Abstract
Upland cotton (Gossypium hirsutum) is one of the most recalcitrant species for in vitro plant regeneration through somatic embryogenesis. Callus from only a few cultivars can produce embryogenic callus (EC), but the mechanism is not well elucidated. Here we screened a cultivar, CRI24, with high efficiency of EC produce. The expression of genes relevant to EC production was analyzed between the materials easy to or difficult to produce EC. Quantitative PCR showed that CRI24, which had a 100% EC differentiation rate, had the highest expression of the genes GhLEC1, GhLEC2, and GhFUS3. Three other cultivars, CRI12, CRI41, and Lu28 that formed few ECs expressed these genes only at low levels. Each of the genes involved in auxin transport (GhPIN7) and signaling (GhSHY2) was most highly expressed in CRI24, with low levels in the other three cultivars. WUSCHEL (WUS) is a homeodomain transcription factor that promotes the vegetative-to-embryogenic transition. We thus obtained the calli that ectopically expressed Arabidopsis thaliana Wus (AtWus) in G. hirsutum cultivar CRI12, with a consequent increase of 47.75% in EC differentiation rate compared with 0.61% for the control. Ectopic expression of AtWus in CRI12 resulted in upregulation of GhPIN7, GhSHY2, GhLEC1, GhLEC2, and GhFUS3. AtWus may therefore increase the differentiation potential of cotton callus by triggering the auxin transport and signaling pathways.
Introduction
Somatic embryogenesis (SE) is a principal model for studying the growth and development of zygotic embryos in higher plants. This process includes callus induction, embryogenic callus (EC) formation, embryo development, and plant regeneration. During the past three decades, much effort has attempted to determine important genes controlling SE [1], [2].
The gene WUSCHEL (Wus) is essential for stem cell formation and maintenance in shoot and root apical meristems [3], [4]. Wus mediates stem cell homeostasis by regulating cell division and differentiation [5]–[7]. In wus mutants, apical meristems are unable to preserve the pool of undifferentiated cells [3]. The maintenance function of WUS can be repressed by inducing AGAMOUS (AG) expression and floral meristem differentiation [8]. Wus was first reported as the key gene promoting SE in pga6 mutants in Arabidopsis, with overexpression of PGA6/Wus causing the vegetative-to-embryonic transition [9]. Wus is crucial for EC renewal during SE in Arabidopsis [10]. Overexpression of Wus in Coffea canephora can also promote SE [11].
Auxin is necessary for SE [12], [13], but the auxin transport and signaling pathways during SE are not well understood. PIN-FORMED (PIN) genes encoding efflux carrier proteins are involved in auxin transport [14], [15]. Genetic analysis indicates that PIN1 is a major regulatory factor for auxin gradients in EC and embryo [16]. Auxin regulates auxin-responsive genes via the Aux/IAA (SHY)-ARF module. At sufficiently low auxin concentration, auxin response factors (ARFs) are repressed by Aux/IAA. At sufficiently high auxin concentration, Aux/IAA is degraded by SCFTIR1, and ARFs are activated [17]–[19]. ARFs positively or negatively control downstream genes, resulting in responses to auxin signaling. Transcript profiling reveals that the auxin signaling pathway may play a vital role during SE in cotton [13].
LEC1, LEC2, and FUS3 are key genes that control SE progression [20], [21]. The capacity for SE is completely repressed in double (lec1 lec2, lec1 fus3, lec2 fus3) or triple (lec1 lec2 fus3) mutants of Arabidopsis thaliana [21]. LEC2 expression changes rapidly during auxin responses [22], suggesting that LEC/FUS may be downstream genes in the auxin signaling pathway [21], [23].
The majority of cotton cultivars are incapable of undergoing SE [24] because of their difficulty in inducing callus differentiation to form EC. Thus, most cultivars are not used for molecular breeding using transgenic technologies with Agrobacterium-mediated transformation via SE. Therefore, it is essential to study the mechanism of SE in cotton so as to improve regeneration of various cotton cultivars.
Here, we report the ectopic expression of A. thaliana Wus (AtWus) in G. hirsutum cv. CRI12, a cultivar that shows poor SE ability under established tissue culture methods. AtWus promoted differentiation of transgenic callus. Furthermore, ectopic expression of AtWus could upregulate GhLEC1 (G. hirsutum LEC1), GhLEC2, and GhFUS3 expression during SE and alter auxin transport and signaling mechanisms. AtWus therefore promotes the efficiency of EC differentiation in cotton callus.
