Abstract
The diurnal rhythm controls many aspects of plant physiology such as flowering, photosynthesis and growth. Rice is one of the staple foods for world's population. Abiotic stresses such as salinity, drought, heat and cold severely affect rice production. Under salinity stress, maintenance of ion homeostasis is a major challenge, which also defines the tolerance level of a given genotype. Salt overly sensitive (SOS) pathway is well documented to play a key role in maintaining the Na+ homeostasis in plant cell. However, it is not reported yet whether the transcriptional regulation of genes of this pathway are influenced by diurnal rhythm. In the present work, we have studied the diurnal pattern of transcript abundance of SOS pathway genes in rice at seedling stage.To rule out the effect of temperature fluctuations on the expression patterns of these genes, the seedlings were grown under constant temperature. We found that OsSOS3 and OsSOS2 exhibited a rhythmic and diurnal expression pattern, while OsSOS1did not have any specific pattern of expression. This analysis establishes a cross-link between diurnal rhythm and SOS pathway and suggests that SOS pathway is influenced by diurnal rhythm in rice.
Keywords: diurnal rhythm, Oryza sativa, salinity, SOS, salt tolerance
Introduction
Soil salinity is one of the most severe abiotic stresses for crops worldwide, affecting several million hectares of agricultural land.1 Therefore, characterization of Na+ transport and its distribution in plants has been a prime area of research for decades.High concentration of sodium ion (Na+) is toxic to plants primarily due to its adverse effects on cellular metabolism and ion homeostasis.2,3 Therefore, maintenance of low level of Na+ in cells is essential for plants.3,4Plants remove Na+ from the cytoplasm by using vacuolar and plasma membrane localized Na+/H+ transporters.4,5 Na+/H+ transporters are membranous proteins that transport protons (H+) across the membrane in exchange for Na+.6,7 This exchange activity requires H+ electrochemical gradient across the membrane which is generated by the H+ pumps such as H+-ATPase present on plasma membrane or vacuole and H+-pyrophosphatase.5 In plants, the exchange activity of the plant vacuolar Na+/H+ transporters has been well studied.5,8,9 In addition, enhanced salinity tolerance has been reported by overexpression of a vacuolar Na+/H+ antiporter in Arabidopsis.10
In plants, three salt overly sensitive genes (SOS1, SOS2 and SOS3) have been found to function in a common pathway that contributes to salt tolerance.11-14SOS1 gene encodes a membrane protein containing 12 putative trans-membrane domains and a long hydrophilic tail at the C-terminal end.15 SOS1 transports sodium ions across the plasma membrane. Expression of SOS1 gene is induced significantly in roots and to a much lesser extent in shoots in seedlings by exposure to high levels of NaCl.15 SOS2 is a Ser/Thr protein kinase, which contains an auto-inhibitory domain.16 SOS3 is a Ca2+ binding protein with strong similarity with the regulatory β subunit of the protein phosphatase calcineurin and with related proteins of the neuronal Ca2+ sensor family.13 Hence, it has been hypothesized that SOS3 perceives the Ca2+ transients elicited by salt stress and activates SOS2 by relieving auto-inhibition.16 These SOS pathway genes are later grouped into large protein families of calcineurin B-like proteins (CBL) and CBL-interacting protein kinases (CIPK), therefore SOS2 and SOS3 are also known as CIPK24 and CBL4, respectively.17 Furthermore, the orthologous- OsSOS1, OsSOS2 (OsCIPK24) and OsSOS3 (OsCBL4) have also been isolated from rice and it has been demonstrated that all OsSOS proteins could coordinately function with AtSOS proteins and nicely complemented mutations in the corresponding sos mutant of Arabidopsis plants.18 Recent work from our lab has also reported various SOS orthologs from Brassica species involved in maintenance of Na+ homeostasis in Brassica.19 Together, these results suggested that the SOS-like salinity tolerance mechanism is conserved in plants.
