brassica campestris ssp. chinensis) | EF216852 | Secrete and combine extracellular CaM | [100] |
| CaMF2 (Capsicum annuum L.) | JF411954 | Pollen development | [27] |
| nsLTP1 (Ginkgo biloba) | DQ836633 | Proteinase inhibitor | [101] |
| LTP (Lilium longiflorum cv. Nellie White) | AF171094 | Pollen tube adhesion | [29] |
| Ace-AMP1 (Allium cepa L.) | AF004946.1 | Antimicrobial | [54] |
| OsC6 (Oryza sativa L.) | AK064672 (GenBank) OSJNBa0060K21 (EMBL) | Postmeiotic anther development | [28] |
| OsDIL (Oryza sativa L.) | Os10g0148000 | Drought tolerance | [42] |
| MtN5 (Medicago truncatula) | MTR_5g094210 | Efficient nodulation | [39] |
| NtLTP1 (Nicotiana tabacum cv. Xanthi) | AB625593 | Lipid secretion from glandular trichomes | [102] |
| TobLTP2 (Nicotiana tabacum cv. Petit Havana SR1) | Q03461 (Swiss-Prot) | Cell wall extension | [34] |
| CALTPI (Capsicum annuum L.) | AF208832 | Pathogen resistance and long-distance systemic signaling | [38] |
| CALTPII (Capsicum annuum L.) | AF208833 | Pathogen resistance and long-distance systemic signaling | [38] |
Discussion
The genus Brassica, which currently comprises 38 species and numerous varieties, is one of the most significant genera contributing to agriculture. Several species and types of Brassicas are cultivated as vegetables, fodder, and sources of oil and condiments [62], [63]. Due to their agricultural importance, Brassica plants have been the subject of much scientific interest. Previous studies have indicated that polyploidy has played a crucial role in the evolution of Brassicaceae. The famous U's triangle theory, determined from cytological analyses of inter-specific hybrids, indicates that three diploid species, B. rapa (A genome), Brassica nigra (B genome), and Brassica oleracea (C genome), were the ancestors of the three amphidiploid species, B. napus (AC genome), B. juncea (AB genome), and Brassica carinata (BC genome) [64]. Further genetic linkage analysis has confirmed the relationship between these six widely cultivated Brassica species via each possible pair-wise combination [65]–[68]. Additionally, B. rapa and Arabidopsis are close relatives [56], and blocks of conserved genome sequence have been identified between these Brassica species [69]–[74]. Therefore, the B. rapa ‘A’ genome is a valuable resource for studying the evolution of polyploid genomes and underpins the genetic improvement of Brassica-related crops. The astonishing current developments in next-generation sequencing technologies provide unprecedented opportunities for decoding genomic information from various plant species. Fortunately, the genome of Chinese cabbage (B. rapa subsp. pekinensis) has been recently released [56]. The availability of whole genome information can enable us to advance research by focusing on the identification and systematical analysis of large gene families with significant functions. Plant nsLtp is a large transporter family composed of 49 members in Arabidopsis, 52 in rice, 156 in wheat, and 24 in Lotus japonicus, all of which are classified as different types [17], [52], [75]. In the present study, we identified 63 putative nsLtps in the genome of B. rapa, including 19 type I, 15 type II, three type III, eight type IV, two type V, four type VI, one type VIII, three type IX, six type XI, and two nsLTPY nsLtps (Table 1). Previous studies indicated that in addition to searching for the proteins with the Pfam domain PF00234 against entire proteins databases, Blast searches also identified some putative nsLtps [17], [52]. Similarly, four additional putative BrnsLtp genes were found using Blast searches (Fig. 1). Therefore, Blast seems to be a necessary and complementary method for identifying putative and previously unknown nsLtps.