Materials and Methods
Plant Materials and Tissue Culture Conditions
We selected four cotton cultivars, CRI24, CRI12, CRI41 and Lu28, as experiment materials. CRI24 has a 100% EC differentiation rate and is the main transgenic material used for Agrobacterium-mediated method in our lab. CRI12 used to be an important basic breeding material because of traits of its relatively high yield and disease resistance, yet it has a low rate of differentiation during SE. CRI41 and Lu28, the main cultivars planted in China, can not undergo SE because of failure in EC induction.
Seeds of the four cotton cultivars were sterilized with 0.1% (w/v) mercuric chloride for 3 min. The seeds were then washed five times with sterilized distilled water and then germinated on modified Murashige and Skoog (MS) medium (25 g l–1 sucrose, 50 ml l–1 MSI, 5.6 g l–1 agar) for hypocotyl induction. Sterilized seeds were cultured at 28°C with a 14 h/10 h light/dark photoperiod. The hypocotyls from 7-day-old sterile seedlings were cut into 2 cm segments. For transgenic experiments using an Agrobacterium-mediated method [25], the hypocotyl cuts of CRI12 were transferred to 250 ml flasks and placed on callus-induction medium (CIM; MS medium plus B5 vitamins, supplemented with 0.05 mg l–1 3-indole acetic acid (IAA), 0.05 mg l–1 kinetin, 0.05 mg l–1 2,4-dichlorophexoxyacetic acid, 25 g l–1 glucose, 2 g l–1 gelrite gellan gum, 50 mg l–1 kanamycin, 100 mg l–1 cefotaxime, pH 5.8). The medium was changed once per month. After 2 months of culture, all calli were transferred onto EC induction medium (EIM; MSB supplemented with 25 g l–1 glucose, 2 g l–1 gelrite, 0.5 g l–1 MgCl2, 0.16 mg l–1 kinetin, 0.08 mg l–1 IAA, 50 mg l–1 kanamycin, 100 mg l–1 cefotaxime, pH 6.5). The medium was changed monthly. After 4 months of culture, ECs were transferred to somatic embryo induction medium (SIM; MSB supplemented with 25 g l–1 glucose, 2 g l–1 gelrite, 0.5 g l–1 MgCl2, 0.08 g l–1 kinetin, 0.12 mg l–1 6-benzylaminopurine, 50 mg l–1 kanamycin, 100 mg l–1 cefotaxime, pH 6.8), and the medium was refreshed monthly. For non-transgenic experiments, the hypocotyl cut explants of CRI24, CRI12, CRI41, and Lu28 were transferred onto NCIM (CIM lacking kanamycin and cefotaxime), and the medium was changed once per month. After 30 days of culture, all calli were transferred onto NEIM (EIM lacking kanamycin and cefotaxime), and the medium was changed monthly. After 90 days of culture, all calli of CRI24 differentiated into EC, most calli of CRI12 and all calli of CRI41 and Lu28 did not differentiate into ECs. To confirm the lack of capacity for EC differentiation among the latter three cultivars, the calli which can not differentiate in ECs were cultured in NEIM for another 120 days, with the medium being refreshed monthly. Indeed, those calli did not differentiate into ECs, and thus no further experiments were conducted with these non-transgenic cultivars.
Gene Cloning and Vector Construction
The nucleotide sequences of GhLEC1, GhLEC2, and GhFUS3 were obtained from the D subgenome database of Gossypium. raimondii by comparing with amino acid sequences of AtLEC1, AtLEC2, and AtFUS3 using the tblastn tool. The three genes were then amplified from a full-length cDNA library of CRI24 with specific primers (Table S1). For ectopic expression of AtWus, the full-length coding regions (CDS) of the gene was cloned from wild-type Arabidopsis (Columbia ecotype) (Table S1). The full-length CDS of AtWus was amplified via PCR with specific primers (Table S1) and ligated into vector pMD18-T. After verifying the sequence, each of the AtWus fragment and Vector pBI121 was digested with BamH I and Sac I. and the AtWus fragment was inserted into pBI121. The nucleotide sequences of GhPIN7, GhSHY2, and GhARF3 were obtained from the D subgenome database of G. raimondii by comparing with amino acid sequences of AtPIN1, AtSHY2, and AtARF1 using the tblastn tool.
