Abstract
Drought is the major cause of crop losses worldwide. Water stress-inducible promoters are important for understanding the mechanisms of water stress responses in crop plants. Here we utilized tobacco (Nicotiana tabacum L.) Bright Yellow 2 (BY-2) cell system in presence of polyethylene glycol, salt and phytohormones. Extension of the system to 85 mM NaCl led to inducibility of up to 10-fold with the water stress and salt responsive soybean GmWRKY53 promoter. Upon ABA and JA treatment fold inducibility was up to 5-fold and 14-fold, respectively. Thus, we hypothesize that GmWRKY53 could be used as potential model candidate for dissecting drought regulatory elements as well as understanding crosstalk utilizing a rapid heterologous system of BY-2 culture.
Keywords: GmWRKY53, tobacco BY-2, water stress, salt stress, abscisic acid
Among abiotic stress conditions, drought is the main culprit that causes huge crop losses worldwide. Therefore, engineering drought tolerance in plants is of major economic importance. Many of the experimental systems that are used in attempts to improve drought tolerance, for example the generation of transgenic plants, are time consuming. For this reason, systems of reduced complexity that allows rapid screening of genes and promoters for potential use in projects aimed at improving drought tolerance are desired. One such system could be the tobacco (Nicotiana tabacum L.) Bright Yellow-2 (BY-2) cell line.1,2 BY-2 cells can be easily transformed to a very high efficiency using Agrobacterium tumefaciens and the cells subsequently can respond to stimuli such as the plant hormone methyl jasmonate (JA).3The use of BY-2 cells also has potential advantages over other cell cultures, such as a highly synchronized growth rate, good transformation after protoplastation or particle bombardment, lack of leaky expression for promoter studies and easy monitoring and analysis. BY-2 cell system is also an excellent tool for sub-cellular localization studies. These characteristics make BY-2 cells an excellent system for many procedures.2 This system of reduced complexity therefore has the potential to be used to identify and better understand regulatory networks in response to water deficit/drought conditions.
A number of reports have described using the BY-2 cell system for promoter analysis utilizing either β-glucuronidase3,4 or luciferase as a reporter,5 for example stably transformed BY-2 cells have successfully been used to dissect JA-inducible promoters.6,7 In addition, this BY-2 system can also be extended to the testing of individual promoter elements and synthetic promoters that respond to JA. Such a system would be a very useful tool for the study of water stress-inducible gene expression for promoters from other plant species. In addition, it would also be useful if this system responded to a number of different water stress inducing treatments, such as salt, polyethylene glycol (PEG) and mannitol. Hence, in the present study a tobacco BY-2 cell system for the rapid analysis of drought-inducible promoters is used that accelerates the identification of stress tolerance mechanisms from different crop plants.
Gene regulation at the mRNA level is considered as one of the key regulatory check in many biological processes. In plants, approximately 7% of the total plant genome constitutes of transcription factors and they are integral part of the gene regulation at mRNA level.8 WRKY transcription factors are one of the ten largest family of transcription factors across the green lineage named after the highly conserved and almost invariant 60 amino acid domain with the signature WRKY sequence at the N-terminus and a zinc finger motif at the C-terminus.9,10 They show specific binding to their cognate DNA-binding site the W-Box (T)(T)TGAC(C/T) that is found in the promoter regions of many stress-inducible promoters of their target genes.9-11 Several studies had reported the role of WRKY transcription factors (TFs) in response to biotic stress but their role in abiotic stress responses was relatively limited.10 Hence, this study was conducted toward understanding role of the GmWRKY53 in different stress and phytohormone signaling network during water stress.
