Abstract
The proliferating cell nuclear antigen (PCNA) is a key component of the eukaryotic DNA replication machinery. It also plays an important role in DNA repair mechanisms. Despite the intense scientific research on yeast and human PCNA, information describing the function of this protein in plants is still very limited. In the previous study Arabidopsis PCNA2 but not PCNA1 was proposed to be functionally important in DNA polymerase η-dependent postreplication repair. In addition to the above study, PCNA2 but not PCNA1 was also shown to be necessary for Arabidopsis DNA polymerase λ-dependent oxidative DNA damage bypass. Taking into account the reported differences between PCNA1 and PCNA2, we tested the idea of a possible cooperation between PCNA1 and PCNA2 in the plant cell. In a bimolecular fluorescence complementation assay an interaction between PCNA1 and PCNA2 was observed in the nucleus, as well as in the cytoplasm. This finding, together with our previous results, indicates that PCNA1 and PCNA2 may cooperate in planta by forming homo- and heterotrimeric rings. The observed interaction might be relevant when distinct functions for PCNA1 and PCNA2 are considered.
Keywords: Arabidopsis thaliana, PCNA, DNA replication, DNA repair, cell cycle
The proliferating cell nuclear antigen (PCNA) is a conserved protein encoded in the genomes of archaebacteria and eukaryotic organisms.1 The lethal phenotype of pcna knockouts indicates the significance of this protein in yeast, animals and plants. PCNA is an important component of the DNA replication machinery. It forms a pseudo-6-fold symmetry ring around DNA and acts as a DNA polymerase δ processivity factor.2-5 The gene sequences coding for PCNA have been identified and reported for diverse species, including human,6 mouse,7 yeast,8 rice,9 common bean10 and runner bean.11 Moreover, in some of them, for instance Drosophila,12,13 Arabidopsis14 and maize,15,16 two pcna genes were discovered. The structural similarities and conservation between PCNA proteins from different eukaryotes including yeast,5 human17 and Arabidopsis18 were confirmed in crystallographic studies.
Thirty years of PCNA research resulted in the identification of a great number of PCNA interacting proteins, which also include enzymes involved in post-translational modifications of PCNA.19-22 One of the broadly studied PCNA post-translational modifications is ubiquitination.20,22 The stalled replication forks induce PCNA ubiquitination at lysine 164 and activate postreplication repair.23 PCNA monoubiquitination leads to error-prone translesion synthesis, and polyubiquitination to error-free template switch combined with recombination.23 In our recent study, using a ubiquitination assay, we observed a similar ubiquitination pattern at lysine 164 for Arabidopsis PCNA1 and PCNA2.22 Based on the fact that PCNA1 and PCNA2 demonstrate 96.6% identity at the amino acid level, this result was not surprising. However, the high similarity between PCNA1 and