Plant diurnal oscillation is a 24 h period cycle. Genes of multiple pathways have been implicated in stress responses and regulated by diurnal rhythm.20-24 Time course transcriptome analysis has revealed that genes involved in several biological pathways like lipid and carbohydrate metabolic processes, photosynthesis, nucleotide binding, translation, amino acid and nucleotide metabolism, nitrogen metabolism and hormone biosynthesis, etc., show rhythmicity.25-28
Rice, a salt sensitive crop, is the staple food for about half of the world’s population. Hence, in order to improve salinity tolerance in this crop plant, it is imperative to develop a “thorough understanding” of the complex molecular mechanisms and gene regulatory networks operative under stress conditions.22 Recently, proteomics approach has revealed that a set of 91 proteins, involved in diverse processes including stress response, is controlled by diurnal cycles in developing endosperm of rice.28 However, there is still no report about the diurnal transcriptional regulation of salt stress related genes like SOS1, SOS2 and SOS3. Rice has been reported to be most sensitive to salinity stress at seedling stage.29,30 The aim of the present study was to see if the expression of SOS pathway members is influenced by the diurnal rhythm in seedlings of rice.
Results
OsSOS3 and OsSOS2 show diurnal rhythmicity
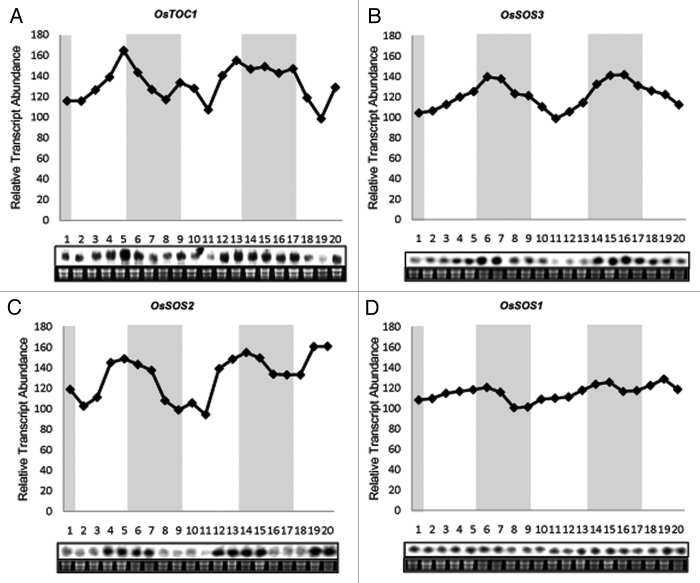
The diurnal expression profiles of OsSOS genes were observed under entraining conditions of 12 h light/12 h dark (LD) with a constant temperature 28 ± 2°C during day and night in IR-64 genotype of rice at seedling stage. OsTOC1 has been reported to be expressed rhythmically during 12 h light and 12 h dark cycle in rice.31 Level of mRNA of OsTOC1 oscillates with light-dark cycles and shows peak in the late day (Fig. 1A). Changes in OsTOC1 levels coincide with dusk transition as reported earlier.31 OsTOC1 expresses with a period of about 24 h and an evening specific phase. This result is consistent with the prior published reports, which in turn validates the experimental conditions used by us.
Figure 1. RNA gel blot analyses showing diurnal rhythm of OsSOS genes in shoots of IR64 rice seedlings. Seedlings were grown under the 12 h light/12 h dark cycle for 14 d. Shoot samples were harvested at 3 h intervals for 2.5 d under the 12 h light/12 h dark. Twenty µg RNA samples was used for northern blot hybridization. Ethidium bromide (EtBr) stained RNA-gel has been shown as the loading control. Y-axis shows relative transcript abundance of genes while numbers on X-axis shows different time points. Shaded area indicates dark period and white area indicates light period. (A) OsTOC1 shows rhythmic expression and has a peak during light to dark transition. (B) OsSOS3 expression peaks during night. (C) mRNA level of OsSOS2 show rhythmicity in 24 h cycles and show higher expression during the transition period of light to dark. (D) Transcript level of OsSOS1 doesn't show any clear rhythmicity.