After careful comparison, we found no VII type nsLtps in B. rapa similar to those in Arabidopsis (Table 2). Meanwhile, only one type VII nsLtp was found in the monocotyledon rice genome [17]. This may be further evidence that the B. rapa is more closely related to the dicotyledon Arabidopsis than to the monocotyledon rice. Moreover, a new type (XI) of nsLtps containing six members was identified in the B. rapa genome, which arose during the triplication that occurred after the divergence from Arabidopsis. A recent study showed that nsLtps evolved very early and novel nsLtps types may also have evolved during land plant evolution [51]. Additionally, nine type II AtLtps were found to have no homologous genes in the three subgenomes of B. rapa (Table 3). Wang et al. [56] identified each of the orthologous blocks in the B. rapa genome corresponding to ancestral blocks using collinearity between orthologs on the genomes of B. rapa and A. thaliana and found significant disparity in the gene loss across the triplicated blocks. Therefore, it was evident that polyploid evolution processes are not simple duplications or triplications but are also likely accompanied by gene mutation and loss. It is an intriguing question as to why B. rapa evolved type XI and lost several type II nsLtps. Is it influenced by the relative importance of the genes and their positions on the chromosomes, or is it simply a random phenomenon? Maybe the upcoming release of the B. napus (AC) and B. oleracea (C) genomes along with further gene function analyses will help to answer these questions. Additionally, the gene duplication analysis demonstrated that 42 of the 63 BrnsLtp genes resulted from duplications, including 17 type I, ten type II, two type III, five type IV, three type VI, two type IX, and three type XI BrnsLtps (Fig. 5). Nineteen of the 42 duplicated genes were tandemly duplicated genes, while the other 23 genes were segmentally duplicated genes. This suggests that these two types of duplication events contributed equally to the expansion of the nsLtp genes in B. rapa. Moreover, we found that only one type VIII and very few type III/V/VI nsLtp genes existed in B. rapa (Table 1), just like those in Arabidopsis and rice [17]. A previous study demonstrated that four of the types (III/V/VI/VIII) were not identified in six Solanaceae species thus far [18]. Our results also showed that type V/VIII BrnsLtps had no duplications (Fig. 5). Why do types III/V/VI/VIII nsLtps possess such few members or none at all? It is still an intriguing question that will need to be answered in the future.
Although many putative nsLtps have been identified in the plant kingdom, until now, only a few of their functions have been illustrated. An increasing amount of evidence has suggested that nsLtps may be involved in many biological processes such as defense reactions against phytopathogens, the adaptation of plants to various environmental conditions, and sexual reproduction such as pollen development, pollen tube adhesion and growth, and fertilization (Table 6). As we know, B. rapa and other Brassica-related crops often suffer various diseases caused by phytopathogens and ultimately lead to yield losses. For example, Sclerotinia stem rot is a world-wide plant disease that results in a 10%–20% yield loss in rapeseed in China; in years of serious illness, this yield loss can be as high as 50%. Previously, Sels et al. [40] had classified some Arabidopsis nsLtps into PR-14 proteins. We found that almost all of these PR-14 proteins belong to type I nsLtps. Amazingly, all these Arabidopsis genes have homologous nsLtp genes and duplications in B. rapa (data not shown). Therefore, ‘A’ genome of B. rapa could help us seek pathogen-resistant genes in B. napus (AC genome) and B. juncea (AB genome). Furthermore, many nsLtps have already been reported to possess antimicrobial properties, such as Arabidopsis LTP4, DIR, Ace-AMP1, CALTPI, and CALTPII (summarized in Table 6). We believed that some nsLtps among the 63 identified BrnsLtps may also have this function, and these elite gene resources may accelerate disease-resistant crop improvement. Additionally, the expression profile of one gene in a particular tissue is an important prerequisite to subsequent elucidation of the corresponding protein required for proper execution of developmental, metabolic and signaling process. Investigation of the expression patterns of the BrnsLtp genes demonstrated that 29 BrnsLtp genes were specifically or highly expressed in a particular tissue (Fig. 7). Interestingly, 11 BrnsLtp genes (BrnsLtpI.3, BrnsLtpI.7/16/18, BrnsLtpII.11, BrnsLtpII.15, BrnsLtpIII.1, BrnsLtpIII.2/3, BrnsLtpVI.1, and BrnsLtpY.1) (including all three type III BrnsLtps) were showed to be specifically expressed in inflorescence (Fig. 7). Liu et al. [18] pointed out that no type III nsLtps were identified in six Solanaceae species so far. Do these three type III BrnsLtps, with inflorescence-specific expression patterns, have special functions? A previous study indicated that a high rate of lipid biosynthesis in pollen grains starts after pollen mitosis I [76]. This is later corroborated by the high expression levels observed in the tapetum for several enzymes related to lipid biosynthesis in B. napus [77]. The lipid biosynthesis during anther development is essential for exine formation [78]. A lipid-rich coat, termed the pollen coat, fills the spaces between the baculae of the exine surface and provides several important functions, such as attachment to pollinators, pollen-stigma interactions, and pathogen attack resistance [79]. Several Ltps are expressed in the tapetum, raising the possibility that these genes may function in the transfer of fatty acids and other lipid precursors during pollen wall deposition. Furthermore, stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis Ltps may have diverse roles in plant growth and reproduction [31]. The Arabidopsis LTP1 protein is highly expressed in the cell walls of stigma and pollen grains [47]. Chae et al. [31] also found that LTP1 was specifically abundant in the stigma, as determined by GUS analysis. Though LTP5 was found to be the most weakly expressed gene in the inflorescence among SCA-like LTPs, RT-PCR and GUS analyses showed that LTP5 is present in pollen and the pistil transmitting tract [32]. Additionally, the E2 gene encoding an Ltp of B. napus is exclusively expressed in tapetal cells and developing microspores [80], and the precursor protein sequence of E2 to be almost identical to that of BrnsLtpI.18, with 98.32% identity (data not shown). Actually,our CREs analsysis also showed that all these 11 BrnsLtps with inflorescence-specific expression profile had a certain number of POLLEN1LELAT52 and GTGANTG10 elements that belong to late pollen genes. Therefore, these 11 BrnsLtps may play important roles in sexual reproduction. After carefully analysis, we found that BrnsLtpI.3 may be homologous to Arabidopsis LTP1 (AtLtpI.5) (Table 3). Previous studies demonstrated that Arabidopsis LTP1 may function as a calmodulin-binding protein in Arabidopsis, and the distribution of these protein epitopes was associated with morphogenic events during somatic embryogenesis (Table 6). Interestingly, except for 14 POLLEN1LELAT52 and 7 GTGANTG10, we also found 13 GT1GMSCAM4 (belongs to soybean calmodulin isoform, SCaM-4) [81], two CANBNNAPA (belongs to B. napus napA storage-protein gene, and is required for embyo- and endosperm-specific transcription) [82] and ten EBOXBNNAPA (belongs to B. napus napA storage-protein gene) [83] CREs existed in the BrnsLtpI.3 promoter region (Fig. 9). So, we inferred that BrnsLtpI.3 may have the same functions like Arabidopsis LTP1.