PCNA2 brings up the question of their functional relevance in the cell. Interestingly, the data from Anderson and coworkers24 pointed to the functional difference between Arabidopsis PCNA proteins in the context of plant postreplication repair. Using a yeast system they have shown that PCNA2, but not PCNA1, could functionally interact with the Arabidopsis translesion DNA polymerase η. To explain the observed differences it was proposed that the ubiquitination of PCNA1 at lysine 164 might be inhibited.24 However, this suggestion was not confirmed in our Arabidopsis PCNA ubiquitination studies.22 In addition to Anderson’s report, Arabidopsis DNA polymerase λ, together with PCNA2 but not PCNA1, was also shown to be required for oxidative DNA damage bypass.25 Taking into account the reported differences between PCNA1 and PCNA224,25 we applied a bimolecular fluorescence complementation assay to examine whether the two proteins can interact in the plant cell. Such cooperation could combine different features of PCNA1 and PCNA2 by the formation of a heterotrimeric ring. Selected combinations of PCNA1, PCNA2 and CycA1 (BiFC control) fused with full length, N-terminal or C-terminal GFP fragments were transiently overexpressed in Nicotiana benthamiana epidermal cells.21,26 The results of this experiment showed an interaction between PCNA1-PCNA1, PCNA2-PCNA2 and most interestingly also between PCNA1-PCNA2, but not between PCNA1/2 and CycA1 (Fig. 1). The fluorescence signal was observed both in the nucleus and the cytoplasm. Based on the current knowledge which restricts the role of PCNA only to the nuclear compartment, the interaction observed in the cytoplasm might result from inefficient transport of PCNA to the nucleus or the overexpression of PCNA. The detected interaction between PCNA1 and PCNA2 in N. benthamiana cells is in agreement with the results of our previous study where we showed that recombinant PCNA1 and PCNA2 were able to form a heterotrimer.18
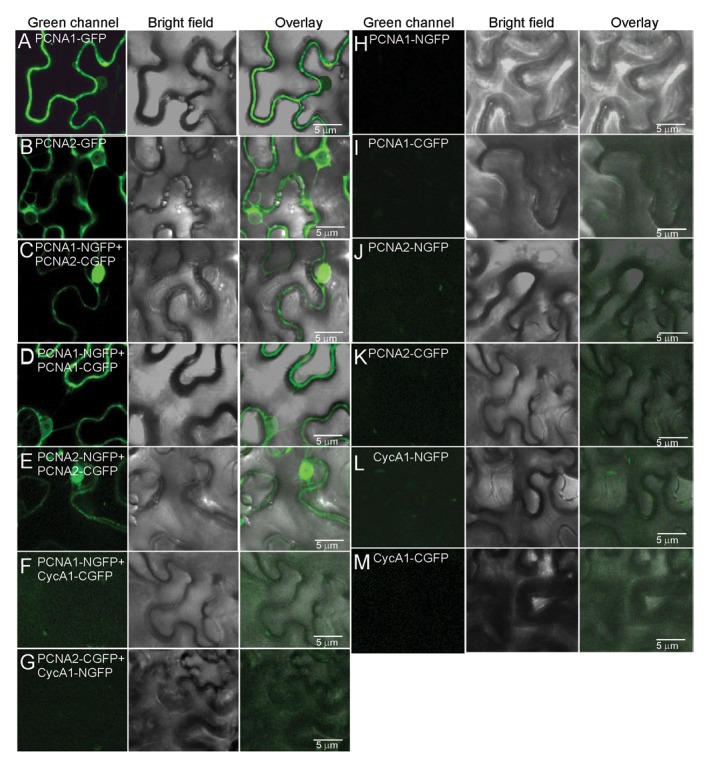
Figure 1. Analysis of PCNA1 and PCNA2 interaction using a bimolecular fluorescence complementation assay. Confocal images showing transient expression of: (A) PCNA1-GFP, (B) PCNA2-GFP, (C) split PCNA1-NGFP and PCNA2-CGFP, (D) split PCNA1-NGFP and PCNA1-CGFP, (E) split PCNA2-NGFP and PCNA2-CGFP, (F) split PCNA1-NGFP and CycA1-CGFP, (G) split PCNA2-CGFP and CycA1-NGFP, (H) PCNA1-NGFP, (I) PCNA1-CGFP, (J) PCNA2-NGFP, (K) PCNA2-CGFP, (L) CycA1-NGFP (M) CycA1-CGFP in epidermal cells of Nicotiana benthamiana. Bar = 5 µm.
In conclusion, the presented data support our idea that PCNA1 and PCNA2 heterotrimerization/cooperation can occur in the plant nucleus. This finding seems to be relevant especially when differential roles of PCNA1 and PCNA2 are considered. Due to the fact that our understanding of PCNA-dependent mechanisms in Arabidopsis is still very limited, a pcna1 and pcna2 knockout analysis would be advisable to study in detail the functional relevance of these two genes.
