Abstract
Brassinosteroids (BRs) and Gibberellins (GAs) are two principal groups of growth-promoting phytohormones. Accumulating evidence supports that there are crosstalks between BR and GA signaling pathways. However, a molecular mechanism for direct signaling crosstalk between BRs and GAs was not revealed until recently. Works from three different groups demonstrated that an interaction between BZR1/BES1 and DELLAs, two groups of key transcriptional regulators from the BR and GA signaling pathways, respectively, mediates the direct signaling crosstalk between BRs and GAs in controlling cell elongation in Arabidopsis. It was shown that DELLA proteins not only affect the protein stability but also inhibit the transcriptional activity of BZR1. Thus, GAs promote cell elongation, at least in part, through releasing DELLA-mediated inhibition of BZR1. This review aims to introduce these recent advances in our understanding of how BRs and GAs coordinate to regulate plant growth and development at the molecular level.
Keywords: brassinosteroids, gibberellins, signaling crosstalk, BRASSINAZOLE- RESISTANT 1, BRI1-EMS-SUPPRESSOR 1, DELLAs, growth and development
Introduction
As sessile organisms, plants need to constantly adapt their growth and development to the changing environment through integrating intrinsic hormonal signals and extrinsic environmental cues. To date, at least eight classes of phytohormones have been identified and characterized in plants, which include auxins, gibberellins (GAs), abscisic acids (ABAs), cytokinins (CKs), ethylene, brassinosteroids (BRs), jasmonates (JAs) and strigolactones.1 Among them, BRs and GAs are two major classes of growth-promoting hormones. BRs, a group of plant-specific polyhydroxylated steroidal hormones that were first discovered in the 1970s, regulate a wide range of growth and developmental processes, including cell elongation, seed germination, stomata formation, vascular differentiation, plant architecture, flowering, male fertility and senescence.2,3 In particular, plants with blocked BR biosynthesis or signaling show a typical dwarf phenotype, providing unambiguous evidence that BRs are essential for normal plant growth and development. GAs, a family of well-known tetracyclic diterpenoid phytohormones, play multiple functions in plant growth and development, including seed germination, stem and hypocotyl elongation, leaf and hypocotyl expansion, pollen maturation and flowering.4-6The GA deficient or insensitive mutants also display dwarfism, indicating the pivotal roles of GAs in controlling cell elongation and regulation of plant growth. Despite these important and overlapping functions of BRs and GAs, how they coordinate to regulate plant growth and development and whether there is a direct crosstalk between their signaling pathways have been the questions of debate and vigorous study. Recently, with the identification and characterization of the core components in both BR and GA signaling pathways, significant progresses have been made in the understanding of BR and GA action and signaling mechanisms. These progresses have enabled us to study the signaling crosstalk between these two hormones. For example, three recent studies from three different laboratories demonstrated that BR and GA signaling pathways integrate at the transcriptional level, which is mediated by a direct interaction between the transcription factors BZR1, BES1 and DELLA proteins.7-9
The Signaling Pathway of BRs
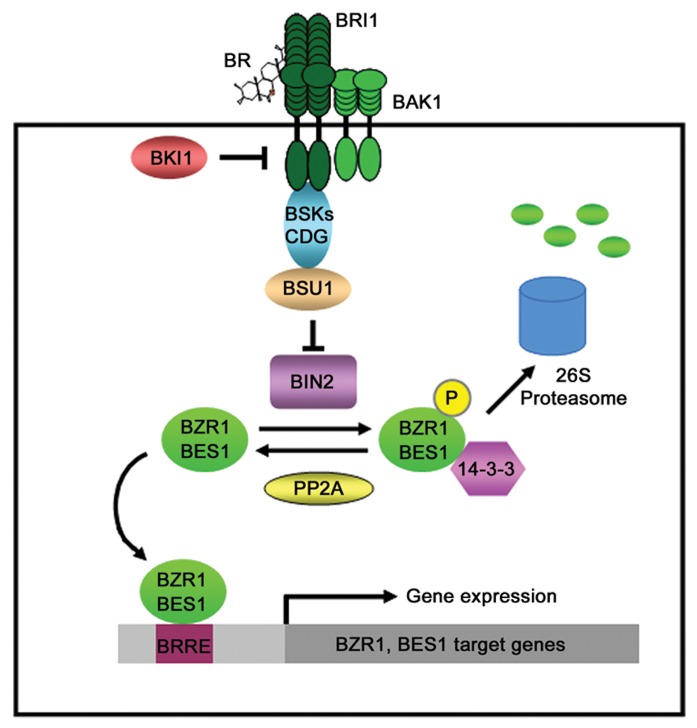
Over the past more than one decade, intensive molecular, genetic and biochemical studies, particularly in the model plant Arabidopsis thaliana, have led to the identification of key BR signaling components and the establishment of a nearly complete signal transduction pathway (Fig. 1). BRs are perceived by the plasma-membrane localized BR receptor Brassinosteroid-Insensitive 1(BRI1), a leucine-rich-repeat containing receptor-like kinase (LRR-RLK).10 The binding of BRs activates the intracellular kinase domain of BRI1 and promotes its association with another LRR-RLK kinase BRI1-Associated Receptor Kinase 1 (BAK1), which enhances BRI1’s kinase activity and BR signaling.11-13 The binding of BRs to BRI1 also