RNA Extraction
All calli of CRI24, CRI12, CRI41 and Lu28 cultured for 90 days in NEIM and of 35S:WUS and CK lines cultured for 4 months in EIM were stored at −80°C. We extracted RNA of the above samples using a modified CTAB method [26]. RNA samples with A260/A280 ratios between 1.8 and 2.0 and A260/A230 ratios >1.5 were considered acceptable.
Quantitative Real Time PCR (QPCR)
Approximately 1 µg total RNA samples were reverse transcribed using the PrimeScript RT reagent kit with gDNA Eraser (Takara). The cDNA templates were diluted three times prior to amplification. The QPCR experiment was conducted according to the guidelines of SYBR Premix Ex Taq™ kit (Takara). QPCR was performed in 96-well plates with a total volume of 20 µL containing 10 µL 2× SYBR Premix Ex Taq™, 6.8 µL PCR-grade water, 2 µL cDNA template, 0.4 µL 50× ROX reference dye I, and 0.4 µL each of forward and reverse primers (10 µM). All QPCRs were run with three technical replicates on an ABI 7900 Real-Time PCR system (Applied Biosystems). The thermal cycling conditions were as follows: an initial denaturation step of 30 s at 95°C, followed by 40 cycles of 95°C annealing for 5 s and 60°C extension for 30 s. The primers used for QPCR are shown in Table S2.
Scanning Electron Microscopy
Scanning electron microscopy was performed on somatic embryos obtained after 1.5 months culture on SIM. Samples were prefixed at room temperature for 12 h in 2.5% (v/v) glutaraldehyde (phosphate buffer, pH 7.2). After dehydration using a graded ethanol series, samples were dried with a CO2 critical-point drying system (HITACHI HCP-2). Subsequently, samples were sputtered with gold dust and observed under a HITACHI S-530 scanning electron microscopy.
Statistics
The rate of EC = number of EC/number of calli. After 45 days of culture in SIM, we determined the weight of the abnormal embryos in 35S:WUS lines and normal cotyledonary embryos in CK lines with three technical replicates. The average weight of each individual somatic embryo = weight of 10 somatic embryos/10. We conducted t-tests to determine significant differences (p<0.05 or p<0.01, depending on the experiment).
Results
Expression of GhLEC and GhFUS3 in Cultivars with Diverse Differentiation Rates
We used four cotton cultivars (G. hirsutum) for tissue culture. The EC differentiation rate of CRI24 was 100% but only 0.59% for CRI12, whereas the calli of Lu28 and CRI41 could not form EC (Table 1). After culturing hypocotyl segments for 30 days on NCIM (callus induction medium lacking kanamycin and cefotaxime), the induced calli were transferred onto NEIM (EC induction medium lackling kanamycin and cefotaxime). After 90 days in NEIM, all the CRI24 calli were non-compacted and had differentiated into EC (Figure 1A ). However, most calli of CRI12 and all calli of CRI41 and Lu28 were compacted, dark green, and did not differentiate (Figure 1B–D ).
Table 1. EC induction in four cotton cultivars.
| CRI12 (WT) | CRI24 (WT) | CRI41 (WT) | Lu28 (WT) | |||||||||
| Replication | Ca | ECb | EC rate (%) | Ca | ECb | EC rate (%) | Ca | ECb | EC rate (%) | Ca | ECb | EC rate (%) |
| 1 | 175 | 1 | 0.57 | 371 | 371 | 100 | 177 | 0 | 0.00 | 209 | 0 | 0.00 |
| 2 | 221 | 3 | 1.36 | 204 | 204 | 100 | 303 | 0 | 0.00 | 242 | 0 | 0.00 |
| 3 | 286 | 0 | 0.00 | 261 | 261 | 100 | 319 | 0 | 0.00 | 138 | 0 | 0.00 |
| Average | 682 | 4 | 0.59 | 836 | 836 | 100 | 799 | 0 | 0.00 | 589 | 0 | 0.00 |
Number of explants forming a callus.
Number of EC forming from a callus. WT, wild type.
Figure 1. Cotton cultivars have different differentiation rates in EC.
