Abstract
Accurate chromosome segregation to progeny cells is a fundamental process ensuring proper inheritance of genetic material. In bacteria with simple cell cycle, chromosome segregation follows replication initiation since duplicated oriC domains start segregating to opposite halves of the cell soon after they are made. ParA and ParB proteins together with specific DNA sequences are parts of the segregation machinery. ParA and ParB proteins in Pseudomonas aeruginosa are important for optimal growth, nucleoid segregation, cell division and motility. Comparative transcriptome analysis of parA null and parB null mutants versus parental P. aeruginosa PAO1161 strain demonstrated global changes in gene expression pattern in logarithmically growing planktonic cultures. The set of genes similarly affected in both mutant strains is designated Par regulon and comprises 536 genes. The Par regulon includes genes controlled by two sigma factors (RpoN and PvdS) as well as known and putative transcriptional regulators. In the absence of Par proteins, a large number of genes from RpoS regulon is induced, reflecting the need for slowing down the cell growth rate and decelerating the metabolic processes. Changes in the expression profiles of genes involved in c-di-GMP turnover point out the role of this effector in such signal transmission. Microarray data for chosen genes were confirmed by RT-qPCR analysis. The promoter regions of selected genes were cloned upstream of the promoter-less lacZ gene and analyzed in the heterologous host E. coliΔlac. Regulation by ParA and ParB of P. aeruginosa was confirmed for some of the tested promoters. Our data demonstrate that ParA and ParB besides their role in accurate chromosome segregation may act as modulators of genes expression. Directly or indirectly, Par proteins are part of the wider regulatory network in P. aeruginosa linking the process of chromosome segregation with the cell growth, division and motility.
Introduction
In eukaryotic cells a defined mitotic apparatus is involved in active segregation of chromosomes to progeny cells during cell division. Studies on numerous low-copy-number plasmids revealed the existence of bacterial counterpart of a mitotic apparatus participating in active partitioning of plasmid molecules to progeny cells, and thereby in their stable maintenance in bacteria [1]. An active plasmid partitioning system consists of two proteins (so called A- and B-type) and an essential cis-acting DNA sequence, designated, by analogy to eukaryotic mitotic apparatus, the centromere-like sequence (parS or parC). The B-type proteins recognize and bind to a specific centromere-like sequence, forming the nucleoprotein complex - segrosome [2]. The A-type proteins are NTPases and provide the dynamic scaffold for segrosome movements. The type of NTPase: Walker-type ATPase, actin-type ATPase and tubulin-type GTPase is the basis for classification of partition systems into three groups: I (variants IA and IB depending on B-component), II and III, respectively [1], [3]. Direct interactions between A and B partners induce the hydrolysis of NTP, which in turn delivers energy for relocation of segrosomes [3], [4].
Recently, many reports have documented the ordered spatial organization of bacterial chromosomes, localization of specific genetic loci to defined regions during the cell cycle, specifically localized replication factories and chromosome segregation controlled in time and space. The majority of existing hypotheses regarding bacterial chromosome segregation are based on active transfer of newly replicated ori domains to the poles of the dividing cell. Representatives of ParA (Walker-type ATPases) and ParB (DNA binding proteins with H-T-H motifs) families, homologs of plasmid partitioning proteins from class IA, are postulated as the main players constituting elements of the prokaryotic chromosomal partitioning apparatus [4,5 6,7].
In the majority of chromosomes (except Enterobacteriaceae and Pasteurellaceae, which are deprived of par genes) the genes encoding Par proteins are located in close vicinity of the chromosome replication initiation site - oriC, next to the gid operon. Together with rnpA, rpmH, dnaA, recF and gyrB they constitute a conserved cluster of genes whose products play key roles in DNA replication, chromosome segregation and cell division [8], [9]. Highly conserved parS sequences have been localized mainly in the so-called ori domain of the primary chromosomes (20% of the chromosome around oriC). More variability among Par proteins and their centromere-like sequences is observed for the secondary chromosomes of species with more than one chromosome [9].
Studies on Bacillus subtilis, Caulobacter crescentus, Streptomyces coelicolor, Vibrio cholerae, Pseudomonas putida, P. aeruginosa, and, most recently, Burkholderia cenocepacia, Helicobacter pylori, Mycobacterium smegmatis, Corynebacterium glutamicum confirmed the participation of chromosomal Par proteins in chromosome segregation to the progeny cells also revealing similarities as well as species-dependent differences. The specific features of the Par proteins in a particular organism are manifested by their involvement in the control of different cellular processes like sporulation, regulation of replication initiation, cell cycle progression, motility or cell-to-cell communication [10], [11], [12], [13], [14], [15], [16], [17], [18].
P. aeruginosa, an opportunistic and medically important human pathogen with a simple cell cycle, has become a model for our studies on bacterial chromosome segregation. In the sequenced P. aeruginosa reference genome (PAO1 strain - NC_002516) the parAB operon is located approximately 7 kb counter clockwise from oriC and ten putative parS sites for ParB binding have been identified [19]. The closest parS sites are located around 4 kb clockwise from oriC in the recF gene. The parAparB operon is transcribed from the weak, parAp, located in the upstream gidB orf (Lasocki and Jagura-Burdzy, unpublished). The predicted promoter regions of gidA and parA were cloned in the promoter-probe vector and tested in E. coli for the regulation by ParA and/or ParB delivered in trans but no regulation was detected (Lasocki and Jagura-Burdzy, unpublished). It cannot be excluded that the nucleoprotein complexes formed at oriC and/or parSs as well as induced changes in DNA topology might alter the expression of parAB genes. Although autoregulation by ParA or ParB protein of par operons is well established feature of plasmid partitioning systems [1], [20], [21], [22], in the case of chromosomally encoded Par systems the autoregulation of par operons has not been determined.
The parA parB genes of P. aeruginosa and a single parS2 sequence are able to stabilize otherwise the unstable replicon in E. coli, which confirms the partitioning functions of the chromosomal parABS system of P. aeruginosa. Functional characterization of the Par proteins showed direct ParA-ParB, ParA-ParA and ParB-ParB interactions in the yeast as well as in bacterial two-hybrid system and in in vitro studies with purified proteins [16], [19], [23], [24]. In vivo experiments in E. coli showed that ParB overproduction causes transcriptional silencing of genes in close proximity to parS2 [19]. This feature may play an important role in the folding of the ori domain, in regulation of gene expression in this region and in regulation of replication in P. aeruginosa [23], [24]. The existence of so-called ori domains created by ParB interactions with parS sequences was confirmed using in situ immunofluorescence. ParB forms a various number (1 to 4) of compact foci on the nucleoid, depending on the stage of the cell cycle and growth conditions [18], [23], [24]. DNA binding activity and polymerization ability of ParB as well as ParA presence determine the distribution and condensation of ParB foci. Our in vitro studies have shown that ParA of P. aeruginosa exhibits a weak ATPase activity (manuscript in preparation) and is able to bind DNA non-specifically, similarly to ParA homologs from other systems [25]. The pattern of ParA-CFP localization (when plasmid-encoded) in parental strain PAO1161 (WT) and in the parA null mutant was dynamic, changing from polar or centrally localized foci to transient haze of fluorescence all over nucleoid. Such ParA patterning disappeared in the parB null mutant cells where ParA-CFP signal was seen as dispersed in the boundaries of the nucleoids confirming that ParB was involved in the dynamic behavior of ParA (manuscript in preparation), also featured in other systems [26], [27].
It was shown that overproduction of ParA as well as ParB in P. aeruginosa leads to the strong inhibition of bacterial growth [16], [19]. Microscopic observations of cells overproducing ParA and ParB proteins demonstrated disturbances in chromosome partitioning - the effect of DNA guillotining and increased number of extended and chromosome-less cells.
The parA and parB mutant cultures grown under various conditions showed a slightly extended generation time in comparison with the wild type P. aeruginosa grown on a rich medium [16], [18]. Microscopic observations of mutant cells from different phases of culture growth demonstrated a 1000-fold increase in number of the cells with defects in chromosome partitioning. Although these defects were observable in the fast growing cells, they were much stronger under slow bacterial growth conditions in the minimal medium [28]. Defects in chromosome partitioning were accompanied by disturbances in the division cycle (the par mutant cells were longer in comparison to the P. aeruginosa wild type [24]), by changes in colony morphology as well as defects in swimming and swarming motility, but not in twitching [16], [18]. The ability to perform movements by bacteria of different species is connected with their ability to colonize various ecological niches, and is frequently related to pathogenesis and biofilm formation. The observed impairment of motility of P. aeruginosa par mutants suggests direct or indirect role of Par proteins in regulation of these processes.
In this work we focused on the transcriptomic analysis of parA null and parB null mutants in comparison with parental P. aeruginosa PAO1161 strain (here/henceforth WT strain) in order to understand their phenotypes. Comparative transcriptome analysis of cells from logarithmically growing cultures exhibited global changes in gene expression in both analyzed par mutants in comparison with the WT strain.
Results and Discussion
Mutations in parA and parB Genes Cause Global Changes in Gene Expression Pattern in P. aeruginosa
Previous analysis of PAO1161 parA null and parB null mutants suggested that ParA and ParB proteins are involved not only in chromosome segregation in P. aeruginosa cells, but may also play a broader role connecting chromosome partitioning with chromosome condensation, replication, cell division, regulation of gene expression and controlling different cellular processes in bacteria, e.g. motility and cell-to-cell communication [16], [18].
To determine the changes in gene expression pattern in parA null and parB null strains of P. aeruginosa, microarray analysis was performed. Three biological replicates of each mutant strain including reference WT PAO1161 strain were cultivated in L-broth with OD600 measurements and CFU ml-1 determination to isolate RNA from logarithmically growing cultures (OD600 = 0.5) prior to the microarray analysis.
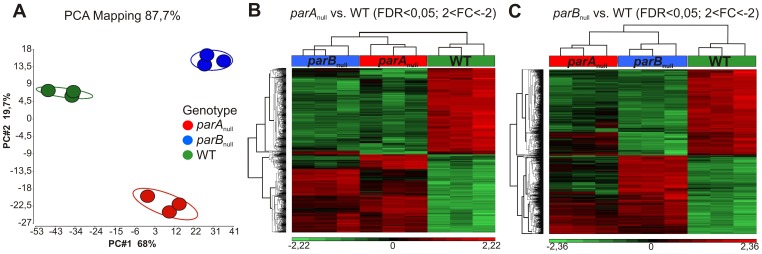
Quality analysis of three biological replicates of studied samples from parA null, parB null and WT strains was done by principal component analysis (PCA) of absolute gene expression. The samples formed clusters for each strain clearly distinct from each other indicating high quality of the transcriptomic data (Figure 1A). In the PCA and hierarchical cluster analysis presented as heat maps in Figure 1B and C, the parA null and parB null samples are closer to each other than to WT samples, but the results also demonstrate substantial differences between two mutants. Both mutant samples show changes in the transcript profiles in comparison to WT samples as a consequence of inactivation of parA and parB genes. There is an obvious clustering of identified genes with similar expression patterns in both mutant strains as compared with WT P. aeruginosa (Figure 1B and Figure 1C).