The nucleotide sequences of the cDNA clones used for probe preparation were taken as query in BLASTN search in Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/) which revealed that regions of clones used for probe preparation were specific to the corresponding genes, thus giving a clear signal on northern blots. In case of OsSOS genes, we found that OsSOS2 and OsSOS3were expressed in diurnal manner. Diurnal rhythms of these two genes have a characteristic waveform, described by peaks and troughs. OsSOS3, with a phase of 24 h show higher expression level during night. Its expression shoots up in night and goes down during day, creating rhythmicity. (Fig. 1B) Similarly, the transcript of OsSOS2 also shows a clear oscillation profile. MRNA levels of OsSOS2 cycles in light-dark periods, showing higher expression around the transition period of light to dark with a period of 24 h (Fig. 1C). The amplitude of OsSOS2 diurnal expression was higher in comparison to that of OsSOS3. mRNA level of OsSOS1 transcript fluctuates with time but do not show a clear rhythmicity. Its expression level drops before dawn (Fig. 1D).
Discussion
The biological clock is an endogenous timing mechanism present in almost all organisms examined to date. This time-keeping mechanism is able to generate 24 h rhythms in a wide variety of biological processes. Regulation of cellular ion homeostasis during stresses is critical for plant survival. One of the responses of plant cell to stresses is the generation of a cytosolic Ca2+ transient and the subsequent activation of Ca2+ sensor protein expression and/or activity.32 The components (SOS1, SOS2 and SOS3) of SOS pathway transduce a salt stress induced Ca2+ signal to reinstate cellular ion homeostasis.33 Although, these three members (proteins) coordinate in sequential manner but do not show diurnal co-expression as OsSOS3 and OsSOS2 have expression peaks at different times of 24 h cycle; during night and dusk transition respectively, while OsSOS1does not have a clear rhythmicity. It has been reported that changes in mRNA levels may not be correlated with changes in protein or enzyme activity levels.34 It has also been suggested that the clock-regulated proteins in rice are modulated at not only transcriptional but also at post-transcriptional and/or post-translational levels.26 Whether SOS2 and SOS3 show diurnal rhythm at protein level and whether specific activities of these proteins also oscillate during the 24 h day-night cycle, these are the important questions to be answered. Nevertheless, expression profiles do provide a useful starting point for more in depth analyses. For instance, in the present study, SOS2 and SOS3 represent good candidates to study the relationship between salinity stress and diurnal rhythm. Plants are always changing and adapting to their changing environments. Thus, experimental changes are always being observed in a background of uncertain variation. Although clocks are temperature compensated, which means that overall clock period is constant over a range of temperatures and temperature changes can act to entrain or reset the clock patterns.
It has also been shown in Brassica juncea that BjSOS3 mRNA is downregulated in presence of calcium chelator EGTA.35 It indicates that Ca2+ has a dual role as signaling agent, controlling the expression of SOS pathway genes at transcript level and initiating signaling at protein level. Diurnal oscillations of cytosolic and chloroplastic Ca2+ have been reported in A. thaliana and Nicotiana plumbaginifolia.36 The relationship between this diurnally regulated Ca2+ and SOS genes is still unexplored. The purpose of the diurnal oscillations of Ca2+ is not known but it has been reportedly involved in regulating numerous signaling events.37 Calcium is suggested to be a part of the light signal transduction chain regulating the rhythm as well as gene expression.37 These reports are in support to our study, as we are also reporting a diurnal expression pattern of genes of SOS pathway which is related to Ca2+ signaling.
As, the expression of SOS pathway genes and Ca2+ ion level show rhythmic oscillation, it would be interesting to study whether the diurnal changes in Ca2+ signatures mediate cross-talk between diurnal rhythm of SOS pathway genes and salinity stress tolerance. In Arabidopsis, elimination of SOS1 leads to the changes in expression of genes related to circadian rhythm.38 It suggests that though SOS1 does not show any diurnal rhythm but it regulates diurnal rhythm of several other genes. It also indicates toward unknown and unexplored connection between SOS pathway and circadian clock.