leads to the disassociation of BRI1 from an inhibitory protein BRI1 Kinase Inhibitor 1 (BKI1), which binds the C-terminal tail of BRI1 to prevent BAK1 binding and thus inhibit BR signaling when BR signal is absent.11,13-15 The activated BRI1 then triggers a series of phosphorylation events, including phosphorylation and activation of two downstream BR-signaling kinases BR-Signaling Kinase 1 (BSK1) and Constitutive Differential Growth 1 (CDG1), which in turn phosphorylate and activate BRI1-Suppressor 1 (BSU1), a Ser/Thr phosphatase.16,17 The activated BSU1 then dephosphorylates and inactivates Brassinosteroid-Insensitive 2 (BIN2), a key negative regulator of BR signaling.18 BIN2 is a cytoplasmic GSK3-like protein kinase and inhibits BR signaling by phosphorylating and inactivating BZR1 and BES1, two transcription factors that positively regulate BR signaling.11,19,20 BIN2 was found to bind to a 12-amino acid docking site in the C-terminus of BZR1 and BES1 and phosphorylate them through multiple GSK3-like phosphorylation sites.21,22 The phosphorylated BZR1 and BES1 are likely to be retained in the cytoplasm by the 14-3-3 phosphopeptide-binding proteins and degraded by the 26S-proteasome or disabled in their DNA binding activity.19,23-25 In the presence of BR signal, BZR1 and BES1 are dephosphorylated and activated by protein phosphatase 2A (PP2A),26 leading to its nuclear translocation and binding to their target genes through the BR-response element (BRRE) and/or E-box sequences.27-30
Figure 1. A current model for brassinosteroid (BR) signaling in Arabidopsis. BRs are perceived by the extracellular domain of the BR receptor BRI1. BR binding activates BRI1 through homodimerization and heterodimerization with its partner BAK1 and releasing it from its inhibitory protein BKI1. The activated BRI1 then sequentially phosphorylates and activates two downstream kinases BSK1 and CDG1 and a Ser/Thr phosphatase BSU1 that will inactivate the BIN2 kinase, a negative regulator of BR signaling. BR signal also activates PP2A, a phosphatase that phosphorylates and activates the transcription factors BZR1 and BES1. The dephosphorylated BZR1 and BES1 will bind the BRRE or E-box motif of their target genes and regulate their expression. When BR signal is absent, BRI1 is bound by BKI1 and cannot interact with BAK1. BIN2 is also in its active form that will phosphorylate and inactivate BZR1 and BES1. The phosphorylated BZR1 and BES1 are retained in the cytoplasm by 14-3-3 proteins and can be degraded by the 26S proteasome.
The Signaling Pathway of GAs
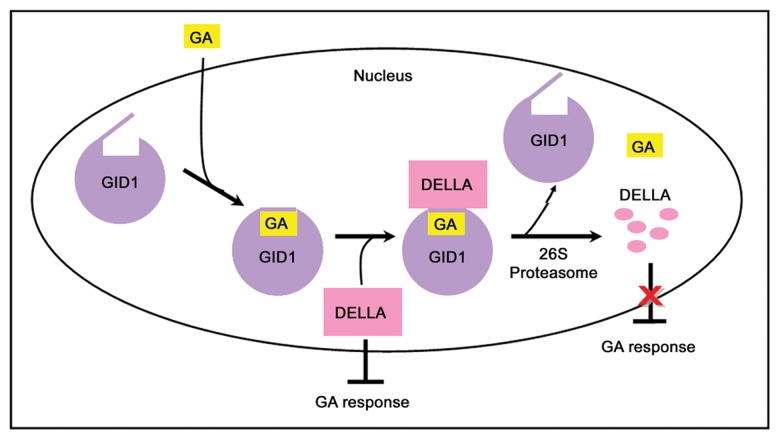
In recent years, considerable progress has been made in the study of molecular mechanisms of GA signaling in plants as well (Fig. 2).4,5,31 GAs are perceived by the GA receptor GIBBERELLIN INSENSITIVE DWARF 1 (GID1) and upon GA binding, the receptor undergoes conformation changes that favor the binding of DELLA proteins, a group of nuclear transcriptional regulators that repress GA signaling and plant growth.4,31-34 The formation of GA-GID-DELLA complex induces the association of DELLAs with the SLEEPY1 (SLY1)/GID2 F-box protein, a SCF-type E3 ubiquitin ligase component that recruits DELLA proteins for ubiquitination and degradation by the 26S proteasome.35,36 Therefore, it is believed that GAs promote plant growth by means of removing the inhibitory DELLA proteins. Different from rice, which contains only one DELLA protein, SLENDER1 (SLR1),37 the Arabidopsis genome encodes five DELLLA members, namely REPRESSOR OF GA1-3 (RGA), GIBBERELLIC ACID INSENSITIVE (GAI), RGA-like 1 (RGL1), RGL2 and RGL3, which accumulate when GA levels are low and directly inactivate a number of growth-promoting transcription factors, such as the bHLH-like phytochrome-interacting factors (PIFs).5
Figure 2. A model for gibberellin (GA) signaling in Arabidopsis. In the absence of GAs, DELLA proteins accumulate in cell nucleus and inhibit GA responses. In the presence of GA signal, the GA receptor GID1 binds the hormone and undergoes a conformational change suitable for DELLAs binding. The resulting GA-GID1-DELLA complex triggers degradation of DELLAs via the 26S proteasome pathway, resulting in release of DELLA-repressed plant growth.