Figure 1. Gene expression analysis in logarithmically growing cultures of parA null, parB null strains versus WT P. aeruginosa.
(A) Quality analysis of three biological replicates of studied strains parA null, parB null and WT of PAO1161 P. aeruginosa by principle component analysis (PCA) of data obtained from expression microarray analysis. The first principle component (PC#1) accounted for 68% and the second principle component PC#2 for 19.7% of the total variation in the dataset. The plot indicates that the transcriptome data are of high quality as the samples cluster together according to the strain: green - WT, red - parA null, blue - parB null. (B) and (C) Cluster analysis of the normalized gene expression for genes that were differentially regulated in parA null and parB null strains as compared to the WT, respectively.
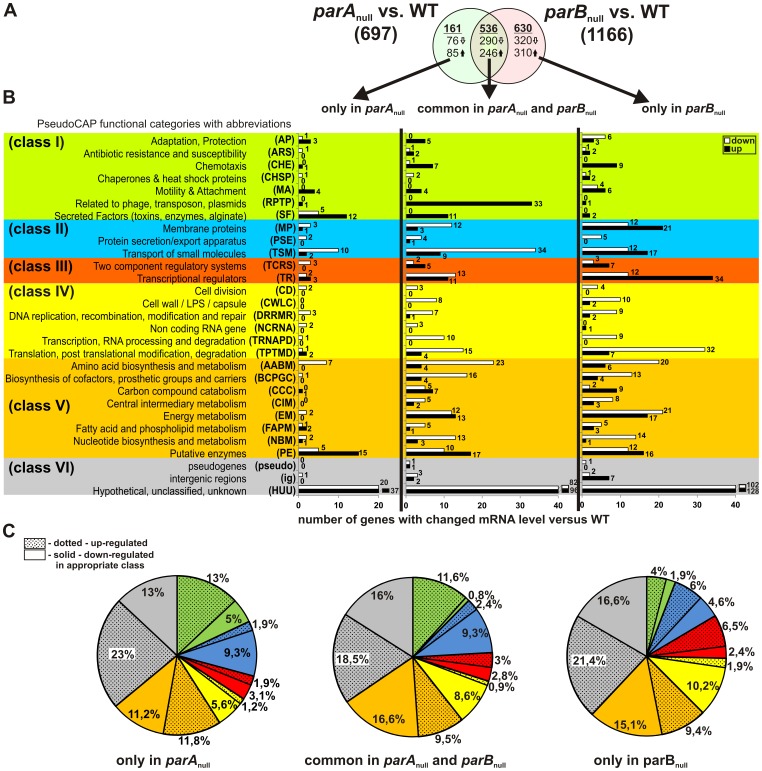
Comparative transcriptome analysis showed statistically significant (p-value, P≤0.05) over two-fold change in expression levels of 697 loci (331 up-regulated and 366 down-regulated), including six intergenic regions and five tRNA-coding genes for parA null samples, and 1166 loci (556 up-regulated and 610 down-regulated), including fourteen intergenic regions and four tRNA-coding genes for parB null samples from logarithmically growing cultures of P. aeruginosa as compared to WT (Figure 2A and Table 1).
Figure 2. Functional classification of genes differentially expressed in logarithmically growing cultures of P. aeruginosa par mutants.
(A) Venn diagram demonstrating the number of genes with changed mRNA level (fold change ≥2; p-value ≤0.05) in parA null and parB null mutant strains as compared to reference WT PAO1161 P. aeruginosa strain. Three gene set lists were created representing genes differentially expressed only in parA null, with different expression in both par mutants (common in parA null and parB null) and with different mRNA level only in parB null. (B) Functional classification of identified genes according to their predicted or known functions. Functional classes are taken from PseudoCAP [29] and are listed on the left with abbreviations in brackets. The original PseudoCAP functional categories were further grouped into six larger classes encompassing: (I) adaptation, protection, motility (green panel); (II) membrane proteins, transport, secretion (blue panel); (III) signal transduction, regulatory functions (red panel); (IV) cellular processes (yellow panel); (V) metabolism (orange panel); (VI) hypothetical, unknown functions (grey panel). (C) The pie charts created for each gene set list illustrating the percentage of genes in each class accounted for the total number of genes with changed expression for: only in parA null, common in parA null and parB null and only in parB null gene set list.
Table 1. Number of genes with changed expression in P. aeruginosa parA null and parB null strains.
| parA null versus WT | ||||||
| Change in mRNA level | >2- to 3-fold | >3- to 5-fold | >5- to 10-fold | >10- to 50-fold | >50-fold | Total |
| Increase | 188 | 75 | 55 | 12 | 1 | 331 |
| Decrease | 291 | 60 | 12 | 2 | 1 | 366 |
| Total | 479 | 135 | 67 | 14 | 1 | 697 |
| parB null versus WT | ||||||
| Change in mRNA level | >2- to 3-fold | >3- to 5-fold | >5- to 10-fold | >10- to 50-fold | >50-fold | Total |
| Increase | 248 | 187 | 73 | 39 | 9 | 556 |
| Decrease | 465 | 128 | 17 | 0 | 0 | 610 |
| Total | 713 | 315 | 90 | 39 | 9 | 1166 |
The loci with altered expression in parA null and parB null as compared to reference PAO1161 P. aeruginosa (WT), indicated by pairwise comparison of microarray data (fold change FC ≥2; p-value ≤0.05). Number of genes (including intergenic regions and tRNA genes) with indicated mRNA level change are shown. Genes were grouped according to the magnitude of differential expression.
As presented in Table 1 most of the genes with changed mRNA level in mutant samples showed differential expression in the range between 2- to 3-fold: 479 genes from total 697 genes for parA null (69%) and 713 genes from total 1166 genes for parB null (61%). Among them more than 60% demonstrated a decreased expression. Changes in gene expression of more than 5-fold (up to >50) were observed for 12% of genes in both mutant strains. Interestingly, the genes that exhibited high level of changes in expression are mostly over-expressed in the mutant strains in comparison to the WT strain (82% genes for parA null and 88% for parB null) suggesting the role of Par proteins as the negative regulators.
The complete list of all genes with altered expression in parA null and parB null in comparison to the WT strain, which exhibited a statistically significant change (fold change, FC ≥2; P≤0.05), is available as Supplementary Information. Two original lists were divided into three gene set lists (according to Venn diagram in Figure 2A), grouping differentially expressed genes in both par mutants (Table S1), only in parA null (Table S2) or only in parB null (Table S3).
Since both mutant strains exhibited similar phenotypes, a substantial amount of overlap in the expression patterns of parA null and parB null strains had been expected. Indeed, 536 of the 697 genes that had altered expression in parA null as compared to WT were also differently regulated in parB null (see Venn diagram in Figure 2A). In the set of 536 genes, expression of which was altered in both mutant strains, all genes but one exhibited the same tendency of change (up- or down-regulation) in expression in both mutants. 290 genes were down-regulated and 246 were up-regulated, usually with the higher fold change in the parB null mutant strain. Only the PA3365 (amiB) gene demonstrated reverse changes in mRNA level when both mutants were compared. It is unclear why PA3365 encoding a probable cytoplasmic chaperone, is 2.1-fold down-regulated in parA null and 4.5-fold overexpressed in the parB null strain.
It is apparent from the transcriptomic data that lack of ParA and ParB proteins has a great impact on gene expression in P. aeruginosa. The 536 genes, indicating expression changes in both mutants and intuitively designated “ParA-ParB regulon”, represent approximately 10% of the PAO1 genome (Table S1). The spectrum of the transcriptomic effects outside of “ParA-ParB regulon” is much broader in the parB null mutant strain (630 genes affected) than in the parA null mutant strain (161 genes affected) (Table S2 and S3). In either mutant strain the absence of one partner protein promotes the proteolytic cleavage of the second partner [16], [18]. Previous experiments [16] demonstrated that in the total sonicates from parA null mutant cells ParB protein was detected in comparable quantities as in the parental strain by Western blotting technique with the use of anti-ParB antibodies (estimated 1000 molecules per cell). However at later stages of the culture growth ParB degradation products were clearly seen and ParB level dropped below detectable (less than 20 molecules per cell). The parB insertion had an identical influence on ParA level although no polar effect might have been expected [18]. It was estimated that ParA level dropped from approximately 400 to less than 40 molecules per cell in the parB insertion mutant. Since insertional and nonsense mutants in par genes behaved similarly ([16] and unpublished) we have decided to use the insertional mutants in parA and parB genes for the transcriptomic analysis as they facilitated irreversible character of the mutations. Although the transcriptomic analysis revealed that mutation in parA gene (insertion of the SmR cassette) has a polar effect on parB expression (parB gene exhibits 75-fold decreased expression in parA null mutant), the visualization of ParB in parA null mutant and the dynamic behaviour of ParA-CFP foci in this mutant (as described in the Introduction) suggest that nevertheless ParB is produced. Both par mutants might be considered as deprived of one Par protein and deficient in the second one.
The difference in number of affected genes between the mutants may suggest either the involvement of ParB in many more functions in the cells than ParA or ParA having stronger effect on gene expression when uncontrolled by ParB since we showed that unbalanced production of Par proteins induces severe toxic effects on the cells [16], [19].
Lack of ParA and ParB Disturbs Expression of Genes Classified to Different Functional Categories
Transcriptomic analysis of parA null and parB null mutants when compared to WT P. aeruginosa from exponential planktonic culture growth exhibited changes in mRNA levels of genes from all functional categories, according to PseudoCAP function classification of Pseudomonas Genome Database ([29]; www.pseudomonas.com). Figure 2B presents an analysis of differentially expressed genes in parA null and parB null mutant strains in comparison with the parental strain, based on their PseudoCAP function classification. The diagram in Figure 2B is divided into three columns: genes with altered expression only in parA null (left part), only in parB null (right part) and a common set of genes exhibiting changes in mRNA level in both par mutants (middle part) (in accordance with Table S1–S3). Numerous genes may be classified into more than one functional category according to PseudoCAP, e.g. the PA4218 is assigned to membrane protein (MP), transport of small molecules (TSM) and antibiotic resistance and susceptibility (ARS) functional categories. For simplicity of presentation in Figure 2B, the closely related functional PseudoCAP categories were arbitrarily grouped into six larger classes from I to VI and a single most likely function for each gene was chosen. All assigned PseudoCAP function categories for the identified genes are presented in Tables S1–S3.
Generally, most of the genes grouped in Class I (Figure 2B, green panel), involved in chemotaxis, motility, attachment, adaptation, protection and secretion functions, were significantly induced in the analyzed mutant cells. Genes classified as plasmid and phage related (RPTP) were all up-regulated. The cluster of genes PA0610-PA0648, significantly overexpressed in both par mutant cells, encodes proteins involved in pyocin production whose expression is often induced in response to stress conditions. Indeed, a part of the observed changes in gene expression pattern in par mutant cells might be the effect of stress response due to the defects in DNA segregation, resulting from incomplete segregation machinery and/or lack of signals coordinating cellular metabolism with chromosome segregation and cell cycle.