Potential promoter elements were identified using MEME.12 Promoter sequences of 1 kb upstream of the predicted ATG start codon were used with the settings as described in Tripathi et al. (2012).13 Promoter constructs were amplified by PCR using genomic DNA from either tobacco or soybean as template with Fail Safe™ PCR system (EPICENTER Inc.). Appropriate restriction enzyme sites were added to the primer sequences to allow cloning. The promoters were introduced into the MS23 vector14 and then sub cloned into the binary vector pGPTV-GUS-KAN or pGPTV-GFP-KAN as previously described.15 Cell suspension cultures of tobacco (Nicotiana tabacum L.) cv Bright Yellow-2 (BY-2) were obtained from the University of Virginia. The suspension cell culture was grown at 26°C on a rotary shaker in modified Murashige and Skoog (MS) medium (Caisson Labs) supplemented with 0.15 mM KH2PO4, 0.2 mg/l 2.4-dichlorophenyoxyacetic acid, 1 mg/l thiamine, 10 mg/l myo-inositol and 3% (w/v) sucrose. Cells were subcultured weekly by transferring 1 ml cell culture into 50 ml of fresh medium. For longer term storage stocks of BY-2 calli were maintained in Petri dishes on MS medium solidified with 1.5% (w/v) agar and supplemented with 0.15 mM KH2PO4, 0.2 mg/l 2.4-dichlorophenyoxyacetic acid, 1 mg/l thiamine, 10 mg/l myo-inositol and 3% (w/v) sucrose and subcultured each month onto fresh plates. Agrobacterium tumefaciens (LBA4404) were transformed with the various promoter constructs and grown in YEB medium containing 50 mg/l rifampicin and 50 mg/l kan.15 One milliliter of overnight culture was diluted with YEB plus antibiotics so that the OD600 was 0.3. Agrobacteria were grown until the OD600 was 0.6. Cells were then pelleted and re suspended in 0.5 ml MS medium. Four days after sub culturing of the BY-2 cells, three transformation events were performed for each construct using different amounts of Agrobacteria (50, 150 and 350 µl) added to the BY-2 cell culture (3 ml). The cell cultures were transformed by co-cultivation with Agrobacteria in a Petri dish in the dark for 2 d. The cells were then washed two to three times using 15 ml fresh MS medium. 100 µl of the cell culture was then plated at low density onto MS agar containing 50 mg/l kanamycin and 500 mg/l cefotaxime. Transformed calli are visible after 3–5 weeks. Single transformed callus was transferred into micro-wells (Greiner Bio-One six-well culture plate) with 5 ml MS medium containing kanamycin and cefotaxime as above. Cells were grown at 28°C for 4 d with shaking. One ml of cells was removed and used to inoculate 5 ml of fresh medium and the cells grown at 28°C for 3 d. For each promoter construct, 12 independent lines were chosen and cultured separately until they were pooled for water stress treatments. Slide mounts of the transgenic cell cultures were observed under Olympus AX70 compound microscope attached to Olympus AX70 digital camera (Olympus). All the images of the cells were taken at 20× lens objective with automatic exposure setting for bright field and 5 sec exposure setting for the fluorescence. Camera sensitivity was set at ISO 200 and the filter used for GFP viewing of the cells was FITC filter set. Transformed cell lines of GmWRKY53 were taken after 6 d of growth and subjected to 10% Polyethylene glycol (PEG-8000, Fisher Scientific USA) and 0.5% or 85 mM NaCl (Fisher Scientific) treatment. 0.1ml of each 5 independent lines was pooled with 4.5 ml of 10% PEG and 85 mM or 0.5% NaCl in a 6-well plate and grown as “treated” while for “untreated” 0.1 ml of each six independent lines were grown in MS/V/C/K media at 28°C with shaking. The cells were harvested with filtration method after 6 d and freezed in liquid nitrogen before using for quantification. A 200 μl of five transgenic lines from the micro-wells were pooled and sub cultured to a 250 ml Erlenmeyer flask with 49 ml MS medium, 50 ml vitamins, 50 μl kanamycin and 100 μl of cefotaxime. Cells were grown at 28°C for 2 d with shaking at 115 rpm. Ten milliliters of cells from each flask were transferred in to new 250 ml flask with 40 ml MS medium containing 40 ml vitamins without hormones (−2,4D). BY-2 cells were grown at 28°C for 1day with shaking at 115 rpm. No antibiotics were added at this time. After 1day BY-2 cells were treated with 100 µM ABA (for ABA treatment purpose) and 100 µM JA (for JA treatment purpose) dissolved in DMSO. 50 µM DMSO was added in control sample used as untreated. Cells were harvested after 24 hrs of treatment and freezed in liquid nitrogen and stored in −80°C before using for quantification.