Figure 9. Schematic representation of the selected CREs in the AtLTP1 and BrnsLtpI.3 promoters.
The current study has led to the discovery of 63 putative nsLtps in the genome of B. rapa, including 19 type I, 15 type II, three type III, eight type IV, two type V, four type VI, one type VIII, three type IX, six type XI, and two nsLTPY nsLtps. Interestingly, type XI appeared as a new type of nsLtps of B. rapa containing six members, while nine type II AtLtps have no homologous genes in B. rapa, indicating that gene loss and mutation are also common events that occur as a consequence of polyploidy. This identification and classification may contribute to increased knowledge regarding the nsLtp gene family in plants. Additionally, the expansion of nsLtp genes in B. rapa was attributed to both segmental and tandem duplications. Based on careful analysis, we hypothesize that some of the type I BrnsLtps may have functions similar to Arabidopsis PR-14 proteins in protecting plants by avoiding phytopathogen attacks. Eleven BrnsLtps with inflorescence-specific expression may play important roles in sexual reproduction. Additionally, BrnsLtpI.3 may have functions similar to Arabidopsis LTP1. Going forward, it is important to experimentally characterize these identified BrnsLtps to facilitate our understanding of their functions.
Materials and Methods
Plant materials
The Chinese cabbage (B. rapa ssp. Pekinensis line Chiifu-401–42) plants were cultivated at 20±2°C with 12-h light/12-h dark cycles in a greenhouse at the Oil Crops Research Institute (OCRI) of the Chinese Academy of Agricultural Sciences (CAAS). Five tissues (roots, stems, leaves, inflorescence, and siliques) were collected from 10-week-old plants and frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Identification of BrnsLtps and bioinformatic analysis
All protein files (.pep files) from B. rapa were downloaded from the Brassica Database (BRAD; http://brassicadb.org/brad/). Proteins with the Pfam domain PF00234 (plant lipid transfer/seed storage/trypsin-alpha amylase inhibitor) were then identified using prediction algorithms (http://pfam.sanger.uk/) [84]. Simultaneously, all the ECMs (C…C…CC…CXC…C…C, where X stands for any amino acid) of AtLtps were used as queries for a BlastP search against all proteins from B. rapa with a cut-off value of e-3 to avoid the loss of nsLtp genes due to mis-annotation and not being annotated. Then, the deduced protein sequences of candidate nsLtp genes were manually checked for the presence of the ECMs, and proteins lacking the essential Cys residues were excluded. Subsequently, the proteins without NSSs (checked by the SignalP 4.0, http://www.cbs.dtu.dk/services/SignalP) [85] and with C-terminal GPI anchor signals (predicted by the big-PI Plant Predictor [86] and PSORT [87]), were also removed. After that, the putative proline-rich proteins were excluded from further analyses. The remaining candidate proteins were submitted to the Batch Web CD-Search Tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to verify the presence of LTP domains. The protein sequences of the RAT1 [88] and At2S1 [89] were then Blast-searched against the rest of the candidate nsLtp proteins to exclude the possible inhibitors and cereal storage proteins. Additionally, the proteins with more 120 amino acids at maturity were discarded. The pI and MM of each BrnsLtp were calculated by Compute pI/Mw tool (http://web.expasy.org/compute_pi/). The coding sequences (CDSs) of the BrnsLtp genes and the protein backbones of BrnsLtps are shown in Tables S1 and S2, respectively. The three-dimensional structures of all putative BrnsLtps were also predicted by Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) [90] and are shown in Figure S1.