Several genes from Class III (Figure 2B, red panel) were significantly induced, especially in parB null mutant. Gene products from TR and TCRS categories play an important role in sensing and responding to signals and perturbations within the cell and to environmental stimuli. They are able to modulate and change cellular metabolism to adapt the organism to changing conditions.
In clear contrast to Class I and III, the majority of genes grouped in other classes were down-regulated, as illustrated by pie charts presented in Figure 2C. Most of the identified genes grouped in Class II (Figure 2B, blue panel) were down-regulated in par mutants, suggesting impairment of some functions connected with cell membrane.
Genes in Classes IV and V play a crucial role in basic cellular processes and metabolism (Figure 2B, yellow and orange panels). All identified genes involved in cell division, transcription and RNA processing were down-regulated in both par mutants (Figure 2B, yellow panel). Essentially, most of the genes from classes IV and V were repressed, suggesting that parA null and parB null mutant cells had changed their metabolism to slow down basic cellular processes. However, there are a few clusters of genes engaged in nitrogen metabolism and denitrification process (nir, nar, nap) that show spectacular overexpression in par mutant cells probably as part of a general stress response (see also below).
Class VI (Figure 2B, grey panel), encompassing genes of hypothetical, unknown functions (HUU PseudoCAP category), together with identified pseudogenes and intergenic regions, constitutes approximately 33–35% of all identified genes with altered expression in par mutants as compared to WT strain (234 genes of total 697 in parA null and 409 genes of total 1166 in parB null). Expression of 132 genes classified as HUU was enhanced, and expression of 102 genes was repressed in parA null strain; in parB null mutant the expression of 224 HUU genes was up-regulated and of 185 down-regulated. The common set of HUU genes with changed expression in both par mutants includes 96 induced genes and 82 down-regulated ones. The large number of genes in this class exceeded the scale of the diagram and is only schematically presented in Figure 2B.
Comparative transcriptome analysis of par mutants cells relative to WT P. aeruginosa demonstrated global changes in gene expression pattern in both analyzed mutants. The list of 536 genes with changed expression in both par mutants constitutes the most probable candidates for “ParA and ParB regulon” genes and further analysis will mainly focus on this group of genes.
Lack of ParA and ParB Affects the Expression of a Number of Transcriptional Regulators
Transcriptional regulators are important factors controlling gene expression and modulating the metabolism and cellular processes as the organism responds to intracellular, as well as environmental signals in order to fulfil its metabolic needs and adapt to the changing growth conditions.
Comparative transcriptomic analysis exhibited a large number of genes encoding known or putative transcriptional regulators or members of two-component regulatory systems with significantly changed mRNA level in par mutant strains as compared to WT strain (37 and 80 genes in parA null and parB null, respectively) suggesting an important role of ParA and ParB proteins in the regulatory network of P. aeruginosa. The target genes of these regulators might be identified in the present study as being regulated by lack of parA and parB, although their regulation may be carried via an indirect mechanism. ParA and ParB influenced the expression of a common set of 29 genes (15 activated, 14 repressed) classified as transcriptional regulators (Table 2).
Table 2. Transcriptional regulators under control of ParA and ParB proteins.
| parA null vs. WT | parB null vs. WT | ||||||
| ID | Gene | Gene product | p-value | FC | p-value | FC | Regulon |
| PA0155 | pcaR | transcriptional regulator PcaR | 0.002 | −2.69 | 0.001 | −3.05 | |
| PA0167 | – | probable transcriptional regulator | 0.003 | −2.05 | 0.000 | −2.94 | Sress, QS |
| PA0236 | – | probable transcriptional regulator | 0.004 | −2.36 | 0.002 | −2.64 | RpoN(KinB), PQS |
| PA0479 | – | probable transcriptional regulator | 0.001 | 2.62 | 0.000 | 5.00 | RpoN(KinB), RpoS |
| PA0610 | prtN | transcriptional regulator PrtN | 0.002 | 3.23 | 0.002 | 3.04 | Stress, QS, RpoN(KinB), PprB |
| PA0612 | ptrB | repressor PtrB | 0.000 | 2.98 | 0.000 | 2.83 | Stress |
| PA0797 | – | probable transcriptional regulator | 0.001 | −3.89 | 0.002 | −2.91 | PprB |
| PA0961 | – | probable cold-shock protein | 0.000 | −2.59 | 0.001 | −2.53 | |
| PA1157 | – | probable two-component response regulator | 0.000 | −5.20 | 0.000 | −5.30 | Stress, PQS |
| PA1182 | – | probable transcriptional regulator | 0.002 | −2.12 | 0.001 | −2.28 | |
| PA1290 | – | probable transcriptional regulator | 0.006 | 2.53 | 0.006 | 2.55 | PQS |
| PA1431 | rsaL | regulatory protein RsaL | 0.002 | 2.22 | 0.001 | 2.54 | QS, PprB |
| PA1504 | – | probable transcriptional regulator | 0.000 | −2.58 | 0.000 | −2.46 | |
| PA2121 | – | probable transcriptional regulator | 0.000 | 3.55 | 0.000 | 4.79 | RpoS, PQS |
| PA2281 | – | probable transcriptional regulator | 0.000 | −2.19 | 0.000 | −2.12 | RpoN(KinB) |
| PA2426 | pvdS | sigma factor PvdS | 0.003 | −2.38 | 0.004 | −2.29 | CORE |
| PA2449 | – | probable transcriptional regulator | 0.006 | −2.02 | 0.001 | −3.10 | |
| PA2572 | – | probable two-component response regulator | 0.001 | 4.57 | 0.000 | 6.76 | RpoS, QS |
| PA2577 | – | probable transcriptional regulator | 0.001 | 2.54 | 0.000 | 3.41 | RpoS |
| PA2622 | cspD | cold-shock protein CspD | 0.000 | 2.39 | 0.001 | 2.04 | |
| PA3027 | – | probable transcriptional regulator | 0.000 | −2.90 | 0.000 | −2.76 | |
| PA3458 | – | probable transcriptional regulator | 0.008 | 2.25 | 0.000 | 9.04 | |
| PA3973 | – | probable transcriptional regulator | 0.013 | 5.02 | 0.004 | 8.34 | RpoS, RpoN(KinB), PprB |
| PA4296 | pprB | two-component response regulator PprB | 0.000 | 2.84 | 0.000 | 6.72 | CORE |
| PA4462 | rpoN | RNA polymerase sigma-54 factor | 0.000 | −2.32 | 0.000 | −2.36 | RpoN(KinB) |
| PA4781 | – | cyclic di-GMP phosphodiesterase | 0.000 | 4.12 | 0.000 | 5.01 | RpoS, RpoN(KinB) |
| PA4843 | – | probable two-component response regulator | 0.000 | 3.28 | 0.000 | 4.14 | RpoS |
| PA4878 | – | probable transcriptional regulator | 0.005 | 3.63 | 0.003 | 4.48 | QS |
| PA5550 | glmR | GlmR transcriptional regulator | 0.000 | −2.30 | 0.000 | −2.73 | Stress |
Genes encoding known or probable transcriptional regulators with changed mRNA level in P. aeruginosa par mutants are listed (p-value ≤0.05; fold change FC ≥2). RpoS, QS, PQS, RpoN(KinB), PprB, stress regulated genes are marked (Regulon column) with marked also genes involved in homeostasis maintenance (CORE) according to Table S1.
Among the 15 significantly activated genes encoding transcriptional regulators only a few have been studied in P. aeruginosa, e.g. PtrB (PA0612), PrtN (PA0610) involved in stress reaction [30], [31]. For most of them only predicted functions are proposed on the basis of sequence analysis, genomic context and experimental data on homologs in other bacterial species (Table 2). Remarkably, large group of the putative transcriptional regulators induced in the absence of Par proteins (PA0479, PA2121, PA2572, PA2577, PA3973, PA4781, PA4843, PA4878) belong to the RpoS and QS (quorum sensing) regulons [32]. RpoS regulon also contains pprB gene significantly induced in both par mutants, encoding two-component response regulator PprB. It possesses a CheY-like receiver domain as well as H-T-H DNA-binding domain and is activated due to the phosphorylation by its partner histidine kinase PprA. PprA and PprB regulate the expression of genes that in turn affect membrane permeability and antibiotic sensitivity of P. aeruginosa [33], [34]. Comparative transcriptional analysis of parA null and parB null mutants identified a number of genes PprB-dependent, including several regulatory genes, up-regulated rsaL, prtN, PA3973 or down-regulated PA0797 [34]. PA1431 (rsaL), up-regulated in parA null and parB null mutants, encodes the regulatory protein RsaL, involved in negative regulation of QS. RsaL binds simultaneously with LasR to the rsaL-lasI bidirectional promoter, thereby preventing the LasR-dependent activation of both genes [35], [36]. Even small changes in the rsaL gene expression might have a huge impact on quorum sensing processes dependent on AHL (N-acyl-L-homoserine lactone). Transcription profiling revealed that RsaL regulates 130 genes independently of its effect on QS signal molecule production, including genes involved in virulence [37].
Among genes down-regulated in the absence of Par proteins there are two genes encoding important general transcriptional sigma factors: PvdS and RpoN (Table 2).
PA2426 encodes PvdS sigma factor involved in the expression of pyoverdine biosynthesis genes cluster and genes important for iron uptake and metabolism as well as a number of virulence factors expressed in response to iron starvation [38], [39]. In addition to pvdS, other genes engaged in iron uptake were down-regulated in par mutant cells, e.g. fpvA, pyochelin synthesis genes or optH, encoding probable TonB-dependent receptor (Table S1).
PA4462 gene encoding RNA polymerase sigma factor, known as RpoN or sigma factor 54, was approximately 2.3-fold down-regulated in both par mutant strains. RpoN controls alginate biosynthesis together with kinase sensor KinB, which phosphorylates AlgB in response to environmental signals. It also controls some regulatory genes and a large number of genes involved in carbohydrate metabolism, quorum sensing, iron regulation, rhamnolipids production, and motility [40], [41]. The expression of a number of genes belonging to the RpoN-KinB regulon was affected in both par mutants, indicating also significant overlap between the RpoN-KinB, RpoS and QS regulons, e.g. nap, nar or glc (Figure 3A).