GUS assays were performed as previously described.15 GFP quantification was performed using a fluorescence assay. Harvested cells were grounded in 1× lysis buffer (Cell Bio Labs Inc.) supplemented with protease inhibitor (Roche). Standard curve production and sample preparation were performed according to the manufacturer's instructions. 100 µL of each lysate was placed in a microplate along with lysis buffer as blank and untransformed wild type BY-2 cell lysate and the fluorescence was determined at 485 nm excitation and 507 emissions with top 50% optics and 100% sensitivity using a Synergy 2 microplate reader (BioTek Instrument Inc.). Protein was estimated using Bradford assay16 for treated, untreated and wild type sample. The fluorescence/mg protein was calculated for treated and untreated samples after subtracting the fluorescence/mg protein of the wild type to subtract any background fluorescence. The values from treated and untreated samples, minus the background fluorescence, were used to calculate fold induction.
We have performed extensive expression profiling at the whole genome level (soybean) or gene space level (tobacco). This included parallel experiments where hydroponically grown soybean and tobacco plants were subjected to dehydration (data not shown). This allowed us to identify several water stress-inducible genes as good candidates for the identification of water stress-responsive promoter elements. These genes included both galactinol synthase (NtGolS; EC 2.4.1.123) and raffinose synthase (NtRS; 1139734; EC 2.4.1.82) from tobacco and several WRKY genes from both tobacco and soybean. These genes all showed an induction of at least 8-fold by water stress in either leaves and/or roots. The promoters of these genes were isolated by PCR and inserted upstream of either the GUS reporter gene in pGPTV-GUS-kan or pGPTV-GFP-kan. The gene that showed the most promise in the BY-2 cell system was the soybean GmWRKY53 (Glyma19 g26400). The tobacco raffinose synthase was used as positive control. They both show strong induction by water stress in both leaves and roots (Table 1).
Table 1. Water stress induction of tobacco NtRS (1139734) and GmWRKY53 (Glyma19 g26400) during dehydration time course in different tissue types.
| Gene | Roots | Leaves | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 20 min | 40 min | 1 h | 2 h | 4 h | 20 min | 40 min | 1 h | 2 h | 4 h | |
| NtRS (1139734) | 1.44 | 1.29 | 3.06 | 14.68 | 16.32 | 1.37 | 1.51 | 2.2.6 | 32.79 | 370.25 |
| 30 min | 1 h | 2 h | 3 h | 5 h | 30 min | 1 h | 2 h | 3 h | 5 h | |
|
GmWRKY53 (Glyma19 g26400) |
3.42 | 4.55 | 8.76 | 9.07 | 11.09 | 1.34 | 1.57 | 4.74 | 20.34 | 25.30 |
We next sought to establish a system of reduced complexity for the rapid analysis of water stress-inducible promoter. We based this system on the BY-2 cell system for the analysis of jasmonate-inducible promoters.6 This BY-2 cell system relies on the observations that BY-2 cells can be stably transformed to a very high level using A. tumefaciens and that these transgenic lines respond well to the plant hormone methyl jasmonate. We attempted to modify this system for the analysis of water stress-responsive promoters by adding polyethylene glycol to the medium. We also added salt to establish whether the system could be suitable for the dissection of salt-responsive promoters. Extending our approach for better optimization, we also added abscisic acid (ABA) and jasmonic acid (JA) to the system. In addition, we employed two different reporter genes, GUS and GFP to establish the most suitable reporter gene for the system. Finally, we also used promoters from both tobacco and soybean to establish whether the system could be suitable for the analysis of promoters from different plant species.
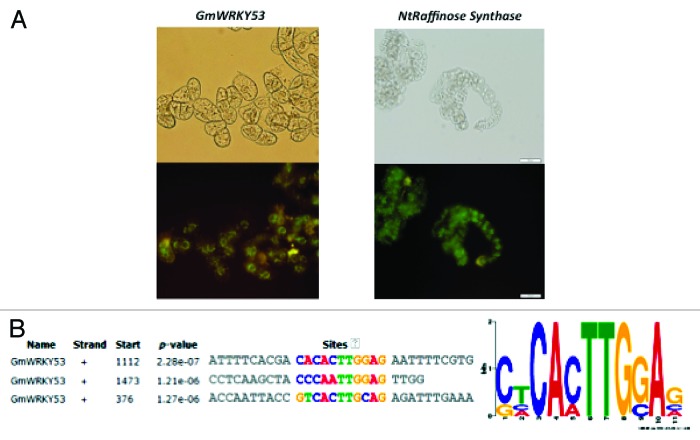
BY-2 cells were transformed with promoter:GUS or promoter:GFP constructs using the pGPTV binary vector pGPTV-GUS-kan or pGPTV-GFP-kan. Hundreds of independent transformed BY-2 cell lines were generated for each construct and 12 were chosen at random for further cultivation as cell cultures. For further analysis, six of the independent cell lines for each construct were pooled into a single 5 ml culture. This combined cell line was then subcultured into medium containing different concentrations of PEG in order to establish a concentration at which the BY-2 cells could respond to the stress conditions but were not either plasmolyzed or killed. Figure 1A shows that at a concentration of 10% PEG, the BY-2 cells showed no signs of plasmolysis or cell death. The promoter:GFP constructs also directed a background level of activity as expected from the oligo array analysis. Optimization experiments showed that this concentration of 10% PEG appears to be best for our purposes, as is 85 mM NaCl.
