Abstract
The initiation of flowering in Arabidopsis is retarded or abolished by environmental stresses. Focusing on salt stress, we provide a molecular explanation for this well-known fact. A protein complex consisting of GI, a clock component important for flowering and SOS2, a kinase activating the [Na+] antiporter SOS1, exists under no stress conditions. GI prevents SOS2 from activating SOS1. In the presence of NaCl, the SOS2/GI complex disintegrates and GI is degraded. SO2, together with the Ca2+-activated sensor of sodium ions, SOS3, activates SOS1. In gi mutants, SOS1 is constitutively activated and gi plants are more highly salt tolerant than wild type Arabidopsis. The model shows GI as a transitory regulator of SOS pathway activity whose presence or amount connects flowering to environmental conditions.
Keywords: GIGANTEA (GI), SOS1, SOS2, SOS3, high salinity, Arabidopsis
To survive, plant species had to adjust to changes in the environment over geological times, or to adapt to niche environments characterized by extreme, unfavorable conditions. Prime examples are crops, most of which originated in subtropical areas of the earth. They have been altered to thrive in a range of geographical latitudes, under different light intensities and light periods, and different temperature, humidity and types of soil. Crucial points determining wild plant reproductive success and farmer’s yield is the seasonal progression guiding growth, flowering and seed or fruit set and ripening. The plant model Arabidopsis, a long-day plant like many cereals (barley, wheat, oat, etc), spinach, potatoes, radish, onion, sugar beet and horticultural crops (carnation, rapeseed for canola oil) blooms as days get longer and flowers when exposed to light in excess of 12 h.1 Plants measure day length to decide on flowering and the transition from the vegetative to the reproductive stage.2 A plant internal biological clock, the circadian clock, is at the basis of this photoperiodism. This clock is also a known requisite for plants to cope with changing environments and to sustain a number of biological functions.3
Arabidopsis GIGANTEA (GI), encoded by a single gene,4 is confined to plant species, and has so far been found in all plants.5-8 The GI protein functions in circadian clock maintenance and the elicitation of photoperiod-dependent flowering.9-12 This is accomplished by day time accumulation of the GI protein followed by proteasome-dependent degradation during the night.13 GI transcript expression is itself under control by the circadian clock.11,12 GI mainly controls flowering by regulating the time of day during which two other crucial components of the photoperiod-dependent flowering pathway are expressed. One is CO (Constans), a nuclear zinc finger protein,14-16 the second being FT (Flowering Locus T), a floral integrator encoding a RAF-kinase-inhibitor-like protein.17gi mutants exhibit lower transcript expressions and changed rhythms of two circadian clock oscillators, CCA1 (circadian clock associated 1) and LHY (late elongated hypocotyl). In addition, gi mutants flower late compared with wild type in long days.9
GI seems to be involved in other important biological functions such as sucrose metabolism18 and cell wall deposition.19 Moreover, gi recessive mutants show defects in abiotic stress responses accompanied by pleiotropic phenotypes and tolerance to paraquat-induced oxidative stress, which is light dependent.20 The longer hypocotyl of gi mutants under constant red light suggests GI involvement in PhyB-mediated red light signaling,4 and the gi mutant is sensitive to low temperature.21,22 Also, gi plants contain high starch levels suggesting starch accumulation and the initiation of flowering to be regulated by GI.23
Although precise biochemical functions for GI have not been defined it appears that GI is part of a network receiving inputs on environmental cues and transmitting them to modulate circadian timing for growth and development. We have recently substantiated this notion based on the recognition that the presence of GI and its absence in gi mutants are crucial for the initiation of flowering in relationship to the plants tolerance to high salinity (Fig. 1).

Figure 1.gi mutants exhibit increased salt tolerance. Three-wk-old plants were irrigated every other day with NaCl solution for two wk.