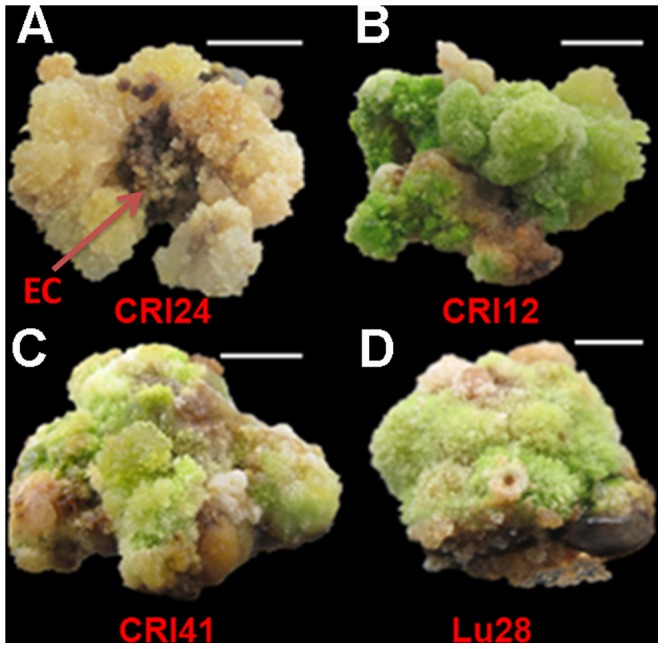
After 90 days of culture in NEIM, all calli of CRI24 produced EC, whereas calli of CRI12, CRI41, and Lu28 were dark green and tight and unable to differentiate into EC. Bar, 1
LEC1, LEC2, and FUS3 are essential for SE [21], but there are few studies on the roles of LEC and FUS3 in SE in plants other than Arabidopsis. Hence, we first isolated the homologs GhLEC1, GhLEC2, GhFUS3 in CRI24 [27]. At the amino acid level, these homologs share high sequence similarity with those of in Arabidopsis (Figure S1). The results implied that GhLEC1, GhLEC2, and GhFUS3 may have functions similar to those of the Arabidopsis homologs that control the capacity for SE [21].
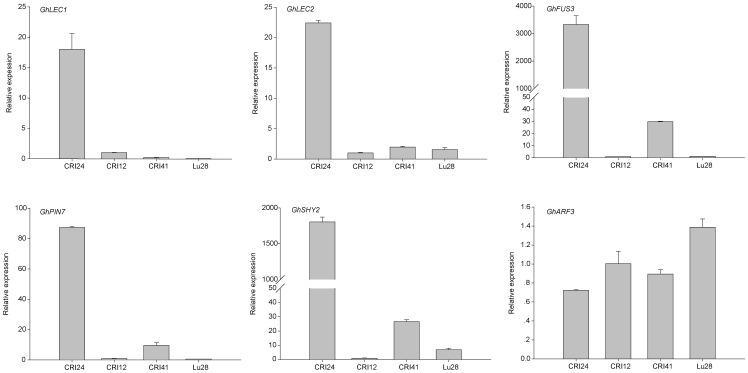
To investigate the expression of GhLEC1, GhLEC2, and GhFUS3 in calli of the four cultivars, we carried out QPCR on calli cultured for 90 days on NEIM. The expression levels of GhLEC1, GhLEC2, and GhFUS3 were significantly higher in the calli of CRI24 than in CRI12, Lu28, and CRI41, with barely detectable expression of GhFUS3 in CRI12 and Lu28 (Figure 2).
Figure 2. Analysis of gene expression in the non-transgenic calli of the four cultivars.
Expression of Genes Involved in Auxin Transport or Signaling Pathways
Auxin plays an important role in SE of cotton [13], [28], but genes involved in auxin transport (PINs) and auxin signaling are not well studied in cotton SE. QPCR analysis of expression levels of these genes in SE used the same samples as for expression patterns of LECs and FUS3. GhPIN7 and GhSHY2 (AUX/IAA2) levels in calli of CRI24 were much higher than in the other three cultivars (Figure 2). In contrast, GhARF3 expression was lower in CRI24 than in other cultivars (Figure 2), suggesting that GhARF3 expression may be inhibited by the GhSHY2 gene product. These results indicated that the auxin transport and signaling pathways were more active in calli with high EC differentiation rates than those with low EC differentiation rates.