Sequence and phylogenetic analysis
The genomic DNA sequences of BrnsLtps and AtLtps were obtained from e!BrassEnsembl (http://www.brassica.info/BrassEnsembl/index.html) and TAIR (http://www.arabidopsis.org), respectively. The number and position of exons and introns were determined by comparing the CDSs with their corresponding genomic DNA sequences, and a map of the gene structure was constructed using a gene structure display server (http://gsds.cbi.pku.edu.cn) [91] and modified by Adobe Photoshop CS3 tool (http://www.photoshop.com). Amino acid sequences were aligned and manually corrected using Clustal X (1.83). The resultant sequence alignments (.fasta file) were converted into a MEGA file (.meg), which then served as input to generate a phylogenetic tree with the Neighbor-Joining algorithm statistical method within MEGA 5.05 software [92]. Bootstrapping was performed 10,000 times to obtain support values for each branch.
Additionally, a Bayesian estimation of phylogeny between BrnsLtps and AtLtps was performed using the MrBayes software (http://mrbayes.csit.fsu.edu/index.php). The MCMC (Markov Chain Monte Carlo) method was used to approximate the posterior probabilities of the trees. The nsLtps.nexus file was generated by MEGA 5.05 using an input file in a fasta format that contained the amino acid sequences of ECMs of BrnsLtps and AtLtps. The commands “execute nsLtps.nexus”, “prset aamodelpr = mixed”, and “mcmc ngen = 1000000 samplefreq = 1000” were consecutively employed in the MrBayes software. The program was stopped after execution of the “sump burnin = 25” command. Ultimately, the phylogram was visualized in TreeView software using file (a.t) as input data.
Multiple sequence alignment of the ECMs
The amino acid sequences of the ECMs of the 63 BrnsLtps were obtained according to the eight Cys residues. The alignment of these sequences was then conducted and manually edited using the DNAMAN program (http://www.lynnon.com).
Chromosome localization and gene duplications
The positions of the BrnsLtps were mapped to the ten B. rapa chromosomes by the BRAD-Brassica Genome Browser (http://brassicadb.org/cgi-bin/gbrowse/cbgdb11/). Conserved collinear blocks on the ten chromosomes of the B. rapa genome were drawn as described previously [56]. These A to X blocks were color-coded based on the inferred ancestral chromosome following an established convention [93]. The duplication of BrnsLtps and the position of each nsLtp gene on the syntenic blocks were checked by searching for homologous genes between Arabidopsis and three subgenomes of B. rapa (LF, MF1, and MF2) [56] (http://brassicadb.org/brad/searchSynteny.php).
Digital expression analysis
The total 153,065 expressed sequence tags (ESTs) of B. rapa subsp. Pekinensis were downloaded from the NCBI website (ftp://ftp.ncbi.nih.gov/repository/UniGene/Brassica_rapa/) as UniGene files (Bra.seq.all.gz). The file (Bra.seq.all) was used to construct a local formatted database by the program (blast-2.2.26-ia32-win32.exe, ftp://ftp:ncbi.nlm.nih.gov/blast/executables/release/LATEST/) in Windows system. After that, all the CDSs of 63 BrnsLtps were saved as fasta format in a query file named “query.fasta”. Then, Basic Local Alignment Search Tool (BLAST) was carried out against the formatted database by using the “query.fasta” file as query. Sequences that satisfied e-value less than 10−10 and score-value more than 100 were considered to find the corresponding UniGene number. Finally, the expression profile was suggested by analyzing the EST counts based on UniGene (http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=3711).
Quantitative RT-PCR analysis
We used TRIzol (Sigma, http://www.sigmaaldrich.com) to extract the total RNA from each sample. An amount of 1 μg of total RNA was first digested with DNase I at 37°C for 30 min to remove DNA contamination, and 1 μl of EDTA (50 mM) was added into the mixture to inactive the DNase I by incubation at 65°C for 10 min. Then, the pretreated RNA was transcribed into first-strand cDNA using a Reverse Transcription System (TOYOBO, http://www.toyobo.co.jp/e/). The synthesized cDNA was diluted ten times by DEPC-treated water and used as template for quantitative RT-PCR. Gene-specific primers or common primers were designed using GeneTool (Table S4, Fig. S2). A housekeeping gene, Actin (Table S4), was used to normalize the expression of each gene in different RNA samples. Quantitative RT-PCR analysis was performed by LightCycler@480 SYBR Green I Master using a LightCycler@480II real-time PCR machine (Roche, http://www.roche-applied-science.com), and the relative expression levels were analysed as described by Yuan et al. [94].