Figure 1. (A) PEG treated BY-2 cells are alive and express GFP after incubation in medium containing 10% polyethylene glycol. (B) Multiple E-box sequences in the promoter region of the GmWRKY53. Potential promoter elements were identified using MEME.
An in-silico analysis of the water stress-inducible promoter from GmWRKY53 was performed for the identification of potential cis-acting elements using MEME.12 GmWRKY53 was found to be enriched with multiple E-box like sequences (5′-CANNTG-3′), the potential binding sites for members of the basic-helix-loop-helix (bHLH) family of transcription factors (Fig. 1B).17 Several genes from the bHLH gene family show strong upregulation by water stress in both tobacco and soybean (expression data not shown), suggesting that regulation of GmWRKY53 by bHLH transcription factors may constitute part of the signaling web involved in modulating water stress responses in soybean.
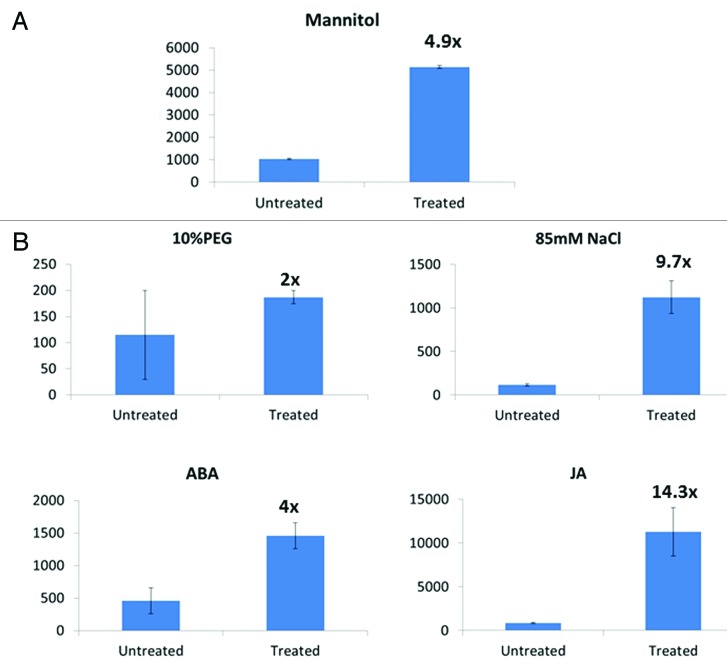
A combined cell culture consisting of six independent ProNtRS:GUS BY-2 lines were treated with 10% mannitol for six days. Each individual experiment led to an inducibility of at least 4-fold by mannitol and the average inducibility over the three experiments was 5.07-fold (Fig. 2A), and hence provides the proof of concept for the system. In addition, this level of induction should be sufficient for both loss of function and gain of function promoter analyses.
Figure 2. (A) GUS activity of Nt RS promoter:GUS in 10% mannitol. The figure shows average of three independent experiments involving five independent transgenic lines. The values on the top of black color treated columns shows the average fold induction. GUS assays were performed as previously described.15 (B) GFP activity of GmWRKY53 promoter: GFP BY-2 in 10% PEG, 0.5% (85 mM) NaCl.,100 µM ABA and 100 µM JA. The figure shows the average of three independent experiments involving five independent transgenic lines and bars represent standard error. The values on the top of black color treated columns shows the average fold induction. The scale bar represents 50 μm for NtRS and GmWRKY53 was used.