Acknowledgments
The project was supported by the Polish Ministry of Science and Higher Education (project Iuventus Plus, No. IP2011 052571). WS was supported by the Polish National Centre for Research and Development (project No. LIDER/28/54/L-/10/NCBiR/2011).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24837
References
- 1.Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann Bot. 2011;107:1127–40. doi: 10.1093/aob/mcq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan CK, Castillo C, So AG, Downey KM. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J Biol Chem. 1986;261:12310–6. [PubMed] [Google Scholar]
- 3.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–7. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 4.Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, et al. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–20. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 5.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–43. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Almendral JM, Huebsch D, Blundell PA, Macdonald-Bravo H, Bravo R. Cloning and sequence of the human nuclear protein cyclin: homology with DNA-binding proteins. Proc Natl Acad Sci USA. 1987;84:1575–9. doi: 10.1073/pnas.84.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi M, Hayashi Y, Hirose F, Matsuoka S, Moriuchi T, Shiroishi T, et al. Molecular cloning and structural analysis of mouse gene and pseudogenes for proliferating cell nuclear antigen. Nucleic Acids Res. 1991;19:2403–10. doi: 10.1093/nar/19.9.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer GA, Burgers PM. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990;18:261–5. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuka I, Daidoji H, Matsuoka M, Kadowaki K, Takasaki Y, Nakane PK, et al. Gene for proliferating-cell nuclear antigen (DNA polymerase delta auxiliary protein) is present in both mammalian and higher plant genomes. Proc Natl Acad Sci USA. 1989;86:3189–93. doi: 10.1073/pnas.86.9.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strzalka W, Ziemienowicz A. Molecular cloning of Phaseolus vulgaris cDNA encoding proliferating cell nuclear antigen. J Plant Physiol. 2007;164:209–13. doi: 10.1016/j.jplph.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Strzalka W, Kaczmarek A, Naganowska B, Ziemienowicz A. Identification and functional analysis of PCNA1 and PCNA-like1 genes of Phaseolus coccineus. J Exp Bot. 2010;61:873–88. doi: 10.1093/jxb/erp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi M, Nishida Y, Moriuchi T, Hirose F, Hui CC, Suzuki Y, et al. Drosophila proliferating cell nuclear antigen (cyclin) gene: structure, expression during development, and specific binding of homeodomain proteins to its 5′-flanking region. Mol Cell Biol. 1990;10:872–9. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruike T, Takeuchi R, Takata K, Oshige M, Kasai N, Shimanouchi K, et al. Characterization of a second proliferating cell nuclear antigen (PCNA2) from Drosophila melanogaster. FEBS J. 2006;273:5062–73. doi: 10.1111/j.1742-4658.2006.05504.x. [DOI] [PubMed] [Google Scholar]
- 14.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 15.López I, Khan S, Vázquez-Ramos J, Hussey PJ. Molecular cloning of a maize cDNA clone encoding a putative proliferating cell nuclear antigen. Biochim Biophys Acta. 1995;1260:119–21. doi: 10.1016/0167-4781(94)00192-6. [DOI] [PubMed] [Google Scholar]
- 16.López I, Khan S, Vázquez J, Hussey PJ. The proliferating cell nuclear antigen (PCNA) gene family in Zea mays is composed of two members that have similar expression programmes. Biochim Biophys Acta. 1997;1353:1–6. doi: 10.1016/S0167-4781(97)00072-9. [DOI] [PubMed] [Google Scholar]
- 17.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/S0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 18.Strzalka W, Oyama T, Tori K, Morikawa K. Crystal structures of the Arabidopsis thaliana proliferating cell nuclear antigen 1 and 2 proteins complexed with the human p21 C-terminal segment. Protein Sci. 2009;18:1072–80. doi: 10.1002/pro.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivona JB, Kelman Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 2003;546:167–72. doi: 10.1016/S0014-5793(03)00622-7. [DOI] [PubMed] [Google Scholar]
- 20.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 21.Strzalka W, Labecki P, Bartnicki F, Aggarwal C, Rapala-Kozik M, Tani C, et al. Arabidopsis thaliana proliferating cell nuclear antigen has several potential sumoylation sites. J Exp Bot. 2012;63:2971–83. doi: 10.1093/jxb/ers002. [DOI] [PubMed] [Google Scholar]
- 22.Strzalka W, Bartnicki F, Pels K, Jakubowska A, Tsurimoto T, Tanaka K. RAD5a ubiquitin ligase is involved in ubiquitination of Arabidopsis thaliana proliferating cell nuclear antigen. J Exp Bot. 2013;64:859–69. doi: 10.1093/jxb/ers368. [DOI] [PubMed] [Google Scholar]
- 23.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–7. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 24.Anderson HJ, Vonarx EJ, Pastushok L, Nakagawa M, Katafuchi A, Gruz P, et al. Arabidopsis thaliana Y-family DNA polymerase η catalyses translesion synthesis and interacts functionally with PCNA2. Plant J. 2008;55:895–908. doi: 10.1111/j.1365-313X.2008.03562.x. [DOI] [PubMed] [Google Scholar]
- 25.Amoroso A, Concia L, Maggio C, Raynaud C, Bergounioux C, Crespan E, et al. Oxidative DNA damage bypass in Arabidopsis thaliana requires DNA polymerase λ and proliferating cell nuclear antigen 2. Plant Cell. 2011;23:806–22. doi: 10.1105/tpc.110.081455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, et al. Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell. 2010;22:1264–80. doi: 10.1105/tpc.109.073635. [DOI] [PMC free article] [PubMed] [Google Scholar]