Salt stress delays flowering in Arabidopsis wild type as a result of reduced transcript levels for CO and FT.24 Flowering of gi in the absence of salt stress is similar to the retardation of flowering in wild type after salt treatment. However, GI protein amount is drastically decreased upon salt treatment. In fact, salt promotes 26S proteasome dependent degradation of the GI protein. The resistance against salt of gi mutants is not due to increased high levels of salt-induced osmoprotectants, such as proline based on expression of, P5CS1 (delta 1-pyrroline-5-carboxylate synthetase) and the ABA independent transcription factor, DREB2A (dehydration-responsive element binding protein 2A). Rather, increased tolerance is based on enhanced activity of SOS1 (salt overly sensitive 1), a Na+/H+ antiporter.
The process is regulated by protein:protein interactions between three proteins: GI, SOS2, a kinase and the SOS1 antiporter protein localized to the plasma membrane. In the absence of NaCl, GI protein directly binds to the SOS2 kinase which is then prevented from phosphorylating and thus activating the antiporter activity of SOS1. The salt stress dependent degradation of GI frees SOS2 to interact with SOS3, a Ca2+ activated/binding protein. SOS1 phosphorylation by the SOS2/SOS3 complex then establishes export of sodium ions from cells that leads to enhanced salt tolerance
We propose a new model for the functioning of GI as a link between the clock and vegetative vs. reproductive growth depending on a changing environment (Fig. 2). The model establishes a precise function for GI that is supported by dynamic changes in protein-protein interactions. This model extends an existing salt stress response network including transcription mediated gene expression and Na+ export from cells by the three components of the well-studied SOS pathway
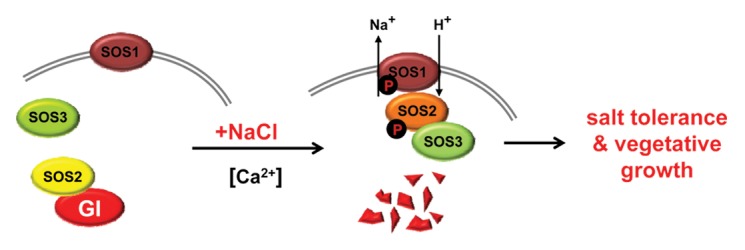
Figure 2. The interaction between GI and components of the SOS pathway. GI acts as a negative regulator of SOS1 activity in the absence of elevated [NaCl]. NaCl-induced degradation of GI leads to SOS2-initiated activation of SOS1 by phosphorylation, while SOS1 is constitutively active in the absence of GI (gi). Salt-dependent degradation of GI or its absence retard or abolish the progression toward flowering.
GI is a very large protein of about 130 kDa, has no known or characterized domains. It is not found in other kingdoms. Functions of GI can so far only be described biochemically by finding interactors. Molecular interactors for GI include F-box proteins, FKF1 (flavin-binding, kelch repeat, F-box 1) in photoperiod-dependent flowering25 and ZTL (zeitlupe) in the circadian clock.26 Another GI interactor is a GA (gibberellic acid) signaling negative regulator, SPY (spindly), O-linked N-acetylglucosamine transferase27 whose mutants spy-1 and spy-3 are resistant to drought and high salinity.27,28 The pleiotropic phenotypes of gi mutants suggest various associating partnerships in diverse biological regulations.
This leads to questions about the significance of GI’s involvement indifferent signaling networks. Sequestration of GI by SOS2 in the form of a negative regulatory circuit interrupts a futile biochemical reaction in the absence of salt stress, while at the same time preserving GI to participate in other functions. The changing environment—salt stress—leads to the degradation of GI, which eliminates its functioning in its other reactions. This may in fact enable other reactions to proceed for which GI is a negative regulator while interrupting circuits for which GI is required as an interacting partner.