Evidence of BR and GA Interaction from Early Studies
The study of BR and GA interaction has received enormous attention from plant biologists because these hormones are not only principle regulators of plant growth but also share several other overlapping functions, including promotion of seed germination and flowering.38 Although a direct crosstalk between BR and GA signaling pathways was not revealed until recently, indirect evidence for their signaling crosstalk can be seen from some early studies concerning BR and GA interaction in processes including cell elongation, seed germination, flowering, hormone biosynthesis and gene expression, etc.
Cell Elongation
Physiological studies from early days favored the view that GAs and BRs act additively or independently on cell elongation. For example, Gregory and Mandava39 and Katsumi40 found that when mung bean epicotyl and cucumber hypocotyl were sequentially treated with BRs and GAs, no synergistic enhancement of cell elongation can be observed. The independent effects of BRs and GAs were also reported in Arabidopsis for their stimulating effects on root growth.41 However, in 2003, synergistic effects of GAs and BRs were reported by Tanaka et al.42 in Arabidopsis in regulating cell elongation. They showed that the relationship between BRs and GAs was actually concentration-dependent. When more than 10−8 M of brassinolide (BL) and of GA3 were administrated to Arabidopsis seedlings, they act synergistically on elongation of light-grown hypocotyls; however, at lower concentrations, they act additively. This is the first evidence demonstrating the synergistic relationship between BRs and GAs and also the first study clarifying the dose-dependence of these hormones for their additive and synergistic effects.
Seed Germination
Although many studies have observed the promoting effects of both GAs and BRs on seed germination, whether they regulate this process coordinately or independently has been elusive. The results of Leubner-Metzger43 suggest that BRs and GAs promote the germination of tobacco seeds through distinct pathways as the photodormancy of dark-imbibed seeds can be released by GA treatment but not by BR treatment. However, Steber and McCourt44 found that BRs could partially rescue the germination of GA deficient and insensitive mutants in Arabidopsis, suggesting the possibility of GA and BR interaction in controlling seed germination by modulating one another’s biosynthesis or response. In support of this assumption, factors that mediate BR-GA interaction during seed germination have been characterized. These include the Arabidopsis GPA1 and GCR1 proteins. GPA1 is a subunit of the heterotrimeric G protein and the seeds of the gpa1 mutant that carries a null mutation in the GPA1 gene have much reduced responsiveness to GAs but are completely insensitive to BR rescue of germination when the level of GAs in seed is reduced, suggesting that GPA1 couples BRs to potentiate GA-stimulated seed germination.45 GCR1 is a putative G protein-coupled cell surface receptor with the predicted seven-transmembrane (7TM) structure.46 Similar to gpa1, the gcr1 mutant seeds are also less sensitive to GAs and BRs in promoting seed germination,46 and when GCR1 is overexpressed, there is a loss of seed dormancy,46,47 suggesting that GCR1 may be a potential interaction node in GA- and BR-regulated seed germination.
Flowering
GAs are major factors that promote flowering in Arabidopsis48,49and BRs were also found to play a positive role in controlling flowering time.50,51 As a consequence, the GA deficient ga1 mutant and BR deficient cpd mutant are delayed in flowering.52,53 Domagalska et al.54 later showed that the double cpd ga1 mutant was further delayed in flowering than the single cpd mutant and that the double 35S::DWF4/35S::GA5 transgenic line expressing both the BR biosynthetic gene DWF4 and GA biosynthetic gene GA5 flowered significantly earlier than the single 35S::GA5 line, implying that BR’s role in promoting flowering depends on the presence and concentration of GAs. This study supports that GA and BR pathways genetically interact to control the flowering time of Arabidopsis.