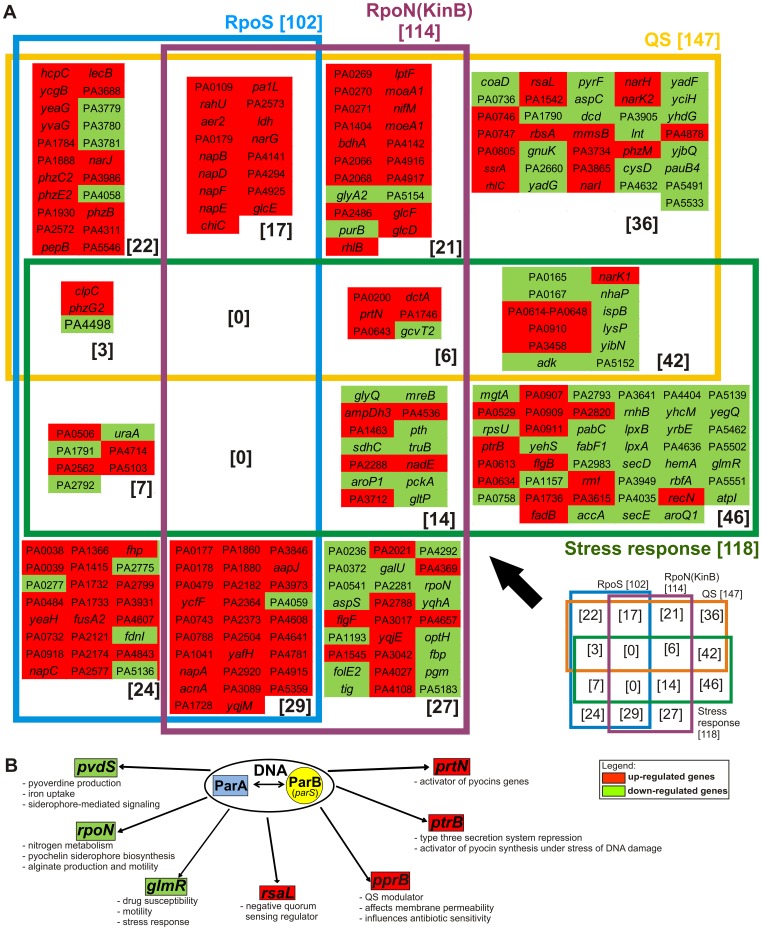
Figure 3. Genes from overrepresented regulons affected by mutations in parA and parB genes in P. aeruginosa.
(A) RpoS, QS, RpoN(KinB) and stress regulated genes with altered expression in par mutants presented as Venn diagram illustrating separate and common genes classified into presented regulons (according to Table S1). (B) The most potent known regulators with changed expression in par mutants. The functions they influence are given.
Among transcriptional regulators repressed in both par mutants there is PA0155 gene encoding the PcaR transcriptional regulator. In P. putida, PcaR positively regulates the pca regulon involved in the chemotactic response to aromatic compounds [42]. PA4974 (opmH), a part of the PcaR regulon, encodes probable outer membrane protein precursor, which is also repressed in both par mutant strains.
The PA1157 gene expression was approximately 5-fold down-regulated in both par mutants. It encodes a putative two-component response regulator, which demonstrates 63% similarity to the transcriptional regulator RstA from E. coli. RstA works in pair with the RstB sensor, mainly in stress response cascade in E. coli [43].
PA5550 gene encoding GlmR transcriptional regulator was repressed 2–3-fold in both par mutant cells. It was demonstrated that mutation in glmR caused drug supersusceptibility, loss of motility, reduced resistance to osmotic and heat shock stress, as well as impaired growth at low temperatures, affecting peptidoglycan and LPS synthesis [44].
The predicted effects of changes in production of listed transcriptional regulators seem to lead to the slowing down of metabolism and also to induce the processes related to transition into stationary phase (RpoS regulon). Because a number of RpoS-dependent genes is also QS-dependent (some activated, others repressed at higher cell culture densities), it is not unexpected that QS-dependent communication and regulation of gene expression in par mutant cells has been altered.
A short summary of processes that ParA and ParB proteins may influence via control of expression level of known regulatory genes is presented in Figure 3B.
Lack of ParA and ParB Alters Expression of Genes Involved in General Stress Response and Maintenance of Cellular Homeostasis
Lack of ParA and ParB causes defects in chromosome partitioning, manifested by appearance of up to 7% of anucleate cells and cells with guillotining chromosomes in P. aeruginosa par mutant cultures [16], [18] and that might induce the stress response. In Figure 3 apart from RpoS-, QS- and RpoN(KinB)-dependent genes with changed expression in par mutant cells (according to gene lists by Schuster et al. [32] and Damron et al. [40]), the genes involved in stress response are presented (according to Cirz et al. [45]). Among them there are also the ptrB and glmR genes described above.
Since lack of functional ParA or ParB leads to incomplete chromosome segregation with visible DNA guillotining effects, it was expected that multiple genes from DRRMR functional category (DNA replication, recombination, modification and repair) would be induced. Interestingly, the only gene from this category activated in both par mutants was PA4763 (recN). It encodes the SOS-inducible DNA repair protein RecN, which in E. coli is indispensable for repair of double-strand breaks in the chromosome when these breaks occur at two or more locations [46].
As a part of general stress response in par mutant cells, a significant decrease in expression of genes encoding ribosomal proteins, e.g. rpsU, rpsA, rpsI, prmA or rpmH, was detected (Table S1–S3). Similarly, genes coding for parts of secretion machinery, secD and secE, were down-regulated in par mutant cells as compared with WT P. aeruginosa. Secretion protein SecE is encoded in the three-cistronic operon nusG-secE-PA4276.1. PA4275 (nusG) encodes an essential transcription antitermination protein NusG, while PA4276.1 encodes tRNA-Trp. All three genes were repressed in par mutants.
In addition, a number of genes with changed expression in both par mutant cells is a part of the core set of genes whose products are important for maintaining homeostasis under different stress conditions in P. aeruginosa (Table S1; [47]). The set of genes engaged in homeostasis maintenance, significantly repressed in par mutants, includes genes encoding for the ferripyoverdine receptor (PA2398), sigma factor PvdS (PA2426), pyochelin (PA4222, PA4223, PA4224, PA4230) as well as the rod shape-determining protein MreC (PA4480), ribose-phosphate pyrophosphokinase (PA4670) and probable xanthine/uracil permease (PA4719).
PA4480 (mreC) and PA4481 (mreB) genes encoding the rod shape-determining proteins MreC and MreB, were 2–3-fold repressed in analyzed par mutants as compared with WT P. aeruginosa. In parB null also the third gene of the mreBCD operon, mreD, exhibited a lower mRNA level relative to the WT strain. MreB is an actin homolog forming dynamic helical filaments or patches beneath the surface of the cell, potentially explored as a scaffold for transporting proteins to different locations throughout the bacterial cell. MreB is essential for maintenance of cell shape, chromosome segregation, and polar localization of several bacterial proteins, e.g. type IV pili [48], [49]. The lower level of expression of mre operon as well as of a number of genes involved in peptidoglycan synthesis (mgtA, mltA, htrB, lpxB, lpxA, lnt and murA) in par mutant cells may impair their growth.
Overrepresentation of RpoS- and QS-dependent Genes with Changed Expression in par Mutants
Our transcriptomic studies identified many genes from RpoS regulon [32] with altered expression in par mutant cells as compared with WT P. aeruginosa (Table S1), although the expression of the rpoS gene was not changed. Interestingly, among the identified RpoS-dependent genes most showed significant activation in both par mutants (Figure 3A). The lack of Par proteins causes visible defects in chromosome segregation in at least 7% of cells in mid-log phase cultures which may explain the slower growth rate [16], [18] and, as demonstrated here, leads to the general stress response and down-regulation of number of genes involved in basic metabolism (class IV and V in Figure 2B) in connection with an earlier induction of RpoS-dependent genes in par mutants. RpoS acts as an alternative sigma factor of genes preferentially expressed in stationary phase of growth as well as a regulator of the general stress response.
The list of RpoS-regulated genes with significantly induced expression in par mutant cells contains also orfs coding for proteins involved in chemotaxis (e.g. PA0176-PA0179, PA1930, PA2573, PA2920, PA4915). Changes in expression level of genes encoding chemotaxis proteins may affect motility of bacteria and explain swimming and swarming defects observed in par mutants [16], [18].
Among genes with changed mRNA levels in P. aeruginosa par mutant cells there was, in addition to RpoS regulon, a number of genes involved in QS function and control [32], [50]. QS-related genes with changed expression in at least one par mutant include those coding for negative regulators of QS: rsaL, rsmA, dksA and rpoN, as well as positive regulators like pprB [51]. QS-dependent genes encoding phenazine/pyocyanin biosynthesis pathway, rhamnolipids, lectin or chitinase, implicated in pathogenesis, adaptation and survival, were significantly activated (Figure 3).
The group of virulence factors includes also genes encoding proteins involved in motility and coding for flagellar elements (PA1077, PA1081) as well as, mentioned above, chemotaxis sensory transducer (PA2573) or two-component response regulator (PA2572), playing a role in the first stages of infection, mainly during adhesion to host cells. Similarly, PA4108, PA4781 and PA2572 are classified as coding virulence factors due to their involvement in regulation of adhesion and biofilm formation by modulation of c-di-GMP level. All these genes were induced in one or both of the analyzed par mutant strains.
One of the metabolic processes regulated by QS is regeneration of the AHL precursors such as methionine and S-adenosylmethionine (SAM) and degradation of adenosine via inosine and hypoxanthine [52]. A number of genes whose products are involved in AHL metabolism, are down-regulated in both or at least one of the analyzed par mutant strains, e.g. PA0390 (metX), PA0430, PA0654 (speD), PA1687 (speE), PA3169 (Table S1–S3).
Among genes under RpoS and QS control with changed mRNA level in both par mutants, the high activation was observed for the cluster of nar genes (10–40-fold induction) and nap genes (4-10-fold up-regulation). Seven nar genes (PA3871- PA3877) are organized in an operon encoding respiratory nitrate reductase components. The nar operon is activated under low-oxygen tension conditions and in the presence of nitrate, and transcriptional regulators Anr, NarXL and Dnr [53].
The PA1172-PA1176 nap genes code for cytochrome c-type, an essential component of the electron transport chain, participating in periplasmic nitrate reduction. In concert with activation of nar, nap genes, the expression of nirS (coding for nitrite reductase precursor), nirQ (encoding regulatory protein NirQ with MoxR-like ATPase motif), norCBD (encoding cytochrome c subunits of nitric oxide reductase and denitrification protein NorD) were induced in parB null mutant. The highest activation in par mutants was exhibited by the PA2664 gene (41-fold in parA null and 296-fold in parB null). PA2664 encodes flavohemoprotein with a role in detoxification of NO (nitric oxide) under aerobic conditions [54]. NO is the intermediate but also an important effector of denitrification pathway. It activates transcriptional regulator Dnr (Dissimilative Nitrate Respiration regulator), acting jointly with NarXL and Anr for activation of nar, nir, nor and nos operons. However dnr was 4.2-fold repressed in parA null, but not affected in parB null. Interestingly, the genes encoding elements of two-component regulatory system the narLX, were also differently expressed in par mutants. The narX gene was 2-fold down-regulated in parA null relatively to WT, while the narL gene was 2.9-fold activated in parB null mutant. It is a peculiar discrepancy since they usually co-activate the nar genes, but it is possible that such situation takes place when Par proteins are present in the cell. It suggests that the effect of the denitrification pathway induction may be achieved by different means in two par mutants.