Salt tolerance is dramatically increased in the presence of NaCl as GI is degraded. As a consequence of the absence of GI (gi) or following its salt-dependent degradation flowering is delayed or abolished. In contrast, salt tolerant gi plants show accelerated growth in high NaCl. This indicates the presence of functional salt exclusion or export mechanisms in a species that is considered highly salt-sensitive, possibly demonstrating higher order decision making processes superseding the biochemical machinery in plants. The mechanism exemplifies an ability of plants to employ synthesized proteins whose accumulation is regulated according to the time of day to predict and deal with changing environmental conditions
Acknowledgments
We thank Dr Hans J. Bohnert for critical reading and insightful comments. This work was supported by grants from the World class University Program (R32-10148) funded by the Ministry of Education, Science and Technology and Next-Generation BioGreen21 Program (SSAC, grant no: PJ009557), Rural Development Administration, Republic of Korea.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24820
References
- 1.Garner WW. Comparative responses of long-day and short-day plants to relative length of day and night. Plant Physiol. 1933;8:347–56. doi: 10.1104/pp.8.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot. 2007;58:3091–7. doi: 10.1093/jxb/erm159. [DOI] [PubMed] [Google Scholar]
- 3.Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–3. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA. 2000;97:9789–94. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayama R, Izawa T, Shimamoto K. Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 2002;43:494–504. doi: 10.1093/pcp/pcf059. [DOI] [PubMed] [Google Scholar]
- 6.Dunford RP, Griffiths S, Christodoulou V, Laurie DA. Characterisation of a barley (Hordeum vulgare L.) homologue of the Arabidopsis flowering time regulator GIGANTEA. Theor Appl Genet. 2005;110:925–31. doi: 10.1007/s00122-004-1912-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhao XY, Liu MS, Li JR, Guan CM, Zhang XS. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol Biol. 2005;58:53–64. doi: 10.1007/s11103-005-4162-2. [DOI] [PubMed] [Google Scholar]
- 8.Curtis IS, Nam HG, Yun JY, Seo K-H. Expression of an antisense GIGANTEA (GI) gene fragment in transgenic radish causes delayed bolting and flowering. Transgenic Res. 2002;11:249–56. doi: 10.1023/A:1015655606996. [DOI] [PubMed] [Google Scholar]
- 9.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 10.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–88. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–82. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 12.Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17:2255–70. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David KM, Armbruster U, Tama N, Putterill J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 2006;580:1193–7. doi: 10.1016/j.febslet.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–57. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 15.Robson F, Costa MMR, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–31. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- 16.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–6. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 17.Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–20. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 18.Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA. 2011;108:5104–9. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards J, Martin AP, Andriunas F, Offler CE, Patrick JW, McCurdy DW. GIGANTEA is a component of a regulatory pathway determining wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana. Plant J. 2010;63:651–61. doi: 10.1111/j.1365-313X.2010.04269.x. [DOI] [PubMed] [Google Scholar]
- 20.Kurepa J, Smalle J, Van Montagu M, Inzé D. Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–64. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- 21.Cao SQ, Song YQ, Su L. Freezing sensitivity in the gigantea mutant of Arabidopsis is associated with sugar deficiency. Biol Plant. 2007;51:359–62. doi: 10.1007/s10535-007-0073-1. [DOI] [Google Scholar]
- 22.Cao S, Ye M, Jiang S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005;24:683–90. doi: 10.1007/s00299-005-0061-x. [DOI] [PubMed] [Google Scholar]
- 23.Eimert K, Wang SM, Lue WI, Chen J. Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell. 1995;7:1703–12. doi: 10.1105/tpc.7.10.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K, Wang Y, Han C, Zhang W, Jia H, Li X. GA signaling and CO/FT regulatory module mediate salt-induced late flowering in Arabidopsis thaliana. Plant Growth Regul. 2007;53:195–206. doi: 10.1007/s10725-007-9218-7. [DOI] [Google Scholar]
- 25.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–5. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W-Y, Fujiwara S, Suh S-S, Kim J, Kim Y, Han L, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–60. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 27.Tseng T-S, Salomé PA, McClung CR, Olszewski NE. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell. 2004;16:1550–63. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin F, Kodaira K-S, Maruyama K, Mizoi J, Tran L-SP, Fujita Y, et al. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 2011;157:1900–13. doi: 10.1104/pp.111.187302. [DOI] [PMC free article] [PubMed] [Google Scholar]