It is also interesting to find out whether the activities of downstream components of SOS2-SOS3 complex oscillates and coincides with diurnally regulated SOS2 and SOS3 transcripts. One of the downstream components, regulated by this complex is CCR1 (cold-circadian rhythm-RNA binding1) which encodes a Gly-rich RNA-binding protein. CCR1 has similar expression profiles in sos1, sos2 and sos3 mutants implicating this as transcriptional output requiring all components of the SOS pathway.39 Expression of CCR1 as well as its homolog CCR2 is regulated by diurnal rhythm.40
Not only downstream genes of SOS pathway, but also the interacting partners of SOS pathway show the diurnal rhythm. It has been reported that SOS2 interacts with catalase 2 (CAT2) and catalase 3 (CAT3) connecting SOS2 to H2O2 metabolism and signaling. Expression of CAT2 is under circadian control, with the highest expression during the light period, consistent with a primary role in detoxifying H2O2 derived from photosynthesis or photorespiration.41 Interestingly, CAT3 is also circadian regulated, but in the opposite manner as CAT2. CAT3 expression is highest in the dark period.41 Circadian oscillation of interacting members and downstream components of SOS pathway revalidate our results.
Our data demonstrates that OsSOS3, OsSOS2 genes show rhythmic expression profile under diurnal condition at the transcription level. This study also strongly establishes a possible molecular link of diurnal rhythm with SOS pathway.
Materials and Methods
Plant materials and growth conditions
Seeds of Oryza sativa L. cv “IR-64” were germinated in half Yoshida medium under hydroponic system for 48h in dark and then grown for 14 d under control conditions (28 ± 2 °C, 12 h light and 12 h dark cycle) in plant growth room.42 Shoot samples were harvested for 2.5 d at an interval of 3 h starting from the dawn of 15 d.
RNA extraction and northern blot analysis
Total RNA was extracted from tissue using TRIzol method as per the manufacturer’s instructions (T9424; Sigma-Aldrich). Northern blots were prepared using 20 μg total RNA. Appropriate probes [partial for OsSOS1(AK065608) corresponding to its N-terminal part and full length for other genes; OsSOS2 (AK102270); OsSOS3 (AK101368); OsTOC1 (AK111828)] were amplified using the primers listed in Table 1 and radiolabeled using the DecaLabel™ DNA labeling kit, (K0621; Fermentas). To avoid any error due to unequal loading of RNA samples, same blot was re-probed each time with different gene probes after washing. Northern blots were hybridized at 65 °C in 5 × SSC, 5 × Denhardt’s reagent, 0.1% SDS and 100 μg/ml denatured salmon sperm DNA for 16–18 h. Membrane was washed twice in 0.5 × SSC, 0.1% SDS and 0.1 × SSC, 0.1% SDS for 15 min each at 65 °C and scanned on phosphorimager using the software Fujifilm Image Reader. The relative transcript abundance was calculated using the Image Gauge (Fuji Photofilm Co. Ltd.).
Table 1. List of primers used in the present study.
| OsTOC1 forward primer | 5′-CGGAATTCATGGTGGGCGCCGGCGAG-3′ |
| OsTOC1 reverse primer | 5′-ACGCGTCGACCTACTCTGGAGAAGAAACCATC-3′ |
| OsSOS1 forward primer | 5′-ATGTGACTGGAAGGGTTTGC-3′ |
| OsSOS1 reverse primer | 5′-TCTAGCCTCCTCTCCCTCAG-3′ |
| OsSOS2 forward primer | 5′-ATGGGAGGGGAGGAGGGAATG-3′ |
| OsSOS2 reverse primer | 5′-CTAGCATGTGGCTGTCCTCAG-3′ |
| OsSOS3 reverse primer | 5′-TCA GTC ATG GGC TTC TGA ATG C-3′ |
Acknowledgments
A.P. acknowledges financial support received from the Department of Biotechnology, Government of India and internal grants of JNU. P.S. and N.S. would like to acknowledge the receipt of Senior Research Fellowship award by CSIR and UGC (India) respectively.