Promoter sequences analysis
The promoter sequences of the BrnsLtps and AtLtps were obtained from e!BrassEnsembl and SeqViewer (http://tairvm09.tacc.utexas.edu/servlets/sv), respectively. If the length of intergenic region between nsLtp and its adjacent upstream gene is more than 2 kb, a 2 kb upstream sequence starts from the ATG initiation code was selected as promoter. The CREs of the nsLtps were predicted though PLACE (http://www.dna.affrc.go.jp/PLACE). Then the number of plant CREs belonging to pollen genes in the promoters of selected BrnsLtps was manually counted.
Supporting Information
Three-dimensional structures of the mature BrnsLtp proteins predicted by Phyre2.
(PDF)
Bayesian phylogenetic analysis of the nsLtp family in B. rapa using MrBayes software. The amino acids of the ECMs region were used for the phylogram construction. The different types of nsLtps were marked by a circle or a triangle with different colors. And the accession number of each gene was showed in the parentheses on the right of the corresponding gene name.
(TIF)
Schematic representation of the primers used in quantitative RT-PCR analysis.
(PDF)
Quantitative RT-PCR analysis for selected BrnsLtp genes in tissues and organs of B. rapa with unspecific expression patterns.
(TIF)
The coding sequences of BrnsLtp genes in B. rapa .
(DOCX)
The protein backbones of BrnsLtps.
(DOCX)
List of ESTs of the BrnsLtp genes in B. rapa .
(XLSX)
Primers used in quantitative RT-PCR.
(XLSX)
Funding Statement
This study was supported by the Chinese National Basic Research and Development Program (2011CB109302), the National Natural Science Foundation of China (31100236), and the Wuhan Youth Science and Technology Chen Guang Program (201271031402). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Blein JP, Coutos-Thevenot P, Marion D, Ponchet M (2002) From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci 7: 293–296. [DOI] [PubMed] [Google Scholar]
- 2. Sleight RG (1987) Intracellular lipid transport in eukaryotes. Annu Rev Physiol 49: 193–208. [DOI] [PubMed] [Google Scholar]
- 3. Sprong H, van der Sluijs P, van Meer G (2001) How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol 2: 504–513. [DOI] [PubMed] [Google Scholar]
- 4. Kaplan MR, Simoni RD (1985) Intracellular transport of phosphatidylcholine to the plasma membrane. J Cell Biol 101: 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vance JE, Aasman EJ, Szarka R (1991) Brefeldin A does not inhibit the movement of phosphatidylethanolamine from its sites for synthesis to the cell surface. J Biol Chem 266: 8241–8247. [PubMed] [Google Scholar]
- 6. Holthuis JC, Levine TP (2005) Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol 6: 209–220. [DOI] [PubMed] [Google Scholar]
- 7. Levine T (2004) Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol 14: 483–490. [DOI] [PubMed] [Google Scholar]
- 8. Kader JC (1975) Proteins and the intracellular exchange of lipids. I. Stimulation of phospholipid exchange between mitochondria and microsomal fractions by proteins isolated from potato tuber. Biochim Biophys Acta 380: 31–44. [PubMed] [Google Scholar]
- 9. Ostergaard J, Vergnolle C, Schoentgen F, Kader JC (1993) Acyl-binding/lipid-transfer proteins from rape seedlings, a novel category of proteins interacting with lipids. Biochim Biophys Acta 1170: 109–117. [DOI] [PubMed] [Google Scholar]
- 10. Kader JC, Julienne M, Vergnolle C (1984) Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur J Biochem 139: 411–416. [DOI] [PubMed] [Google Scholar]
- 11. Arondel V, Vergnolle C, Tchang F, Kader JC (1990) Bifunctional lipid-transfer: fatty acid-binding proteins in plants. Mol Cell Biochem 98: 49–56. [DOI] [PubMed] [Google Scholar]
- 12. Tsuboi S, Osafune T, Tsugeki R, Nishimura M, Yamada M (1992) Nonspecific lipid transfer protein in castor bean cotyledon cells: subcellular localization and a possible role in lipid metabolism. J Biochem 111: 500–508. [DOI] [PubMed] [Google Scholar]
- 13. Jose-Estanyol M, Gomis-Ruth FX, Puigdomenech P (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42: 355–365. [DOI] [PubMed] [Google Scholar]
- 14. Douliez JP, Michon T, Elmorjani K, Marion D (2000) Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J Cereal Sci 32: 1–20. [Google Scholar]
- 15. Vergnolle C, Arondel V, Jolliot A, Kader JC (1992) Phospholipid transfer proteins from higher plants. Methods Enzymol 209: 522–530. [DOI] [PubMed] [Google Scholar]
- 16. Shin DH, Lee JY, Hwang KY, Kim KK, Suh SW (1995) High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings. Structure 3: 189–199. [DOI] [PubMed] [Google Scholar]
- 17. Boutrot F, Chantret N, Gautier MF (2008) Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu W, Huang D, Liu K, Hu S, Yu J, et al. (2010) Discovery, Identification and Comparative Analysis of Non-Specific Lipid Transfer Protein (nsLtp) Family in Solanaceae. Genomics, Proteomics & Bioinformatics 8: 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kader JC (1996) Lipid-Transfer Proteins in Plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654. [DOI] [PubMed] [Google Scholar]