Hormone Biosynthesis
BRs and GAs also regulate each other’s biosynthesis. The BR regulation of GA biosynthesis has been consistently reported in plants including tomato,55 pea56 and sunflower57 and the results generally suggest a negative role of BRs in regulating GA biosynthesis. For GA regulation of BR biosynthesis, the evidence mainly came from studies in rice. SPINDLY (SPY), an O-linked N-acetylglucosamine transferase, is a negative regulator of GA signaling.58 The SPY knockdown rice (spy) not only has increased lamina joint bending, a brassinosteroid-related phenotype, but also has increased levels of BRs, suggesting that OsSPY may play roles in both GA signaling and BR biosynthetic pathways.58 In addition to OsSPY, OsGSR1, a GAST (GA-stimulated transcript) gene-encoded protein in rice, was shown to activate BR biosynthesis by directly interacting with a BR biosynthetic enzyme DIM/DWF1.59 In Arabidopsis, our recent study9 showed that the expression of GA20ox2 and GA3ox1, two rate-limiting GA biosynthetic genes, was significantly increased by BR treatment and the bzr1-1D mutation. Also, the expression of CPD and DWF4, two key BR biosynthetic genes, was increased in the gai mutant although they were not affected by GA treatment. These results indicate that BRs and GAs can modulate each other’s biosynthesis and this modulation is mediated by pathways involving BZR1 and DELLA proteins.
Regulation of the Expression of BR and GA Signaling Genes
A recent study reported that BRs and GAs acted antagonistically in the root immunity of rice.60 It was shown that the pathogen Pythium graminicola exploits BRs as virulence factors and hijacks the rice BR machinery to inflict disease, which challenges the prevailing view that BRs positively regulate plant innate immunity.61,62 Moreover, this immunosuppressive effect of BRs was demonstrated, at least in part, due to an antagonistic crosstalk with GAs by enhancing the stability of OsSLR1, the only DELLA protein in rice that functions as a key negative regulator in the resistance to P. graminicola. Vleesschauwer et al.60 also found that both pathogen infection and exogenous BR treatment could increase the expression of OsSLR1. These data suggest that BRs may attenuate the GAs-induced defense responses in rice through interference with GA signaling.60 In contrast to the positive role of BRs in stabilizing DELLA protein in rice, BRs were found to reduce the expression of four DELLA genes in cotton (Gossypium hirsutum) fiber cells, including GhGAI1, which is engaged in fiber cell initiation.63 In a chromatin immunoprecipitation (ChIP) study in Arabidopsis, four of the five DELLA-encoding genes (RGA, GAI, RGL1 and RGL3) were identified as direct targets of BZR1, the transcription factor in the BR pathway, suggesting that BRs may modulate GA responses by directly controlling the expression of DELLA-encoding genes.29 However, in a recent study9 we demonstrated by qualitative real-time PCR (qRT-PCR) analysis9 that the expression of these DELLA genes was not affected by either BR treatment or by the bzr1-1D mutation that enhances BR signaling, suggesting that BZR1 and DELLAs coordinate the interaction between BRs and GAs in controlling cell elongation primarily through protein-protein interaction rather than through BZR1 regulation of DELLA gene expression. Whether BRs regulate DELLA gene expression during other biological processes remains an open question to answer.
Common Target Genes
GA and BR pathways also interact at the level of gene expression. Previous studies have shown that BRs and GAs co-regulate the expression of a number of growth and development related genes either antagonistically or consistently, including γ-TIP,64 MERI-5,65 GASA1 and GA566 in Arabidopsis. The application of high throughput microarray approaches identified more common target genes of both pathways. For instance, the analysis of ~4,000 expressed sequence tags (ESTs) in rice treated with GA and BR revealed some specific genes coordinately regulated by both hormones.67 In Arabidopsis, an early comparative genome expression analysis of BR- and GA-responsive genes has identified co-regulated genes by BRs and GAs, although the number was rather small.68 Recently, by comparing the microarray data sets from BR-insensitive mutant bri1-116 and GA-deficient mutant ga1-3, Bai et al.7 found that GAs and BRs co-regulate a large number of common genes. Of the 1,194 genes affected by ga1-3, 419 genes (35%) were also affected in the bri1-116 mutant. In addition, our comparison of RGA-regulated genes from a published microarray data set69 and the published BZR1 target genes29 also revealed that up to 30% of the RGA-responsive genes are also direct targets of BZR1.9 All these results suggest that BRs and GAs may control a common transcriptional module and this is likely mediated by the BZR1 and DELLA proteins.
The Recent Advances in Uncovering the Molecular Mechanisms of BR and GA Signaling Crosstalk
Although previous physiological, transcriptomic and genetic studies have provided mounting evidence for a cooperative and interdependent relationship between BRs and GAs, a direct crosstalk between their signaling pathways was not revealed until recently. According to Vert and Chory,1 a direct crosstalk between two signaling pathways refers to the sharing of common signaling components or direct interactions between components from their signaling pathways. For examples, the direct signaling crosstalk between BR and auxin signaling was reported to be mediated by an interaction between the BR-regulated kinase BIN2 and the auxin-regulated transcription factor ARF2.70 BIN2 directly phosphorylates and inactivates ARF2, a repressor of auxin signaling, thus resulting in increased transcription of auxin-responsive genes.70 In late 2012, studies from three independent research groups reported that an interaction between BZR1 and DELLA proteins mediates the direct signaling crosstalk between BRs and GAs in controlling cell elongation in Arabidopsis.7-9 This breakthrough finding provided a molecular framework for better understanding of how BRs and GAs coordinate to control plant growth and development.