It is unclear why there is such a high overexpression of nar, nap and nif genes in cultures of exponentially growing par mutants cells where oxygen concentration and nutrients availability should not yet be limiting factors for cells to grow and divide. Most likely, it can be explained by activation of the denitrification process as a part of the stress-response cascade in reaction to the cellular signals mimicking the necessity of cells to turn to the less-active metabolically state, characteristic for stationary phase cultures where lack of oxygen is a real problem. The denitrification processes in P. aeruginosa are controlled not only by low-oxygen tension but also by the cell-to-cell communication signals [55]. In P. aeruginosa two chemically distinct signaling molecules have been characterized: AHLs (produced by LasI and RhlI) and PQS (Pseudomonas Quinolone Signal: 2-heptyl-3-hydroxy-4-quinolone, produced by pqs operon). The AHL-dependent regulators LasR (through RhlR) and RhlR repress the denitrification operons so it is likely that derepression of the denitrification pathway is the result of the decrease in AHLs synthesis (see above). The second effector PQS acts posttranslationally inhibiting activities of NAR, NOR and NOS reductases, but stimulating NIR (NO2 - reductase) that produces NO. High amounts of NO induce genes important for NO detoxification e.g. PA2664 fhp (activated in both mutants to the extreme levels), PA2665 encoding transcriptional activator FhpR and PA2663 involved in activation of the production of polysaccharides virulence-related factors, such as pyoverdine, PQS, elastase [56], at the same time reducing swimming and swarming motility (both genes are over-expressed in the parB mutant). NO at non-toxic levels regulates the social behaviour through regulation of level of another secondary messenger c-di-GMP by inducing enzymes involved in its degradation (see next section).
Lack of ParA and ParB Induces Expression of Genes Involved in c-di-GMP Turnover and Signalling
Cyclic-di-GMP (cyclic diguanylate) is an important messenger ubiquitous in bacterial cells controlling various processes, e.g. switch between the motile planktonic and biofilm lifestyles of bacteria, virulence of animal and plant pathogens, antibiotic production, progression through the cell cycle and other cellular functions [57]. The level of c-di-GMP in the cell is tightly controlled by the opposite actions of two classes of enzymes. Proteins encoding HD-GYP or EAL domains are specific phosphodiesterases (PDEs) involved in hydrolysis of c-di-GMP, whereas diguanylate cyclases (DGCs) possessing the GGDEF domain are engaged in the production of c-di-GMP from two GTP molecules [57]. The GGDEF, EAL and HD-GYP domains are usually linked to various N-terminal sensory input domains, often transmembrane, suggesting that numerous environmental and cellular signals are integrated into the c-di-GMP signalling network. Cyclic-di-GMP is bound by transcriptional regulators, proteins containing PilZ domains, proteins carrying degenerate GGDEF or EAL domains, as well as RNA in riboswitches modulating protein-protein interactions, protein-DNA interactions, protein enzymatic activity, protein-RNA interactions and by transcription, translation and other cellular processes [58].
Among the genes involved in c-di-GMP turnover with altered expression in at least one par mutant, three genes were found encoding diguanylate cyclases (DGCs) with GGDEF domain (PA0169, PA2771, PA4843), five genes encoding proteins with both GGDEF and EAL domains (PA0861, PA2567, PA3311, PA4367, PA5017) and three genes coding for phosphodiesterases with HD-GYP motif (PA2572, PA4108, PA4781). For some genes, RpoS-dependent transcription was suggested (Table S1; [32]).
Significant overexpression (2-7-fold) of three genes coding for phosphodiesterases may cause a decrease in c-di-GMP level in par mutants cells promoting a higher expression of flagellar components and virulence factors genes. Indeed, we observed overexpression of a number of genes encoding flagellar proteins and this overproduction could lead to motility dysfunction. Some genes encoding virulence factors also seemed to be overexpressed in par mutant strains, e.g. lecA, lasA, lasB, hcpC, apr, rhl and phz.
The PA4843 gene up-regulated 3-4-fold in both par mutant strains encodes a protein classified into TCRS containing response regulator with a CheY-like receiver domain important in sensing signals from the environment and a GGDEF domain. It shows 50% similarity to pleD gene product responding to c-di-GMP level required for the swarmer-to-stalked-cell transition in C. crescentus [59].
Because so many genes whose products are involved in c-di-GMP turnover exhibited spectacular changes in expression in par mutant cells (especially in the parB null mutant) it was particularly interesting to find the effectors of modulated level of c-di-GMP action in P. aeruginosa. Recent studies of Duvel et al. [60] described the proteomic method allowing for identification of c-di-GMP binding proteins in P. aeruginosa. By comparing the list of genes encoding proteins identified in c-di-GMP pull-down experiments [60] with the list of genes whose expression was affected by mutations in parA and parB, we found 25 genes in parA null, 54 in the parB null mutant, and common 20 genes in both mutant strains, whose activity might be regulated by c-di-GMP. Among those with significantly up-regulated mRNA level in both par mutant cells were genes encoding: PA4781 c-di-GMP phosphodiesterase (described above); PA0176 aerotaxis transducer Aer2; PA2573, PA2920, PA4915 and PA2788, predicted membrane chemotaxis transducers; PA2799 and PA4608, hypothetical proteins possessing the PilZ domain (a known receptor for the second messenger c-di-GMP, with homology to type IV pilus assembly protein); PA3458, probable transcriptional regulator. These few examples show that the spectrum of c-di-GMP effectors is broad and the impact of altered expression of some c-di-GMP efector proteins from one side and differences in expression of genes encoding DGC and PDE from another side in par mutant cells may trigger a large cascade of effects/defects. Some of them seem to be connected with chemotaxis, motility, signal transduction and regulatory functions, but for others the role in P. aeruginosa regulatory network and metabolism is waiting to be elucidated.
Validation of Microarray Results by RT-qPCR
RT-qPCR (reverse transcription followed by quantitative PCR) was performed to confirm the observed changes in gene expression in parA null and parB null strains as compared with WT. The same RNA samples were used in RT-qPCR analysis as those used in microarray analysis. In the first step RNA from three biological replicates of each strain was used in reverse transcription reaction to obtain cDNA. For selected genes specific primers were designed (Table S4) and used in qPCR with cDNA as a template.
Genes listed in Table 3 were chosen for RT-qPCR analysis because their fold change in expression in par mutants varied significantly across a relatively broad range and some of them were physiologically relevant candidates for further analysis. The constitutively expressed housekeeping gene nadB (PA0761), which encodes an L-aspartate oxidase and expression of which was not altered in par mutants, was used as an internal control in qPCR reactions. Essentially all the RT-qPCR results correlated with the microarray alterations (promotion or repression) in gene expression. PA2398 (fpvA), PA4307 (pctC), PA4675 (optH) and PA5139 showed reduced expression, while PA0586, PA1081, PA1930, PA2570, PA2572, PA2573, PA2920, PA3520, PA3688, PA3973, PA4108 and PA4843 demonstrated increased expression in both par mutants as compared with WT (Table 3). For the PA1196 and PA2567 genes, mild overexpression in parA null mutant (<1.8-fold) was detected using the qPCR method that was not included in the microarrays data with cut-off of fold change >2. Using RT-qPCR analysis significant overexpression of PA3006 gene was detected in both par mutants relatively to the WT strain whereas microarray analysis indicated changes only in the parB mutant. On the basis of RT-qPCR analysis, the PA3006 gene is also considered as a part of the ParA/ParB regulon.
Table 3. Validation of microarray data by RT-qPCR analysis.
| parA null versus WT | parB null versus WT | |||||||||
| fold change | fold change | |||||||||
| ID | Gene | Product | MC | RT | SD RT | change | MC | RT | SD RT | change |
| PA0586 | ycgB | conserved hypothetical protein | 5.81 | 5.85 | 0.43 | up | 6.36 | 4.32 | 0.16 | up |
| PA1081 | flgF | flagellar basal-body rod protein FlgF | 3.90 | 4.70 | 0.11 | up | 3.26 | 2.59 | 0.47 | up |
| PA1196 | – | probable transcriptional regulator | nd | 1.37 | 0.19 | up | 7.25 | 7.59 | 0.46 | up |
| PA1930 | – | probable chemotaxis transducer | 6.60 | 2.86 | 0.80 | up | 17.53 | 3.47 | 1.61 | up |
| PA2398 | fpvA | ferripyoverdine receptor | −4.26 | −1.51 | 0.01 | down | −8.31 | −8.10 | 0.42 | down |
| PA2567 | – | cyclic di-GMP phosphodiesterase class I | nd | 1.73 | 0.26 | up | 5.38 | 4.44 | 0.90 | up |
| PA2570 | pa1L | PA-I galactophilic lectin | 6.95 | 6.14 | 1.06 | up | 8.26 | 6.53 | 1.19 | up |
| PA2572 | – | probable two-component response regulator | 4.57 | 3.93 | 0.37 | up | 6.76 | 3.27 | 0.04 | up |
| PA2573 | – | probable chemotaxis sensory transducer | 4.37 | 6.75 | 0.57 | up | 7.36 | 6.51 | 1.23 | up |
| PA2920 | – | probable chemotaxis transducer | 2.49 | 4.55 | 0.95 | up | 9.39 | 8.01 | 1.23 | up |
| PA3006 | psrA | transcriptional regulator PsrA | nd | 5.23 | 0.14 | up | 7.80 | 5.78 | 0.60 | up |
| PA3520 | – | – | 14.82 | 8.04 | 1.50 | up | 21.12 | 7.74 | 1.65 | up |
| PA3688 | – | – | 7.39 | 5.68 | 0.34 | up | 9.48 | 2.61 | 0.35 | up |
| PA3973 | – | probable transcriptional regulator | 5.02 | 5.78 | 0.13 | up | 8.34 | 6.22 | 0.39 | up |
| PA4108 | – | cyclic di-GMP phosphodiesterase class II | 3.63 | 3.78 | 0.10 | up | 6.93 | 4.39 | 0.94 | up |
| PA4307 | pctC | chemotactic transducer PctC | −5.78 | −3.61 | 0.25 | down | −3.83 | −4.86 | 0.57 | down |
| PA4675 | optH | probable TonB-dependent receptor | −5.08 | −2.94 | 1.32 | down | −3.58 | −4.67 | 0.69 | down |
| PA4843 | – | probable two-component response regulator | 3.28 | 3.79 | 0.46 | up | 4.14 | 3.88 | 0.47 | up |
| PA5139 | – | – | −5.08 | −2.43 | 0.45 | down | −5.27 | −5.65 | 0.23 | down |
The RT-qPCR on RNA samples applied for microarrays analysis for chosen genes with changed mRNA level in P. aeruginosa par mutants (p-value ≤0.05; fold change ≥2). Abbreviations: MC - microarray data; RT - RT-qPCR data; SD RT - standard deviation for RT-qPCR analysis; nd - change not detected. Standard deviation from at least three independent experiments is presented.
parA and parB act as repressors and activators of gene expression
Ten putative promoter regions of PA0459, PA0588, PA1196, PA1930, PA2567, PA3973, PA4108, PA4542, PA4596 and PA4915 genes, selected on the basis of transcriptomic data, were amplified by PCR and cloned into the broad host-range promoter-probe vector pCM132 [61]. The expression of the transcriptional fusions of studied promoter with lacZ was analyzed in transformants of three E. coli strains, namely: DH5Δlac (pGBT30), DH5Δlac (pKLB1 tacp-parA) and DH5Δlac (pKLB2 tacp-parB) in overnight cultures without induction of tacp (Figure 4A). All cloned regions contained promoter sequences, that led expression of the reporter lacZ gene in E. coli DH5Δlac (pGBT30) strain. The lowest β-galactosidase activity was detected for PA0588p, PA1196p, PA1930p, PA4915p fusions (less than 500 U), the highest activity was observed for PA4108p-lacZ fusion (above 2000 U).