- 20. Carvalho Ade O, Gomes VM (2007) Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 28: 1144–1153. [DOI] [PubMed] [Google Scholar]
- 21. Kim TH, Park JH, Kim MC, Cho SH (2008) Cutin monomer induces expression of the rice OsLTP5 lipid transfer protein gene. J Plant Physiol 165: 345–349. [DOI] [PubMed] [Google Scholar]
- 22. Cameron KD, Teece MA, Smart LB (2006) Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol 140: 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee SB, Go YS, Bae HJ, Park JH, Cho SH, et al. (2009) Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola . Plant Physiol 150: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC (1991) Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3: 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coutos-Thevenot P, Jouenne T, Maes O, Guerbette F, Grosbois M, et al. (1993) Four 9-kDa proteins excreted by somatic embryos of grapevine are isoforms of lipid-transfer proteins. Eur J Biochem 217: 885–889. [DOI] [PubMed] [Google Scholar]
- 26. Potocka I, Baldwin TC, Kurczynska EU (2012) Distribution of lipid transfer protein 1 (LTP1) epitopes associated with morphogenic events during somatic embryogenesis of Arabidopsis thaliana . Plant Cell Rep 31: 2031–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen C, Chen G, Hao X, Cao B, Chen Q, et al. (2011) CaMF2, an anther-specific lipid transfer protein (LTP) gene, affects pollen development in Capsicum annuum L. Plant Sci. 181: 439–448. [DOI] [PubMed] [Google Scholar]
- 28. Zhang D, Liang W, Yin C, Zong J, Gu F, et al. (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park SY, Jauh GY, Mollet JC, Eckard KJ, Nothnagel EA, et al. (2000) A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12: 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park SY, Lord EM (2003) Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Mol Biol 51: 183–189. [DOI] [PubMed] [Google Scholar]
- 31. Chae K, Gonong BJ, Kim SC, Kieslich CA, Morikis D, et al. (2010) A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J Exp Bot 61: 4277–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chae K, Kieslich CA, Morikis D, Kim SC, Lord EM (2009) A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 21: 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chae K, Lord EM (2011) Pollen tube growth and guidance: roles of small, secreted proteins. Ann Bot 108: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nieuwland J, Feron R, Huisman BA, Fasolino A, Hilbers CW, et al. (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17: 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guiderdoni E, Cordero MJ, Vignols F, Garcia-Garrido JM, Lescot M, et al. (2002) Inducibility by pathogen attack and developmental regulation of the rice Ltp1 gene. Plant Mol Biol 49: 683–699. [DOI] [PubMed] [Google Scholar]
- 36. Jung HW, Kim W, Hwang BK (2003) Three pathogen-inducible genes encoding lipid transfer protein from pepper are differentially activated by pathogens, abiotic, and environmental stresses. Plant Cell Environ 26: 915–928. [DOI] [PubMed] [Google Scholar]
- 37. Lin P, Xia L, Ng TB (2007) First isolation of an antifungal lipid transfer peptide from seeds of a Brassica species. Peptides 28: 1514–1519. [DOI] [PubMed] [Google Scholar]
- 38. Sarowar S, Kim YJ, Kim KD, Hwang BK, Ok SH, et al. (2009) Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep 28: 419–427. [DOI] [PubMed] [Google Scholar]
- 39.Pii Y, Molesini B, Pandolfini T (2013) The involvement of Medicago truncatula non-specific lipid transfer protein N5 in the control of rhizobial infection. Plant Signal Behav 8. [DOI] [PMC free article] [PubMed]
- 40. Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46: 941–950. [DOI] [PubMed] [Google Scholar]
- 41. Schweiger W, Steiner B, Ametz C, Siegwart G, Wiesenberger G, et al. (2013) Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa-5A, identifies novel candidate genes. Mol Plant Pathol 14: 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo C, Ge X, Ma H (2013) The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol Biol 82: 239–253. [DOI] [PubMed] [Google Scholar]
- 43. Guo L, Yang H, Zhang X, Yang S (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp Bot 64: 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giordani T, Buti M, Natali L, Pugliesi C, Cattonaro F, et al. (2011) An analysis of sequence variability in eight genes putatively involved in drought response in sunflower (Helianthus annuus L.). Theor Appl Genet 122: 1039–1049. [DOI] [PubMed] [Google Scholar]
- 45. Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403. [DOI] [PubMed] [Google Scholar]
- 46.Pii Y, Pandolfini T, Crimi M (2010) Signaling LTPs: A new plant LTPs sub-family? Plant Signal Behav 5. [DOI] [PMC free article] [PubMed]