BRs Mediate GA Responses in Etiolated Seedlings
Previous physiological studies have demonstrated that BRs could improve the germination of severe GA biosynthetic and insensitive mutants.44 In a recent study, Bai et al.7 also showed that BRs could partially recover the growth phenotypes of GA insensitive mutants and wild type seedlings treated with the GA biosynthetic inhibitor paclobutrazol (PAC), but GA treatment could not restore the hypocotyl elongation of either BR deficient or insensitive mutants. Therefore, it was concluded that BR signaling is required for GA function in promoting hypocotyl elongation. This notion was supported by the results of microarray analyses that aimed to study the responses to GAs and BRs in dark-grown BR- and GA-deficient mutants.7,8,29,71In particular, Gallego-Bartolome et al.8 showed that BR treatment could cause a partial or complete reversion of the expression of 40% of genes affected by GA deficiency, whereas GA treatment only reversed 16% of the genes affected by BR deficiency. This difference reflects a hierarchy in BR and GA regulation of gene expression, which is consistent with the results from physiological studies.7 Also, the big number of genes affected by both GAs and BRs from these studies contrasts the small number of common target genes of BRs and GAs identified from an earlier study by Nemhauser et al.,68 which might be due to the different experimental conditions and/or plant materials used. For example, in Nemhauser et al.’s experiment,68microarray was performed with BR- or GA-treated wild type seedlings grown in the light, whereas the microarray experiments of Gallego-Bartolome et al.8and Bai et al.7 were conducted on BR- or GA-deficient seedlings grown in the dark. This difference reflected an impact of light on GA and BR responses.
BZR1 Directly Interacts with DELLA Proteins
Since BRs and GAs control a common transcription module through DELLA and BZR1 activities, it is possible that DELLAs may directly interact with BZR1. Indeed, interaction studies using different in vitro and in vivo approaches, including yeast two-hybrid, pull-down, bimolecular fluorescence complementation (BiFC) and co-immunoprecipitation (Co-IP) assays, demonstrated that BZR1 interacts with RGA, GAI and other DELLA members in Arabidopsis.7-9 BES1 (also known as BZR2), a close homolog of BZR1, also interacts with the DELLA proteins.7,9Detailed protein domain analysis further revealed that the LHR1 domain of RGA or GAI and the BIN2 phosphorylation domain (including the PEST motif) of BZR1 are responsible for their interaction.7-9 These results suggested a role of BZR1, BES1 and DELLA proteins in mediating BR and GA signaling crosstalk and the subsequent functional studies using transcriptional transient assays confirmed that a direct interaction between BZR1 and DELLLAs is required for their antagonistic effects in regulating transcription of genes important for growth.7-9
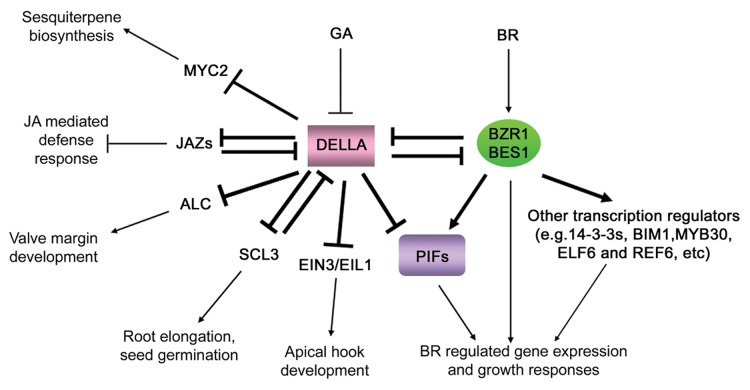
In recent years, DELLA proteins have merged as central regulators of crosstalk among different signaling pathways5,31and our new findings of their interaction with BZR1 and BES1 further strengthened this notion. BZR1 and BES1, two key transcription factors for BR signaling, fulfill their functions by directly controlling thousands of target genes29,30or interacting with other transcriptional regulators, such as DELLAs, PIFs, IWS1, BIM1, MYB30, ELF6 and REF,6etc (Fig. 3).7-9,28,72-75 Thus, like DELLA proteins, BZR1 and BES1 may also function as integrators of BR crosstalk with other signaling pathways.
Figure 3. A hypothetical model of transcription network in plants centered by DELLAs and BZR1/BES1. DELLAs interact with many different transcription factors, including BZR1, BES1, PIFs, MYC2, JAZs, ALC, SCL3 and EIN3/EIL1, to regulate specific developmental processes. BZR1 and BES1 regulate plant growth and development by directly binding to their target genes or interacting with other transcription regulators such as 14-3-3s, BIM1, MYB30, ELF6 and REF,6 etc. Arrows indicate activation and bar-ended lines mean inhibition. Bold lines represent direct protein-protein interactions.