Ectopic Expression of AtWus Improves EC Induction
CRI12 is one of the most recalcitrant cotton cultivars for plant regeneration via SE. To study the function of AtWus in SE, we overexpressed AtWus in CRI12 (35S:WUS). An empty vector was used as the control (CK) for parallel transformation. Hypocotyl segments transformed with 35S:WUS or the control CK were cultured on CIM containing kanamycin and cefotaxime to induce resistant calli. One month later, these segments were transferred to fresh CIM. There were no apparent differences between 35S:WUS and CK within the first 2 months. Then, calli that formed on segments were transferred to EIM containing kanamycin and cefotaxime for inducing EC. Calli were transferred to fresh EIM monthly. After culture for 1.5 months on EIM, most 35S:WUS calli were non-compacted and light green, and some began to differentiate into EC (Figure 3A, B ). However, the CK calli were compact and dark green and did not undergo EC differentiation (Figure 3C, D ). After culturing for 4 months on EIM, the differentiation rate of 35S:WUS transformants was 47.75% compared with 0.61% for CK (Table 2).
Figure 3. The callus of 35S:WUS and CK cultured for 1.5 months in EIM.

A, B: Calli of 35S:Wus lines at the beginning of EC formation, C, D: Calli of CK lines were unable to differentiate. Bar, 1 cm.
Table 2. EC induction in CRI12.
| 35S:WUS | Empty vector (CK) | |||||
| Replication | Ca | ECb | EC rate (%) | Ca | ECb | EC rate (%) |
| 1 | 173 | 88 | 50.66 | 151 | 2 | 1.32 |
| 2 | 201 | 94 | 46.77 | 114 | 0 | 0.00 |
| 3 | 93 | 41 | 44.09 | 223 | 1 | 0.45 |
| Average | 467 | 223 | 47.75** | 488 | 3 | 0.61 |
Number of explants forming callus.
Number of EC forming from a callus. **p<0.01.
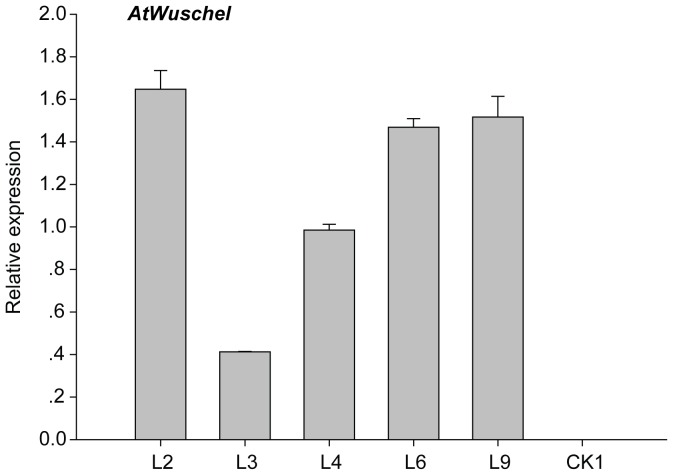
QPCR was used to determine AtWus expression level in transformed calli. The calli of transgenic cultures (L2, L3, L4, L6, and L9) cultured for 4 months in EIM were selected for analysis, whereas the calli of CK that did not differentiate into EC served as the control (CK1). The data revealed AtWus was overexpressed in 35S:WUS cultures (Figure 4).
Figure 4. AtWus expression in calli of CRI12.
Ectopic Expression of AtWus Regulates Auxin Transport and Signal Transduction
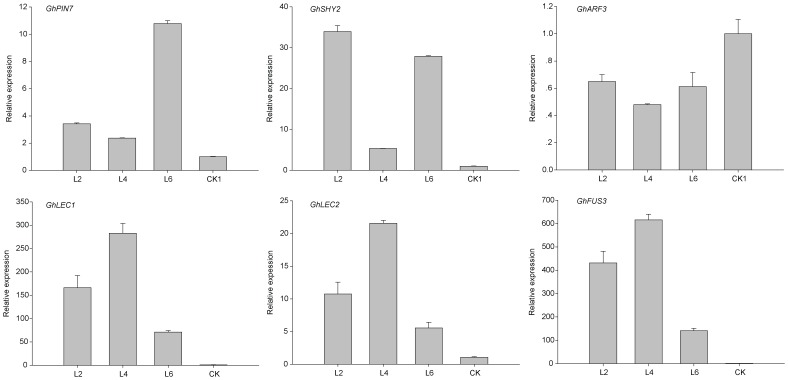
AtWus induction by IAA can regulate AtPIN expression during SE in Arabidopsis [10]. Hence, the role of AtWus in auxin transport and signal transduction in cotton was studied. Transgenic calli of L2, L3, L4, L6, and L9 cultured for 4 months in EIM were analyzed by QPCR, with CK1 calli serving as a control. GhPIN7 expression was higher in 35S:WUS transformed callus lines than in CK (Figure 5).