Figure 4. Regulation of gene expression by ParA and ParB of P. aeruginosa.
(A) The β-galactosidase activity in E. coli DH5Δlac transformants bearing pCM132 derivatives with analyzed promoter regions-lacZ fusions and pGBT30 derivatives [72] expressing ParA, ParB or no protein as a control (vector). (B) Fold changes determined by comparative microarray analysis of P. aeruginosa parA null, parB null versus WT strains (p-value ≤0.05; fold-change ≥2) for chosen genes which promoter regions were analyzed using promoter-lacZ fusions in (A).
When the activities of the promoters were tested in the presence of ParA or ParB produced from the compatible high-copy-number plasmid, the level of lacZ expression was clearly affected for most of the tested sequences (Figure 4A). Although the par genes were inserted under control of tacp, no IPTG was used to further induce the expression in order to avoid the possible “toxic” effect of overproduced proteins. Our previous studies confirmed production of the appropriate protein detected by Western blot analysis at the basal level expressed from the tacp promoter without IPTG induction in transformant cultures (data not shown).
The direct regulation of lacZ expression by Par proteins complying with the transcriptomic data was shown for three out of ten tested promoters PA4108p, PA4596p and PA1930p (Figure 4). Two promoters were not affected by ParA and ParB when tested in the heterologous host (PA0459p and PA4915p).
Interestingly, in clear contrast with transcriptomic data and observed induction in P. aeruginosa par mutants, the expression of PA0588p-, PA1196p-, PA2567p-, PA3973p- and PA4542p-lacZ fusions revealed stimulatory effect of ParA, ParB or both proteins under tested conditions (Figure 4). The lack of regulation or reverse regulatory effect of Par proteins on P. aeruginosa promoters in the E. coli may indicate the complex regulation of expression of the studied promoters in their host. Indeed, expression of some of them depends on the potent P. aeruginosa regulators: RpoS, RpoN or PprB (PA0588p, PA2567p, PA3973p) [32], [40].
Performed regulatory experiments in E. coli for promoter regions of chosen P. aeruginosa genes in the presence of ParA and ParB demonstrated that both proteins are able to modulate gene expression acting directly or indirectly as repressors or activators. Further studies are needed to explain obtained results in the context of complex regulatory network influencing gene expression in P. aeruginosa.
Conclusions
The role of ParA and ParB in chromosome segregation and organization in P. aeruginosa has already been documented [16], [18], [19], [23], [24], [28], but their influence on gene expression has not yet been studied. This is the first report considering ParA and ParB proteins as regulators of gene expression. The parA null and parB null mutations do not lead to the lethality [16], [18] although they disturb the proper segregation of the newly replicated genomes. The populations of mutants which produce up to 7% of anucleate cells and many more cells with aberrantly segregated chromosomes grow on a rich medium with only slightly prolonged division time (36 min for parA null, 33 min for parB null versus 30 min for WT) [16], [18], [24], [28]. Phenotypic characterization of P. aeruginosa parA null and parB null mutants exhibited defects in swarming and swimming motility [16], [18]. These defects might be correlated with altered expression of genes involved in motility, chemotaxis, or signal transduction functions (Table S1–S3; Figure 2B, 3, 4). Additionally, genes encoding products involved in c-di-GMP signalling [57] might influence motility functions in par mutant cells, e.g. PA2567, PA4108. The par mutant cells are impaired in motility and form colonies with altered morphology. The question arose how the cells might cope with disturbances in chromosome segregation and survive as the population.
The comparative transcriptome analysis of parA null, parB null mutant populations from the mid-log phase of growth of planktonic cultures versus the parental PAO1161 strain of P. aeruginosa demonstrated global changes in gene expression pattern. Despite the good nutrients and oxygen supply, genes of the RpoS regulon were mostly induced suggesting entry of bacteria into a less metabolically active state. Delaying the cell growth and division might provide the bacteria with a chance to segregate the chromosomes by mechanisms alternative to par system. Functional categorization of identified genes demonstrated an increase in expression of genes whose products are engaged in adaptation, protection and motility function with clear down-regulation of genes that encode proteins involved in basic metabolism and cellular processes (Figure 2). In addition, a number of genes encoding parts of two-component regulatory systems and transcriptional regulators exhibited changes in mRNA level in par mutant cells.
The intriguing question is the signal prompting such delay and RpoS regulon activation. Our transcriptomic studies point out the importance of two signalling molecules: NO, the intermediate and effector of denitrification pathway, which regulates the cell-to-cell signalling, here induced as the result of stress response and c-di-GMP involved also in quorum sensing. The levels of both molecules might be affected in par mutants since the expression of genes involved in their synthesis and decay is significantly altered. There is another signalling molecule, PQS, which is considered to be an essential mediator of the survival strategies for bacterial population and one of these strategies is to enter a dormant state that slows down metabolism. The 38 genes dependent on PQS show an altered expression in ParA-ParB regulon (Table S1), however, the mechanism of PQS action is still not fully understood. PQS is postulated to act not only as the transcriptional regulator, but also as the posttranscriptional modulator [62]. Further studies may identify the signal responsible for slowing down the metabolism in mid-log phase of culture growth and inducing adaptive responses.
Another open question is the mode of action of Par proteins as the potential regulators in the WT P. aeruginosa. In vivo regulatory experiments showed direct regulation of some genes by Par proteins, playing the role of repressors (PA4108p, PA4596p, PA1930p) as well as activators (PA0588p, PA2567p, PA4542p, PA1196p, PA3973p) (Figure 4A). The altered expression of a large number of genes encoding known or predicted transcriptional regulators in parA null and parB null mutants suggests also the indirect regulation mediated by regulatory genes under ParA and ParB control.
It has been postulated for the chromosomal homologs that ATP bound-ParA interacts non-specifically with DNA and ParB by stimulating ATPase activity releases ParA from the nucleoid [26], [27], [63]. In the absence of ParB, ParA is most probably in its dimer state bound with ATP proficient to bind non-specifically DNA. One of the reasons of the global changes in genes expression in P. aeruginosa parB null mutant might be the effect of uncontrolled action of ParA on DNA in the absence of ParB. On the other hand ParB interacts with parS sequences and is able to spread on flanking DNA regions possibly influencing gene expression [18], [19]. Creation of large nucleoprotein complexes greatly influences DNA topology and this might be an additional level of control of genes expression.
A hierarchical cascade of direct and indirect regulation by ParA and ParB may be enhanced by signal molecules mentioned above. Among them, the secondary messenger c-di-GMP emerges as an important factor. The c-di-GMP signalling is involved in the regulation of change of the lifestyle from planktonic to biofilm or controlling the virulence determinants in bacteria [57]. Recently, Baraquet et al. [64] demonstrated that FleQ/FleN/DNA interactions are modulated by c-di-GMP, changing the mode of action of FleQ from repressor to activator, which in turn influenced exopolysaccharide production. Moreover, the cell cycle dependent fluctuations of c-di-GMP were visualized in bacteria, showing the asymmetrical distribution of c-di-GMP correlated with the time of cell division and polar localization [65]. The local concentration of c-di-GMP might be crucial for macromolecular complexes allowing independent and parallel control of different output reactions.
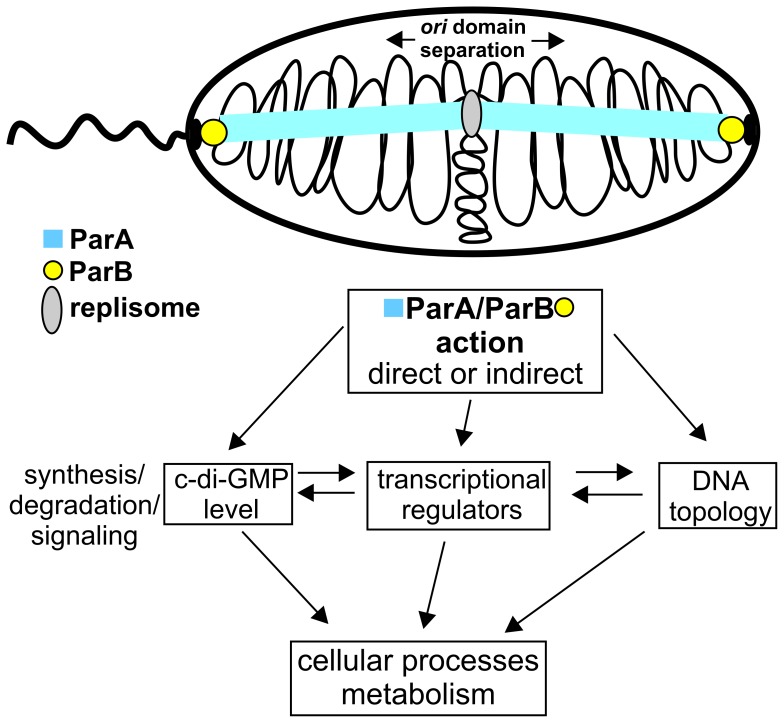
On the basis of performed studies and present knowledge we propose the model of ParA and ParB action in P. aeruginosa cells (Figure 5). The ParA and ParB proteins play a major role in chromosome segregation in bacterial cells. ParB interacts with parS sequences and helps to organize, condense and orient the newly replicated oriC regions. ParB interactions with ParA stimulate ParA ATPase activity and redistribution of the large nucleoprotein complexes to opposite halves of the cell prior to cell division. An additional role of ParA and ParB action on DNA is modulation of gene expression with an opportunity to coordinate different processes within the bacterial cell cycle including chromosome condensation, organization, segregation, chromosome replication, cell division and growth rate. The mode of action of ParA and ParB is complex. It may involve direct interactions with promoter regions of certain genes, as well as it might be the consequence of specific (parS sequences) and unspecific interactions with DNA and induction of topological changes. All these mechanisms may influence expression of specific targets or regulatory genes with a potential to regulate subsequent genes, as a part of the regulatory network. ParA and ParB interactions with partner proteins may also modulate the process (unpublished data). The molecular mechanisms explaining the mode of ParA and ParB action in gene regulation need further investigations. This study provides useful information about the possible links between chromosome segregation, the progression of the cell cycle, control of the growth rate by intertwining with RpoS regulon, QS-regulatory networks and regulation of genes expression.