Expression of the GmWRKY53 gene has been described as being inducible by salt stress.18 The BY-2 systems have the potential to be utilized as a system for rapid dissection of regulatory elements.19 Although GmWRKY53 is induced at least 10-fold in soybean tissues by water stress, the promoter only directed a maximum of about 2-fold induction by 10% PEG in the BY-2 system when the GFP reporter genes was employed (Fig. 2B). This was similar to several other tested promoters (data not shown). By contrast, addition of 85 mM NaCl led to inducibilities of about 10-fold in expression from the GmWRKY53 promoter, despite the use of the GFP reporter gene (Fig. 2B). This suggests that the BY-2 cell system, when used with 85 mM NaCl, is a useful system for the testing and dissection of salt stress-inducible promoters. An induction of 10-fold is sufficient for the dissection of complex promoter architecture with multiple cis-acting elements because knock out of a single element that results in a 50% drop in inducibility still leaves a 5-fold induction that is sufficient for the analysis of remaining elements. Additionally, the use of the GUS reporter gene rather than GFP may also result in even higher levels of inducibility.
To extend the applicability of the BY-2 cell system and have an idea regarding the potential cross talk during water stress the soybean promoter GFP constructs was monitored after the two major phytohormones, ABA and JA treatments. After working on different concentrations of phytohormone, 100 μM of ABA and JA were used for quantifying the promoter activity after 24 hrs of treatment. The GmWRKY53 performs better in both treatments with explicit induction of approximately 5-fold after ABA treatment and 14-fold after JA treatment (Fig. 2B).
Figure 2 supports that the BY-2 cell system can be used to dissect some water stress-inducible promoters, such as NtRS, and salt stress-inducible promoters such as GmWRKY53 (Glyma19 g26400). The system is relatively rapid, with the production of transgenic cell lines taking just a few weeks and the testing of inducibility taking just one week. This is considerably quicker than the time taken to generate transgenic plants and the system therefore has the potential to save considerable time in the dissection and analysis of plant stress-responsive promoters.
However, although the raffinose synthase promoter showed 5-fold inducibility by PEG with the GUS reporter gene, while with the GFP reporter gene, 1.3-fold inducibility was obtained and the best inducibility with any promoter was just 1.8-fold (data not shown). The GUS reporter gene is more suitable for this system than GFP. One of the reasons for this may be the level of auto fluorescence found in cells. By contrast, the level of GUS activity in cells is normally negligible.
One of the major observations that we present here is that the promoter from salt inducible soybean GmWRKY53 directs about 10-fold inducibility by salt in the BY-2 cell system. Hence, it shows that the GmWRKY53 promoter being a heterologous promoter has potential for being used as a model for rapid dissection of regulatory elements. Likewise, the performance upon ABA treatment and JA treatment shows that GmWRKY53 is an ABA-responsive promoter and could be a better candidate to explore on crosstalk with JA as well as with salt stress. The expression of GmWRKY53 at the mRNA level and its promoter activity on ABA treatment provides evidence for its regulation in an ABA-dependent manner. This observation was also confirmed by examining the promoter through Plant Cis-acting Regulatory DNA Elements database (www.dna.affrc.go.jp/PLACE/).20 The GmWRKY53 promoter was found enriched with abscisic acid responsive elements (ABRE), the potential binding sites of basic leucine zipper (bZIP) transcription factors.21 ABRE motifs are well known for their involvement in regulation of drought or water stress signaling in ABA-dependent manner.22 Thus the enrichment of ABRE-like sequences, E-box like elements and W-Box in the promoter region of the GmWRKY53 suggests the potential of protein-protein interaction to confer tolerance against abiotic stress. Recently, Babitha et al. (2012) have shown that co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis.23 The transgenic lines show enhanced tolerance to multiple abiotic stresses like drought, salt and oxidative stress via regulation of more number of targets. Thus, we hypothesize utilizing the current knowledge reported in this study for GmWRKY53, which is a salt and water-stress inducible gene has full potential for being used as suitable candidate that will provide novel insights toward functionality in response to abiotic stress. The detailed functional analysis of promoter using this rapid system of BY-2 culture will help in understanding the co-operative behavior of the major transcription factors in regulation of stress responses in crop species.