DELLAs Negatively Regulate the BR Pathway and BZR1 Positively Regulates the GA Pathway
Pharmacological analyses showed that both bzr1-1D and bes1-D mutants have reduced sensitivity to the GA biosynthetic inhibitor PAC7,9 and the rga gai double knockout mutant has reduced sensitivity to the BR biosynthetic inhibitor BRZ, as compared with their respective wild types.9 Consistently, the della pentuple mutant lacking all the five members of the DELLA family genes showed a considerably enhanced BR response and the GA-insensitive mutant gai-1, which accumulates high levels of GAI, had reduced BR responses.7 These data suggest that BZR1 serves as a positive regulator of the GA pathway and DELLAs function as negative regulators of the BR pathway. Further genetic studies supported this conclusion. The bzr1-1D mutant was able to partially suppress the short hypocotyl phenotypes of the GA deficient ga1-3 mutant7 but not of the GA insensitive gai mutant.9 Overexpression of a non-degradable RGA protein in the bzr1-1D background also resulted in a dwarf phenotype,9 implying that DELLAs may be epistatic to BZR1 in controlling cell elongation. The reason why bzr1-1D could suppress the phenotype of ga1-3 but not of the gai mutant is likely because BR treatment or bzr1-1D mutation could induce the expression of GA biosynthetic genes,9 which will rescue the GA deficient mutant phenotypes but not that of the GA-insensitive mutants.
GAs and BRs Co-regulate a Large Number of Genes Through DELLAs and BZR1
Since BRs and GAs co-regulate a large number of common target genes7,8 and BZR1 directly interacts with DELLA proteins,7-9 it is important to examine whether the co-regulation of these common target genes was mediated by the BZR1-DELLA interaction module. It was apparent from the transcriptional transient assays that deletions of domains required for BZR1-DELLA interaction (LHR1 of RGA or BIN2 phosphorylation domain of BZR1, respectively) abolished the antagonistic effect of RGA and BZR1.8,9 Also, the results of microarray studies demonstrated that genes affected in the BR-insensitive mutant bri1-116 and GA-deficient mutant ga1-3 overlap significantly.29,71Of the 1,194 genes differentially expressed in ga1-3 when compared with the wild type, 419 genes (35%) were also affected in the bri1-116 mutant.7 Among these co-regulated genes, 387 genes (92.3%) were affected in the same way by bri1-116 and ga1-3. For 276 (71%) of these genes, the effects of bri1-116 were reversed by the bzr1-1D mutation and the effects of ga1-3 were reversed by loss of DELLA proteins,7 suggesting that GAs and BRs exert similar effects on a large number of common genes through DELLA and BZR1 activities.7,8
DELLAs Restrain Plant Growth by Destabilizing and Inactivate BZR1
Since BZR1 and RGA could modulate both BR and GA signaling pathways, we hypothesized that BRs may regulate DELLA protein accumulation and GAs regulate BZR1 accumulation. However, it was shown by three different groups that the accumulation of DELLA proteins was not affected by BR treatment nor by mutations that disrupt BR biosynthesis or signaling (e.g., the BR deficient mutant det2-1 and the BR insensitive mutant bri1-116),7-9 suggesting that unlike GAs, BRs do not induce degradation of DELLA proteins. On the other hand, GAs were found to slightly induce the dephosphorylation of the BZR1 protein and the GA biosynthetic inhibitor PAC worked oppositely.9Consitently, transgenic plants with ectopic expression of undegradable DELLAs (RGA or GAI lacking the 17-aa DELLA domain) showed a drastic diminishment of the BZR1 protein.9 The GA-induced BZR1 stability is likely mediated by PP2A, a protein phosphatase that was shown to dephosphorylate and stabilize BZR1,26 as block of the PP2A function by its inhibitor okadaic acid (OA) abolished GA-induced dephosphorylation of BZR1.9 Thus, it is conceivable to believe that DELLA proteins restrain plant growth, at least in part, by destabilizing the BZR1 protein.
In addition to affecting BZR1 stability, DELLA proteins such as RGA and GAI preferentially interact with the dephosphorylated BZR1.7-9 Since the dephosphorylated BZR1 is the active form of BZR1, its binding with DELLAs may mitigate its transcriptional activity. Indeed, experiments using electrophoretic mobility shift assay (EMSA), protein-DNA pull down assay and ChIP analysis all confirmed that RGA conjugation prevented BZR1’s binding to its target genes,7,8 and results of transcriptional transient assays further demonstrated that DELLAs and BZR1 attenuate each other’s transcriptional activity and target gene expression and the antagonizing effect depends on their physical interaction.7-9 Thus, DELLAs appear to modulate the output of BR signaling pathway, at least in part, by interacting with the active form of BZR1 and inhibiting its transcriptional activities.