Figure 5. Model of ParA and ParB action in P. aeruginosa.
The chromosome is illustrated as thin black line, the replisome as grey ellipse, ParA structures as overlapping blue squares, ParB as yellow circle, cell envelope and single polar flagella as thick black line (see text for description).
Materials and Methods
Bacterial Strains and Growth Conditions
Pseudomonas aeruginosa PAO1161 (leu- r -), derivative of PAO1, was kindly provided by B. M. Holloway (Monash University, Clayton, Victoria, Australia). P. aeruginosa PAO1161 RifR (WT), PAO1161 RifR parA1-39::smh (parA null) and PAO1161 RifR parB1-18::TcR (parB null) strains were obtained as described previously [16], [18], [71]. E. coli K12 strain DH5α was used for standard cloning procedures whereas E. coli DH5Δlac {Nalr; deoR thi1 relA1 supE44 endA1 gyrA96 recA1 hsdR17 Δ(argF lac) U169} was used for regulatory studies.
Bacteria were grown in L broth [66] at 37°C or on L agar (L-broth with 1.5%, w/v, agar) supplemented with antibiotics as appropriate: 10 µg ml-1 for chloramphenicol resistance and 50 µg ml-1 for kanamycin resistance in E. coli.
Cells of P. aeruginosa PAO1161 strains were taken from a deep-frozen stock, spread on L-agar plates and grown overnight at 37oC. Bacteria from single colonies were then used to inoculate L-broth liquid cultures and grown overnight with shaking at 37oC. Three independent overnight cultures for each strain were diluted 1∶100 in fresh L-broth and propagated with shaking at 37oC. Samples were collected from the cultures at regular intervals to measure the optical density at 600 nm (OD600) and to determine the CFU/ml. Aliquots of 4 ml for exponential phase culture (OD600 = 0.5; in total 2×109 cells) were subjected to RNA extraction.
RNA Isolation
Three independent replicates of total RNA were isolated from each strain using an RNeasy mini-kit with on-column DNase digestion (Qiagen) according to the manufacturer’s instructions. The DNase digestion using TURBO DNase kit (Ambion) was used to eliminate DNA contamination. The RNA quality and integrity was checked using Bioanalyzer (Agilent Technologies) and the concentration was estimated using Nano Drop ND-1000 Spectrophotometer.
Affymetrix Genechip Microarrays
For DNA microarrays, three biological replicates of total RNA (10 µg) from each strain were used for cDNA synthesis, fragmentation, and labeling according to the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA, USA). Briefly, random hexamer primers (final concentration, 25 ng µl-1; Invitrogen) were added to the total RNA (10 µg) along with in vitro synthesized B. subtilis control spikes. cDNA was synthesized using Superscript II (final concentration of 25 U µl-1 (Invitrogen) according to the manufacturer’s instructions under the following conditions: 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min. RNA was removed by alkaline treatment and subsequent neutralization. cDNA was purified using a MiniElute PCR Purification Kit (Qiagen) and was eluted in 12 µl of EB Buffer. cDNA was fragmented with DNase I (0.6 U per µg of cDNA; Amersham) at 37°C for 10 min and then end-labeled with biotin-ddUTP using a GeneChip® DNA Labeling Reagent (Affymetrix) at 37°C for 60 min. Fragmented labeled cDNA samples were hybridized to the array (GeneChip P. aeruginosa Genome Array) and scanned with Affymetrix GeneChip Scanner 3000.
Microarray Data Analysis
Microarray gene expression data were analyzed using Partek Genomic Suite 6.6 beta software (Partek Inc., St. Louis, MO). Raw data (.cel files) were imported and processed using GeneChip Robust Multiarray Averaging (GC RMA) background correction, quantile normalization, Log2 transformation and median polish summarization. Principal component analysis (PCA) was used to check the batch effect and to identify the outliers. Analysis of variance (ANOVA) using REML (restricted maximum likelihood) was performed in order to identify differentially expressed genes for a particular genotype versus wild type (parA null vs. WT and parB null vs. WT). Gene lists were created using a cut off of p-value with FDR (False Discovery Rate) ≤0.05, with a fold change 2 (−2≥ FC ≥2). Hierarchical clustering of significantly and differentially expressed genes was performed to group samples with similar expression patterns into clusters.
Microarray as well as data analysis were performed in the Laboratory of Microarray Analysis, Department of Systems Biology, Warsaw University and Institute of Biochemistry and Biophysics PAS, Warsaw, Poland (www.corelab.pl).
RT-qPCR Analysis
The same RNA samples were used for RT-qPCR analysis in order to verify microarray data for chosen genes. Three biological replicates of total RNA (2 µg) from each strain served as a template for cDNA synthesis with SuperScipt VILO Master Mix reverse transcriptase (Invitrogen). The cDNA was purified using QiaQuick PCR purification Kit (Qiagen) and then used as a template in qPCR performed with SYBR® Green JumpStart™ Taq ReadyMix kit (Sigma). Three biological replicates with three technical replicates per each were used for each gene. The specific qPCR primers, were used to amplify reference and target genes. Sequences of primers used for RT-qPCR analysis are available in Table S4. Before use, primers were tested for equal efficiency of the qPCR reactions. The efficiency of the quantitative PCR reaction with each primer pair was calculated and used to calculate the ratio of each studied gene to the reference gene. Only efficiency values of about 0.95 or more were accepted. For each cDNA sample, three reactions were carried out using two template amounts of 20–60 ng, each in duplicate. The quality of results was evaluated based on expected Ct differences between the two cDNA amounts as well as product melting curves. Changes in individual gene expression between the WT and mutant strain were calculated with normalization of Ct values to mean Ct value for nadB (PA0761) reference housekeeping gene using the Pfaffl method [67], [68]. RT-qPCR analysis using P. aeruginosa housekeeping gene proC (PA0393) as an internal normalizer confirmed no changes in expression of nadB gene (ratio 1) in parA null, parB null versus WT strains of P. aeruginosa (data not shown).
qPCR was performed using the Light Cycler 480 (Roche). PCR products were detected with SYBR green fluorescent dye and amplified according to the following protocol: one cycle at 95°C for 5 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min. The melting curve was 65 to 95°C with increments of 0.5°C/s. Each PCR mixture contained the following: 5 µl SYBR Green JumpStart Taq ReadyMix for quantitative PCR (Sigma), 2 µl of diluted cDNA, and each of the forward and reverse primers at 0.4 µM; nuclease-free water was added to obtain a final volume of 10 µl. In each run, negative controls (no cDNA) for each primer set were included.
Regulatory Experiments with Promoter-lacZ Fusions and parA or parB Expressed in trans in E. coli
The putative promoter regions of chosen genes were PCR amplified on the genomic DNA of the PAO1161 as the template using appropriate pairs of primers listed in Table S4. Amplified regions after EcoRI-BamHI digestion were inserted into the broad-host-range pCM132 promoter probe vector after EcoRI-BglII cleavage upstream to the promoter-less lacZ reporter gene (see Table S5). The empty vector pCM132 with the deletion of EcoRI-BglII fragment (as a negative control) and its derivatives with inserted promoter regions were transformed into competent cells of E. coli DH5Δlac (pGBT30), DH5Δlac (pKLB1 tacp-parA) and DH5Δlac (pKLB2 tacp-parB) strains. Competent cells of E. coli were prepared by the standard CaCl2 method [69]. The β-galactosidase activity in liquid overnight cultures of transformants was analyzed as previously described [70]. Three independent assays for at least three independent transformants in one set of experiment were performed. The average of three independent experiments is presented.
Microarray Data Accession Number
The raw microarray data supporting the results of this article were deposited in the NCBI‘s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE47031 (release after publication acceptance).
Supporting Information
The P. aeruginosa ParAB regulon genes. The list of genes differentially expressed in parA null and parB null (common in parA null and parB null list) as compared with reference PAO1161 P. aeruginosa, indicated by pairwise comparison of microarray data (fold change FC ≥2; p-value ≤0.05). All assigned PseudoCAP function categories [29] for the identified genes are presented as abbreviations (see legend). RpoS, QS, PQS, RpoN(KinB), PprB, stress regulated genes are marked (Regulons column) with marked also genes involved in homeostasis maintenance (CORE in Regulons column), according to appropriate references [32], [50], [62], [40], [34], [45], [47]. The last column indicates genes from Venn diagram presented in Figure 3A with genes set marked by appropriate number in bracket.
(XLS)
Genes differentially expressed only in parA null mutant. The list of genes differentially expressed only in parA null mutant versus reference WT PAO1161 strain (fold change FC ≥2; p-value ≤0.05), not present on the list for parB null mutant microarray data. All assigned PseudoCAP function categories [29] for the identified genes are presented as abbreviations (see legend).
(XLS)
Genes differentially expressed only in parB null mutant. The list of genes differentially expressed only in parB null mutant versus reference WT PAO1161 strain (fold change FC ≥2; p-value ≤0.05), not present on the list for parA null mutant. All assigned PseudoCAP function categories [29] for the identified genes are presented as abbreviations (see legend).
(XLS)
Primers used in this work.
(DOCX)
Plasmids used in this work.
(DOCX)
Acknowledgments
We gratefully acknowledge the technical assistance with microarray analysis of Roksana Iwanicka-Nowicka from the Laboratory of Microarray Analysis, IBB PAS, Warsaw, Poland. We thank Iwona Czaban for the technical assistance and the construction of pCM132 derivatives.