- 47. Thoma S, Hecht U, Kippers A, Botella J, De Vries S, et al. (1994) Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol 105: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang NJ, Lee CC, Cheng CS, Lo WC, Yang YF, et al. (2012) Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB). BMC Genomics 13 Suppl 1S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jose-Estanyol M, Puigdomenech P (2000) Plant cell wall glycoproteins and their genes. Plant Physiology and Biochemistry 38: 97–108. [Google Scholar]
- 50. Dvorakova L, Cvrckova F, Fischer L (2007) Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genomics 8: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Edstam MM, Viitanen L, Salminen TA, Edqvist J (2011) Evolutionary history of the non-specific lipid transfer proteins. Mol Plant 4: 947–964. [DOI] [PubMed] [Google Scholar]
- 52. Wang HW, Hwang SG, Karuppanapandian T, Liu A, Kim W, et al. (2012) Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum. DNA Res 19: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lehmann MS, Pebay-Peyroula E, Cohen-Addad C, Odani S (1989) Crystallographic data for soybean hydrophobic protein. J Mol Biol 210: 235–236. [DOI] [PubMed] [Google Scholar]
- 54. Tassin S, Broekaert WF, Marion D, Acland DP, Ptak M, et al. (1998) Solution structure of Ace-AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry 37: 3623–3637. [DOI] [PubMed] [Google Scholar]
- 55. Long M, Rosenberg C, Gilbert W (1995) Intron phase correlations and the evolution of the intron/exon structure of genes. Proc Natl Acad Sci U S A 92: 12495–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X, Wang H, Wang J, Sun R, Wu J, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa . Nat Genet 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- 57. Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana . BMC Plant Biol 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37: 859–869. [DOI] [PubMed] [Google Scholar]
- 59. Rogers HJ, Bate N, Combe J, Sullivan J, Sweetman J, et al. (2001) Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10 . Plant Mol Biol 45: 577–585. [DOI] [PubMed] [Google Scholar]
- 60. Champigny MJ, Shearer H, Mohammad A, Haines K, Neumann M, et al. (2011) Localization of DIR1 at the tissue, cellular and subcellular levels during Systemic Acquired Resistance in Arabidopsis using DIR1:GUS and DIR1:EGFP reporters. BMC Plant Biol 11: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, et al. (2013) Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front Plant Sci 4: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cardoza V, Stewart CN (2004) Invited review: Brassica biotechnology: Progress in cellular and molecular biology. In Vitro Cellular & Developmental Biology-Plant 40: 542–551. [Google Scholar]
- 63. Cheng F, Mandakova T, Wu J, Xie Q, Lysak MA, et al. (2013) Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa . Plant Cell 25: 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7: 389–452. [Google Scholar]
- 65. Parkin IA, Sharpe AG, Keith DJ, Lydiate DJ (1995) Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131. [DOI] [PubMed] [Google Scholar]
- 66. Bohuon EJ, Keith DJ, Parkin IA, Sharpe AG, Lydiate DJ (1996) Alignment of the conserved C genomes of Brassica oleracea and Brassica napus . Theor Appl Genet 93: 833–839. [DOI] [PubMed] [Google Scholar]
- 67. Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, et al. (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Suwabe K, Morgan C, Bancroft I (2008) Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa . Genome 51: 169–176. [DOI] [PubMed] [Google Scholar]
- 69. Lukens L, Zou F, Lydiate D, Parkin I, Osborn T (2003) Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana . Genetics 164: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Parkin IA, Sharpe AG, Lydiate DJ (2003) Patterns of genome duplication within the Brassica napus genome. Genome 46: 291–303. [DOI] [PubMed] [Google Scholar]
- 71. Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, et al. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana . Genetics 171: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lim GA, Jewell EG, Li X, Erwin TA, Love C, et al. (2007) A comparative map viewer integrating genetic maps for Brassica and Arabidopsis. BMC Plant Biol 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae . Genome Res 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lysak MA, Cheung K, Kitschke M, Bures P (2007) Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol 145: 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tapia G, Morales-Quintana L, Parra C, Berbel A, Alcorta M (2013) Study of nsLTPs in Lotus japonicus genome reveal a specific epidermal cell member (LjLTP10) regulated by drought stress in aerial organs with a putative role in cutin formation. Plant Mol Biol 82: 485–501. [DOI] [PubMed] [Google Scholar]
- 76. Evans DE, Taylor PE, Singh MB, Knox RB (1992) The interrelationship between the accumulation of lipids, protein and the level of acyl carrier protein during the development of Brassica napus L. Planta. 186: 343–354. [DOI] [PubMed] [Google Scholar]