Conclusions and Future Perspectives
The recent three publications revealed a molecular mechanism for the direct crosstalk between the BR and GA signaling pathways, which have considerably advanced our understanding of how different signals are integrated and coordinated during plant growth and development. The finding that an interaction between the two groups of transcription factors (BZR1 family members and DELLA proteins) functions as the integration point of the BR and GA signaling pathways to coordinate the expression of downstream target genes provides a clear molecular interpretation for the long-standing question regarding how BRs and GAs synergistically regulate plant growth. Despite this striking progress that has been made, many important questions remain to be answered. For example, how GAs induce BZR1 dephosphorylation and how DELLAs cause BZR1 destabilization is yet unclear. Although the work of Li et al.9 suggested a role of the phosphatase PP2A in these processes, the GA-induced BZR1 dephosphorylation is relatively subtle compared with that induced by BRs and the DELLA-induced PP2A destabilization was only examined by pharmacological treatments with PAC; no in vivo detection of PP2A abundance or activity was performed. In addition, the possibility for whether DELLA proteins mediate BZR1 degradation by interacting with a BZR1-specific ubiquitin E3 ligase has not been tested and this awaits the identification of the E3 ligase. Second, since any components in a signaling pathway may have their spatial and temporal expression patterns, it is necessary to explore how the crosstalk between the BR and GA signaling pathways is spatially and temporally regulated. As such, the effects of crosstalk could be depicted as dynamic outputs instead of “snapshots” of static phenotypes. Third, the recent three papers only reported the interaction between DELLAs and BZR1 in controlling cell elongation. Given the central roles of DELLA proteins in integrating different signaling pathways,73,76-82it is of highly interest to explore whether DELLAs interact with BZR1 to regulate other physiological or developmental processes. Finally, whether other protein components are involved in the crosstalk between GA and BR signaling pathways is also an important question to answer.
Acknowledgments
We apologize for incomplete citation of the references related to the topic due to a space limitation. This work is financially supported by the Hong Kong RGC General Research Funds (CUHK codes 465009, 465410 and 464412), a grant from National Natural Science Foundation of China (no. 91125027) and the Direct Grants from the Chinese University of Hong Kong (to J.-X.H.).
Glossary
Abbreviations:
- BRs
Brassinosteroids
- GAs
Gibberellins
- BZR1
BRASSINAZOLE-RESISTANT 1
- BES1
BRI1-EMS-SUPPRESSOR 1
- ABAs
abscisic acids
- CKs
cytokinins
- JAs
jasmonates
- BRI1
Brassinosteroid-Insensitive 1
- LRR-RLK
leucine-rich-repeat containing receptor-like kinase
- BAK1
BRI1-Associated Receptor Kinase 1
- BKI1
BRI1 Kinase Inhibitor 1
- BSK1
BR-Signaling Kinase 1
- CDG1
Constitutive Differential Growth 1
- BSU1
BRI1-Suppressor 1
- BIN2
Brassinosteroid-Insensitive 2
- PP2A
Protein Phosphatase 2A
- BRRE
BR-Response Element
- GID1
GIBBERELLIN INSENSITIVE DWARF 1
- SLY1
SLEEPY1
- SLR1
SLENDER1
- RGA
REPRESSOR OF GA1-3
- GAI
GIBBERELLIC ACID INSENSITIVE
- RGL1
RGA-like 1
- PIFs
Phytochrome-Interacting Factors
- BL
brassinolide
- 7TM
seven-transmembrane
- SPY
SPINDLY
- GAST
GA-stimulated transcript
- ChIP
chromatin immunoprecipitation
- BiFC
bimolecular fluorescence complementation
- Co-IP
co-immunoprecipitation
- BRZ
brassinazole
- PAC
paclobutrazol
- OA
okadaic acid
- EMSA
electrophoretic mobility shift assay
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24686
References
- 1.Vert G, Chory J. Crosstalk in cellular signaling: background noise or the real thing? Dev Cell. 2011;21:985–91. doi: 10.1016/j.devcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–30. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–24. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- 4.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–98. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- 5.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–45. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 6.Ragni L, Nieminen K, Pacheco-Villalobos D, Sibout R, Schwechheimer C, Hardtke CS. Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell. 2011;23:1322–36. doi: 10.1105/tpc.111.084020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, et al. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14:810–7. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, et al. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:13446–51. doi: 10.1073/pnas.1119992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal. 2012;5:ra72. doi: 10.1126/scisignal.2002908. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–38. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 12.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–12. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–35. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–22. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 15.Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:658–63. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–60. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell. 2011;43:561–71. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–60. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–90. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–91. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 21.Peng P, Zhao J, Zhu Y, Asami T, Li J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J Biol Chem. 2010;285:24646–53. doi: 10.1074/jbc.M110.142547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 23.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 24.Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA. 2007;104:13839–44. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–89. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13:124–31. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–8. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–59. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–77. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–46. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 31.Sun TP. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154:567–70. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;19:1209–20. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirano K, Ueguchi-Tanaka M, Matsuoka M. GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 2008;13:192–9. doi: 10.1016/j.tplants.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 35.McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–30. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16:1392–405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–73. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Gregory LE, Mandava NB. The activity and interaction of brassinolide and gibberellic acid in mung bean epicotyls. Physiol Plant. 1982;54:239–43. doi: 10.1111/j.1399-3054.1982.tb00253.x. [DOI] [Google Scholar]