Funding Statement
This work was supported by MNiSW/NCN grant N N303 457038(4570/B/P01/2010/38) awarded to AAB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gerdes K, Moller-Jensen J, Jensen RB (2000) Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol 37: 455–466. [DOI] [PubMed] [Google Scholar]
- 2. Hayes F, Barilla D (2006) The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol 4: 133–143 10.1038/nrmicro1342 [DOI] [PubMed] [Google Scholar]
- 3. Gerdes K, Howard M, Szardenings F (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141: 927–942 10.1016/j.cell.2010.05.033 [DOI] [PubMed] [Google Scholar]
- 4. Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, et al. (2010) A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12: 791–798 10.1038/ncb2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toro E, Hong S-H, McAdams HH, Shapiro L (2008) Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA 105: 15435–15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fogel MA, Waldor MK (2006) A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20: 3269–3282 10.1101/gad.1496506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mierzejewska J, Jagura-Burdzy G (2012) Prokaryotic ParA–ParB–parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67: 1–14 10.1016/j.plasmid.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 8. Ogasawara N, Yoshikawa H (1992) Genes and their organization in the replication origin region of the bacterial chromosome. Mol Microbiol 6: 629–634 10.1111/j.13652958.1992.tb01510.x [DOI] [PubMed] [Google Scholar]
- 9. Livny J, Yamaichi Y, Waldor MK (2007) Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189: 8693–8703 10.1128/JB.0123907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee PS, Grossman AD (2006) The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis . Mol Microbiol 60: 853–869 10.1111/j.13652958.2006.05140.x [DOI] [PubMed] [Google Scholar]
- 11. Mohl DA, Easter J, Gober JW (2001) The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus . Mol Microbiol 42: 741–755. [DOI] [PubMed] [Google Scholar]
- 12. Jakimowicz D, Chater K (2002) The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol Microbiol 45: 1365–1377. [DOI] [PubMed] [Google Scholar]
- 13. Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM (2002) Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148: 537–548. [DOI] [PubMed] [Google Scholar]
- 14. Venkova-Canova T, Baek JH, Fitzgerald PC, Blokesch M, Chattoraj DK (2013) Evidence for two different regulatory mechanisms linking replication and segregation of Vibrio cholerae chromosome II. PLoS Genet 9: e1003579 10.1371/journal.pgen.1003579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubarry N, Pasta F, Lane D (2006) ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J Bacteriol 188: 1489–1496 10.1128/JB.188.4.14891496.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lasocki K, Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G (2007) Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J Bacteriol 189: 5762–5772 10.1128/JB.0037107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwinska J, Chater KF (2007) Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae . Mol Microbiol 65: 625–641 10.1111/j.13652958.2007.05815.x [DOI] [PubMed] [Google Scholar]
- 18. Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G (2009) ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology 155: 1080–1092 10.1099/mic.0.0246610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G (2004) ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J Bacteriol 186: 6983–6998 10.1128/JB.186.20.69836998.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jensen RB, Dam M, Gerdes K (1994) Partitioning of plasmid R1. The parA operon is autoregulated by ParR and its transcription is highly stimulated by a downstream activating element. J Mol Biol 236: 1299–1309 doi 10.1016/0022-2836(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 21. Dunham TD, Xu W, Funnell BE, Schumacher MA (2009) Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J 28: 1792–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwong SM, Yeo CC, Poh CL (2001) Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS . Mol Microbiol 40: 621–633. [DOI] [PubMed] [Google Scholar]
- 23. Kusiak M, Gapczynska A, Plochocka D, Thomas CM, Jagura-Burdzy G (2011) Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa . J Bacteriol 193: 3342–3355 10.1128/JB.0032811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mierzejewska J, Bartosik AA, Macioszek M, Plochocka D, Thomas CM, et al. (2012) Identification of C-terminal hydrophobic residues important for dimerization and all known functions of ParB of Pseudomonas aeruginosa . Microbiology 158: 1183–1195 10.1099/mic.0.0562340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leonard TA, Butler PJ, Lowe J (2005) Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J 24: 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marston AL, Errington J (1999) Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol Cell 4: 673–682. [DOI] [PubMed] [Google Scholar]
- 27. Murray H, Errington J (2008) Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135: 74–84 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- 28. Vallet-Gely I, Boccard F (2013) Chromosomal organization and segregation in Pseudomonas aeruginosa . PLoS Genet 9: e1003492 10.1371/journal.pgen.1003492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, et al. (2010) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39: D596–D600 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsui H, Sano Y, Ishihara H, Shinomiya T (1993) Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175: 1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu W, Jin S (2005) PtrB of Pseudomonas aeruginosa suppresses the type III secretion system under the stress of DNA damage. J Bacteriol 187: 6058–6068 10.1128/JB.187.17.60586068.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schuster M, Hawkins AC, Harwood CS, Greenberg EP (2004) The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51: 973–985 10.1046/j.13652958.2003.03886.x [DOI] [PubMed] [Google Scholar]
- 33. Wang Y, Ha U, Zeng L, Jin S (2003) Regulation of membrane permeability by a two-component regulatory system in Pseudomonas aeruginosa . Antimicrob Agents Chemother 47: 95–101 10.1128/AAC.47.1.95101.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong Y-H, Zhang X-F, Soo H-ML, Greenberg EP, Zhang L-H (2005) The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa . Mol Microbiol 56: 1287–1301 10.1111/j.13652958.2005.04612.x [DOI] [PubMed] [Google Scholar]
- 35. Rampioni G, Bertani I, Zennaro E, Polticelli F, Venturi V, et al. (2005) The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter. J Bacteriol 188: 815–819 10.1128/JB.188.2.815819.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rampioni G, Polticelli F, Bertani I, Righetti K, Venturi V, et al. (2006) The Pseudomonas quorum-sensing regulator RsaL belongs to the tetrahelical superclass of H-T-H proteins. J Bacteriol 189: 1922–1930 10.1128/JB.0155206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, et al. (2007) RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa: RsaL and quorum sensing homeostasis. Mol Microbiol 66: 1557–1565 10.1111/j.13652958.2007.06029.x [DOI] [PubMed] [Google Scholar]
- 38. Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML (2002) Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 99: 7072–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML (2002) GeneChip® expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol 45: 1277–1287. [DOI] [PubMed] [Google Scholar]
- 40. Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, et al. (2011) Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol 194: 1317–1330 10.1128/JB.0610511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin S, Ishimoto K, Lory S (1994) Nucleotide sequence of the rpoN gene and characterization of two downstream open reading frames in Pseudomonas aeruginosa . J Bacteriol 176: 1316–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Romero-Steiner S, Parales RE, Harwood CS, Houghton JE (1994) Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol 176: 5771–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ogasawara H, Hasegawa A, Kanda E, Miki T, Yamamoto K, et al. (2007) Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J Bacteriol 189: 4791–4799 10.1128/JB.0031907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramos-Aires J, Plesiat P, Kocjancic-Curty L, Kohler T (2004) Selection of an antibiotic-hypersusceptible mutant of Pseudomonas aeruginosa: identification of the GlmR transcriptional regulator. Antimicrob Agents Chemother 48: 843–851 10.1128/AAC.48.3.843851.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE (2006) Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188: 7101–7110 10.1128/JB.0080706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG (2005) RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol Microbiol 57: 97–110 10.1111/j.13652958.2005.04677.x [DOI] [PubMed] [Google Scholar]
- 47. Balasubramanian D, Mathee K (2009) Comparative transcriptome analyses of Pseudomonas aeruginosa . Hum Genomics 3: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cowles KN, Gitai Z (2010) Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa . Mol Microbiol 76: 1411–1426 10.1111/j.13652958.2010.07132.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Typas A, Banzhaf M, Gross CA, Vollmer W (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10: 123–136 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schuster M, Lostroh CP, Ogi T, Greenberg EP (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185: 2066–2079 10.1128/JB.185.7.20662079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Venturi V (2006) Regulation of quorum sensing in Pseudomonas . FEMS Microbiol Rev 30: 274–291 10.1111/j.15746976.2005.00012.x [DOI] [PubMed] [Google Scholar]
- 52. Heurlier K, Denervaud V, Haas D (2006) Impact of quorum sensing on fitness of Pseudomonas aeruginosa . Int J Med Microbiol 296: 93–102 10.1016/j.ijmm.2006.01.043 [DOI] [PubMed] [Google Scholar]
- 53. Schreiber K, Krieger R, Benkert B, Eschbach M, Arai H, et al. (2007) The Anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189: 4310–4314 10.1128/JB.0024007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arai H, Hayashi M, Kuroi A, Ishii M, Igarashi Y (2005) Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa . J Bacteriol 187: 3960–3968 10.1128/JB.187.12.39603968.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, et al. (2002) Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3: 593–603. [DOI] [PubMed] [Google Scholar]
- 56. Attila C, Ueda A, Wood TK (2007) PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Appl Microbiol Biotechnol 78: 293–307 10.1007/s002530071308-y [DOI] [PubMed] [Google Scholar]
- 57. Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7: 263–273 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- 58. Ryan RP, Tolker-Nielsen T, Dow JM (2012) When the PilZ don’t work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol 20: 235–242 10.1016/j.tim.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 59. Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, et al. (2011) Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell 43: 550–560 10.1016/j.molcel.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duvel J, Bertinetti D, Moller S, Schwede F, Morr M, et al. (2012) A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa . J Microbiol Methods 88: 229–236 10.1016/j.mimet.2011.11.015 [DOI] [PubMed] [Google Scholar]
- 61. Marx CJ, Lidstrom ME (2001) Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147: 2065–2075. [DOI] [PubMed] [Google Scholar]
- 62. Haussler S, Becker T (2008) The Pseudomonas Quinolone Signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4: e1000166 10.1371/journal.ppat.1000166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vecchiarelli AG, Han Y-W, Tan X, Mizuuchi M, Ghirlando R, et al. (2010) ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition: ATP control of ParA in plasmid partition. Mol Microbiol 78: 78–91 10.1111/j.13652958.2010.07314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baraquet C, Murakami K, Parsek MR, Harwood CS (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40: 7207–7218 10.1093/nar/gks384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, et al. (2010) Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328: 1295–1297 10.1126/science.1188658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kahn M, Kolter R, Thomas C, Figurski D, Meyer R, et al. (1979) Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol 68: 268–280. [DOI] [PubMed] [Google Scholar]
- 67. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 doi 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 68. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29(9): 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 70.Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 71. Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29: 303–313 10.1016/03781119(84)900593 [DOI] [PubMed] [Google Scholar]
- 72. Jagura-Burdzy G, Ibbotson JP, Thomas CM (1991) The korF region of broad-host-range plasmid RK2 encodes two polypeptides with transcriptional repressor activity. J Bacteriol 173: 826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The P. aeruginosa ParAB regulon genes. The list of genes differentially expressed in parA null and parB null (common in parA null and parB null list) as compared with reference PAO1161 P. aeruginosa, indicated by pairwise comparison of microarray data (fold change FC ≥2; p-value ≤0.05). All assigned PseudoCAP function categories [29] for the identified genes are presented as abbreviations (see legend). RpoS, QS, PQS, RpoN(KinB), PprB, stress regulated genes are marked (Regulons column) with marked also genes involved in homeostasis maintenance (CORE in Regulons column), according to appropriate references [32], [50], [62], [40], [34], [45], [47]. The last column indicates genes from Venn diagram presented in Figure 3A with genes set marked by appropriate number in bracket.
(XLS)
Genes differentially expressed only in parA null mutant. The list of genes differentially expressed only in parA null mutant versus reference WT PAO1161 strain (fold change FC ≥2; p-value ≤0.05), not present on the list for parB null mutant microarray data. All assigned PseudoCAP function categories [29] for the identified genes are presented as abbreviations (see legend).
(XLS)
Genes differentially expressed only in parB null mutant. The list of genes differentially expressed only in parB null mutant versus reference WT PAO1161 strain (fold change FC ≥2; p-value ≤0.05), not present on the list for parA null mutant. All assigned PseudoCAP function categories [29] for the identified genes are presented as abbreviations (see legend).
(XLS)
Primers used in this work.
(DOCX)
Plasmids used in this work.
(DOCX)