- 77. Piffanelli P, Ross JH, Murphy DJ (1997) Intra- and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus . Plant J 11: 549–562. [DOI] [PubMed] [Google Scholar]
- 78. Zhou Z, Dun X, Xia S, Shi D, Qin M, et al. (2012) BnMs3 is required for tapetal differentiation and degradation, microspore separation, and pollen-wall biosynthesis in Brassica napus . J Exp Bot 63: 2041–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Boavida LC, Becker JD, Feijo JA (2005) The making of gametes in higher plants. Int J Dev Biol 49: 595–614. [DOI] [PubMed] [Google Scholar]
- 80. Foster GD, Robinson SW, Blundell RP, Roberts MR, Hodge R (1992) A Brassica napus mRNA encoding a protein homologous to phospholipid transfer proteins, is expressed especially in the tapetum and developing microspores. Plant Sci 84: 184–192. [Google Scholar]
- 81. Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, et al. (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135: 2150–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ellerstrom M, Stalberg K, Ezcurra I, Rask L (1996) Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol Biol 32: 1019–1027. [DOI] [PubMed] [Google Scholar]
- 83. Stalberg K, Ellerstom M, Ezcurra I, Ablov S, Rask L (1996) Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199: 515–519. [DOI] [PubMed] [Google Scholar]
- 84. Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34: D247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 86. Eisenhaber B, Wildpaner M, Schultz CJ, Borner GH, Dupree P, et al. (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36. [DOI] [PubMed] [Google Scholar]
- 88. Alam N, Gourinath S, Dey S, Srinivasan A, Singh TP (2001) Substrate-inhibitor interactions in the kinetics of alpha-amylase inhibition by ragi alpha-amylase/trypsin inhibitor (RATI) and its various N-terminal fragments. Biochemistry 40: 4229–4233. [DOI] [PubMed] [Google Scholar]
- 89. Guerche P, Tire C, De Sa FG, De Clercq A, Van Montagu M, et al. (1990) Differential Expression of the Arabidopsis 2S Albumin Genes and the Effect of Increasing Gene Family Size. Plant Cell 2: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371. [DOI] [PubMed] [Google Scholar]
- 91. Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29: 1023–1026. [PubMed] [Google Scholar]
- 92. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11: 535–542. [DOI] [PubMed] [Google Scholar]
- 94. Yuan J, Chen D, Ren Y, Zhang X, Zhao J (2008) Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol 146: 1637–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Z, Xie W, Chi F, Li C (2005) Identification of non-specific lipid transfer protein-1 as a calmodulin-binding protein in Arabidopsis. FEBS Lett 579: 1683–1687. [DOI] [PubMed] [Google Scholar]
- 96. Brotman Y, Lisec J, Meret M, Chet I, Willmitzer L, et al. (2012) Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana . Microbiology 158: 139–146. [DOI] [PubMed] [Google Scholar]
- 97. Molina A, Garcia-Olmedo F (1997) Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J 12: 669–675. [DOI] [PubMed] [Google Scholar]
- 98. Molina A, Diaz I, Vasil IK, Carbonero P, Garcia-Olmedo F (1996) Two cold-inducible genes encoding lipid transfer protein LTP4 from barley show differential responses to bacterial pathogens. Mol Gen Genet 252: 162–168. [DOI] [PubMed] [Google Scholar]
- 99. Tian A, Cao J, Huang L, Yu X, Ye W (2009) Characterization of a male sterile related gene BcMF15 from Brassica campestris ssp. chinensis . Mol Biol Rep 36: 307–314. [DOI] [PubMed] [Google Scholar]
- 100. Wang C, Xie W, Chi F, Hu W, Mao G, et al. (2008) BcLTP, a novel lipid transfer protein in Brassica chinensis, may secrete and combine extracellular CaM. Plant Cell Rep 27: 159–169. [DOI] [PubMed] [Google Scholar]
- 101. Sawano Y, Hatano K, Miyakawa T, Komagata H, Miyauchi Y, et al. (2008) Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiol 146: 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Choi YE, Lim S, Kim HJ, Han JY, Lee MH, et al. (2012) Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. Plant J 70: 480–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three-dimensional structures of the mature BrnsLtp proteins predicted by Phyre2.
(PDF)
Bayesian phylogenetic analysis of the nsLtp family in B. rapa using MrBayes software. The amino acids of the ECMs region were used for the phylogram construction. The different types of nsLtps were marked by a circle or a triangle with different colors. And the accession number of each gene was showed in the parentheses on the right of the corresponding gene name.
(TIF)
Schematic representation of the primers used in quantitative RT-PCR analysis.
(PDF)
Quantitative RT-PCR analysis for selected BrnsLtp genes in tissues and organs of B. rapa with unspecific expression patterns.
(TIF)
The coding sequences of BrnsLtp genes in B. rapa .
(DOCX)
The protein backbones of BrnsLtps.
(DOCX)
List of ESTs of the BrnsLtp genes in B. rapa .
(XLSX)
Primers used in quantitative RT-PCR.
(XLSX)