Glossary
Abbreviations:
- d
days
- LD
light /dark cycles
- h
hours
- min
minutes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24738
References
- 1.Rengasamy P. Soil processes affecting crop production in salt-affected soils. Funct Plant Biol. 2010;37:613–20. doi: 10.1071/FP09249. [DOI] [Google Scholar]
- 2.Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109:735–42. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacoby B. Mechanisms involved in salt tolerance of plants. In: Pessarakli M, ed. Handbook of plant and crop stress, 2. New York: Marcel Dekker,1999: 97-123. [Google Scholar]
- 4.Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–99. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 5.Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–51. doi: 10.1016/S0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 6.Padan E, Venturi M, Gerchman Y, Dover N. Na(+)/H(+) antiporters. Biochim Biophys Acta. 2001;1505:144–57. doi: 10.1016/S0005-2728(00)00284-X. [DOI] [PubMed] [Google Scholar]
- 7.Wiebe CA, Dibattista ER, Fliegel L. Functional role of polar amino acid residues in Na+/H+ exchangers. Biochem J. 2001;357:1–10. doi: 10.1042/0264-6021:3570001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA. 1999;96:1480–5. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darley CP, van Wuytswinkel OCM, van der Woude K, Mager WH, de Boer AH. Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem J. 2000;351:241–9. doi: 10.1042/0264-6021:3510241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–8. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 11.Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–27. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Zhu JK. An Arabidopsis mutant with increased calcium requirement for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA. 1997;94:14960–4. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu JK. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000;124:941–8. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–91. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007;143:1001–12. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar G, Purty RS, Sharma MP, Singla-Pareek SL, Pareek A. Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J Plant Physiol. 2009;166:507–20. doi: 10.1016/j.jplph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Itoh H, Izawa T. A study of phytohormone biosynthetic gene expression using a circadian clock-related mutant in rice. Plant Signal Behav. 2011;6:1932–6. doi: 10.4161/psb.6.12.18207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 2011;157:1128–37. doi: 10.1104/pp.111.181792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nongpiur R, Soni P, Karan R, Singla-Pareek SL, Pareek A. Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal Behav. 2012;7:1230–7. doi: 10.4161/psb.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–3. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 24.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–41. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, et al. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE. 2011;6:e16907. doi: 10.1371/journal.pone.0016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang H, Cho MH, Hahn BS, Lim H, Kwon YK, Hahn TR, et al. Proteomic identification of rhythmic proteins in rice seedlings. Biochim Biophys Acta. 2011;1814:470–9. doi: 10.1016/j.bbapap.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Yang R, Li M, Xing Z, Yang W, Chen G, et al. Transcriptome phase distribution analysis reveals diurnal regulated biological processes and key pathways in rice flag leaves and seedling leaves. PLoS ONE. 2011;6:e17613. doi: 10.1371/journal.pone.0017613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu HT, Xu SB, Zheng CH, Wang T. Comparative proteomic study reveals the involvement of diurnal cycle in cell division, enlargement, and starch accumulation in developing endosperm of Oryza sativa. J Proteome Res. 2012;11:359–71. doi: 10.1021/pr200779p. [DOI] [PubMed] [Google Scholar]
- 29.Zeng L. Exploration of relationships between physiological parameters and growthperformance of rice (Oryza sativa L.) seedlings under salinity stress using multivariate analysis. Plant Soil. 2005;268:51–9. doi: 10.1007/s11104-004-0180-0. [DOI] [Google Scholar]
- 30.Zeng L, Shannon MC. Salinity effects on seedling growth and yield components of rice. Crop Sci. 2000;40:996–1003. doi: 10.2135/cropsci2000.404996x. [DOI] [Google Scholar]
- 31.Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44:1229–36. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- 32.Knight H, Trewavas AJ, Knight MR. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997;12:1067–78. doi: 10.1046/j.1365-313X.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–73. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kushwaha HR, Kumar G, Verma PK, Singla-Pareek SL, Pareek A. Analysis of a salinity induced BjSOS3 protein from Brassica indicate it to be structurally and functionally related to its ortholog from Arabidopsis. Plant Physiol Biochem. 2011;49:996–1004. doi: 10.1016/j.plaphy.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Johnson CH, Knight MR, Kondo T, Masson P, Sedbrook J, Haley A, et al. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–5. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- 37.Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- 38.Oh DH, Ali Z, Hyeong CP, Bressan RA, Dae JY, Bohnert HJ. Consequences of SOS1 deficiency: intracellular physiology and transcription. Plant Signal Behav. 2010;5:766–8. doi: 10.4161/psb.5.6.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–75. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–25. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong HH, McClung CR. The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol Gen Genet. 1996;251:196–203. doi: 10.1007/BF02172918. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory manual for physiological studies of rice, 3. Manila: International Rice Research Institute, 1972:1–83. [Google Scholar]