- 40.Katsumi M. Interaction of a brassinosteroid with IAA and GA3 in the elongation of cucumber hypocotyl sections. Plant Cell Physiol. 1985;26:615–25. [Google Scholar]
- 41.Müssig C, Shin GH, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133:1261–71. doi: 10.1104/pp.103.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka K, Nakamura Y, Asami T, Yoshida S, Matsuo T, Okamoto S. Physiological roles of brassinosteroids in early growth of Arabidopsis: brassinosteroids have a synergistic relationship with gibberellin as well as auxin in light-grown hypocotyl elongation. J Plant Growth Regul. 2003;22:259–71. doi: 10.1007/s00344-003-0119-3. [DOI] [Google Scholar]
- 43.Leubner-Metzger G. Brassinosteroids and gibberellins promote tobacco seed germination by distinct pathways. Planta. 2001;213:758–63. doi: 10.1007/s004250100542. [DOI] [PubMed] [Google Scholar]
- 44.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–9. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullah H, Chen JG, Wang S, Jones AM. Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 2002;129:897–907. doi: 10.1104/pp.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, et al. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–15. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ. GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA. 2002;99:4736–41. doi: 10.1073/pnas.072087699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson RN, Heckman JW, Somerville CR. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992;100:403–8. doi: 10.1104/pp.100.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998;118:773–81. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–59. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–30. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun T, Goodman HM, Ausubel FM. Cloning the Arabidopsis GA1 Locus by Genomic Subtraction. Plant Cell. 1992;4:119–28. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–82. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 54.Domagalska MA, Sarnowska E, Nagy F, Davis SJ. Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana. PLoS ONE. 2010;5:e14012. doi: 10.1371/journal.pone.0014012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nadhzimov UK, Jupe SC, Jones MG, Scott IM. Growth and gibberellin relations of the extreme dwarf dx tomato mutant. Physiol Plant. 1988;73:252–6. doi: 10.1111/j.1399-3054.1988.tb00594.x. [DOI] [Google Scholar]
- 56.Jager CE, Symons GM, Ross JJ, Smith JJ, Reid JB. The brassinosteroid growth response in pea is not mediated by changes in gibberellin content. Planta. 2005;221:141–8. doi: 10.1007/s00425-004-1454-8. [DOI] [PubMed] [Google Scholar]
- 57.Kurepin LV, Joo SH, Kim SK, Pharis RP, Back TG. Interaction of brassinosteroids with light quality and plant hormones in regulating shoot growth of young sunflower and Arabidopsis seedlings. J Plant Growth Regul. 2012;31:156–64. doi: 10.1007/s00344-011-9227-7. [DOI] [Google Scholar]
- 58.Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1 and modulating brassinosteroid synthesis. Plant J. 2006;48:390–402. doi: 10.1111/j.1365-313X.2006.02875.x. [DOI] [PubMed] [Google Scholar]
- 59.Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, et al. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009;57:498–510. doi: 10.1111/j.1365-313X.2008.03707.x. [DOI] [PubMed] [Google Scholar]
- 60.De Vleesschauwer D, Van Buyten E, Satoh K, Balidion J, Mauleon R, Choi IR, et al. Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 2012;158:1833–46. doi: 10.1104/pp.112.193672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, et al. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33:887–98. doi: 10.1046/j.1365-313X.2003.01675.x. [DOI] [PubMed] [Google Scholar]
- 62.Bajguz A, Hayat S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol Biochem. 2009;47:1–8. doi: 10.1016/j.plaphy.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 63.Hu MY, Luo M, Xiao YH, Li XB, Tan KL, Hou L, et al. Brassinosteroids and Auxin Down-Regulate DELLA Genes in Fiber Initiation and Elongation of Cotton. Agric Sci China. 2011;10:1168–76. [Google Scholar]
- 64.Schünmann PH, Ougham HJ. Identification of three cDNA clones expressed in the leaf extension zone and with altered patterns of expression in the slender mutant of barley: a tonoplast intrinsic protein, a putative structural protein and protochlorophyllide oxidoreductase. Plant Mol Biol. 1996;31:529–37. doi: 10.1007/BF00042226. [DOI] [PubMed] [Google Scholar]
- 65.Medford JI, Elmer JS, Klee HJ. Molecular cloning and characterization of genes expressed in shoot apical meristems. Plant Cell. 1991;3:359–70. doi: 10.1105/tpc.3.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–8. doi: 10.1104/pp.010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang G, Komatsu S. Molecular cloning and characterization of a novel brassinolide enhanced gene OsBLE1 in Oryza sativa seedlings. Plant Physiol Biochem. 2004;42:1–6. doi: 10.1016/j.plaphy.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–75. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 69.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–57. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–34. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheminant S, Wild M, Bouvier F, Pelletier S, Renou JP, Erhardt M, et al. DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell. 2011;23:1849–60. doi: 10.1105/tpc.111.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu X, Li L, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:7618–23. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–86. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci USA. 2010;107:3918–23. doi: 10.1073/pnas.0909198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–9. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–9. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–4. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 78.Hou X, Lee LY, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–94. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 79.Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, et al. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010;24:2127–32. doi: 10.1101/gad.593410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:2160–5. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, et al. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22:915–27. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24:2635–48. doi: 10.1105/tpc.112.098749. [DOI] [PMC free article] [PubMed] [Google Scholar]