In conclusion, although the tobacco raffinose synthase promoter showed sufficient inducibility by 10% mannitol for promoter dissection using the GUS reporter gene (5-fold), no tested gene showed similar induction using GFP. This suggests that the BY-2 cell system preferably be based on the use of the GUS reporter gene as previously shown for jasmonate-inducible promoters.7 Having established a working system for promoter analysis, further optimization should now be possible for GmWRKY53 promoter as a positive control for salt stress-inducible promoters. With this positive control further variables such as cell density, the presence or absence of plant hormones such as auxin in the medium, the length of treatment. Subsequently, further optimization of the amount of PEG or salt can occur. In addition, examination of the system with plant hormone abscisic acid (ABA), which is known to be a major player in plant responses to abiotic stresses such as water stress shows that use of ABA than mannitol would be better to answer water stress-related questions. However, further optimization of BY-2 cell system on gene-to-gene or promoter-to-promoter basis is highly recommended. (Supplemental Material)
Supplementary Material
Acknowledgments
Authors acknowledge the instrumentation support from South Dakota State University Functional Genomics Core Facility supported in part by the National Science Foundation/EPSCoR Grant No. 0091948 and by the State of South Dakota. This project was supported by funds from Center of Excellence for Drought Tolerance Research, South Dakota State University to PT and RCR. Research in the Rushton laboratory is also supported by The United National Research Initiative grants 2008-35100-04519 and 2008-35100-05969 from the USDA National Institute of Food and Agriculture, The United Soybean Board, The South Dakota Soybean Research and Promotion Council and The North Central Soybean Research Program.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/24097
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24097
References
- 1.Inzé D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 2.Geelen DN, Inzé DG. A bright future for the bright yellow-2 cell culture. Plant Physiol. 2001;127:1375–9. doi: 10.1104/pp.010708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishida S, Takahashi Y, Nagata T. The mode of expression and promoter analysis of the arcA gene, an auxin-regulated gene in tobacco BY-2 cells. Plant Cell Physiol. 1996;37:439–48. doi: 10.1093/oxfordjournals.pcp.a028965. [DOI] [PubMed] [Google Scholar]
- 4.Mathieu M, Neutelings G, Hawkins S, Grenier E, David H. Cloning of a pine germin-like protein (GLP) gene promoter and analysis of its activity in transgenic tobacco Bright Yellow 2 cells. Physiol Plant. 2003;117:425–34. doi: 10.1034/j.1399-3054.2003.00050.x. [DOI] [PubMed] [Google Scholar]
- 5.Ono S, Tanaka T, Watakabe Y, Hiratsuka K. Transient assay system for the analysis of PR-1a gene promoter in tobacco BY-2 cells. Biosci Biotechnol Biochem. 2004;68:803–7. doi: 10.1271/bbb.68.803. [DOI] [PubMed] [Google Scholar]
- 6.Xu B, Timko M. Methyl jasmonate induced expression of the tobacco putrescine N -methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol Biol. 2004;55:743–61. doi: 10.1007/s11103-004-1962-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang HB, Bokowiec MT, Rushton PJ, Han SC, Timko MP. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol Plant. 2012;5:73–84. doi: 10.1093/mp/ssr056. [DOI] [PubMed] [Google Scholar]
- 8.Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, et al. Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 2007;144:538–49. doi: 10.1104/pp.107.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7:491–8. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 12.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi P, Rabara RC, Langum TJ, Boken AK, Rushton DL, Boomsma DD, et al. The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics. 2012;13:270. doi: 10.1186/1471-2164-13-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprenger-Haussels M, Weisshaar B. Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 2000;22:1–8. doi: 10.1046/j.1365-313x.2000.00687.x. [DOI] [PubMed] [Google Scholar]
- 15.Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell. 2002;14:749–62. doi: 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–70. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 19.Rabara RC, Tripathi P, Lin J, Rushton PJ. Dehydration-induced WRKY genes from tobacco and soybean respond to jasmonic acid treatments in BY-2 cell culture. Biochem Biophys Res Commun. 2013;431:409–14. doi: 10.1016/j.bbrc.2012.12.156. [DOI] [PubMed] [Google Scholar]
- 20.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–7. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–7. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 23.Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, Udayakumar M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013;22:327–41. doi: 10.1007/s11248-012-9645-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.