Figure 5. Analysis of gene expression in the transgenic calli carrying AtWus.
Transcript profiling during SE in cotton has been used to establish the association between the auxin signaling pathway and callus differentiation [13]. The above-mentioned data also demonstrated an interaction between the auxin signaling pathway and EC induction whereby GhSHY2 transcripts were increased in 35S:WUS lines compared with the CK1 line. However, GhARF3 transcripts were reduced in 35S:WUS, possibly owing to suppression by GhSHY2 (Figure 5).
AtWus Activates the Expression of LEC and FUS3 during SE in Cotton
GhLEC1, GhLEC2, and GhFUS3 were expressed in calli of CRI24 but only barely detected in CRI12. To characterize the regulatory relation between AtWus and LEC/FUS3, we selected L2, L4, and L6 callus lines cultured for 4 months in EIM for further study. QPCR analysis revealed that GhLEC1, GhLEC2, and GhFUS3 transcript levels were low in calli of CK1. In calli of L2, L4 and L6, however, higher expression levels of the three genes were detected owing to upregulation by AtWus (Figure 5). Hence, AtWus promoted the expression of GhLEC1, GhLEC2, and GhFUS3 during SE in cotton, similar to Arabidopsis [10].
AtWus Overexpression Results in Abnormal Development of Somatic Embryos
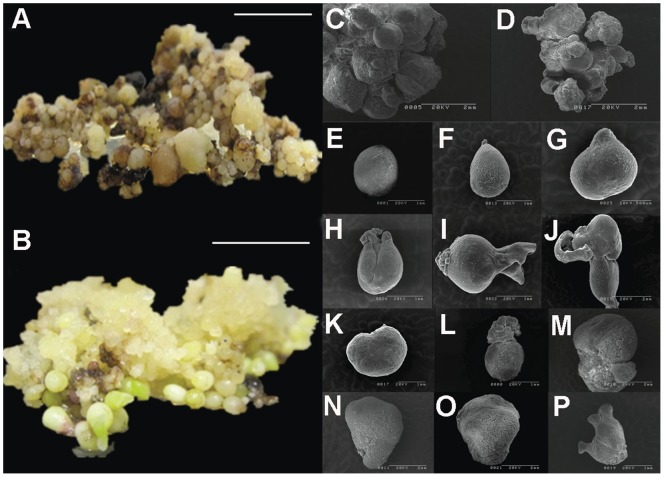
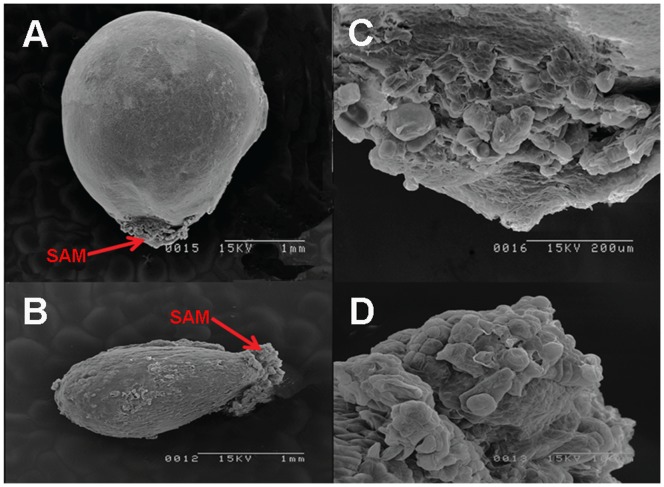
For somatic embryo induction, 4-month-old EC of 35S:WUS and CK lines were transferred to SIM containing kanamycin and cefotaxime for somatic embryo induction. ECs were transferred onto a fresh SIM once per month and cultured for 50 days on SIM. EC formed from calli of CK lines grew into normal-looking globular, heart-shaped, and cotyledonary embryos (Figure 6 B ). However, ECs that developed from 35S:WUS callus lines grew into various abnormal translucent embryos (Figure 6A ) that could not form the normal-looking cotyledonary embryos except for some leaf-like or multi-cotyledon embryos, although they produced more somatic embryos than CK (Figure 6C, D ). Scanning electron microscopy was used to investigate the structural abnormalities of these embryos. Somatic embryos from 35S:WUS lines were much larger and heavier than those from CK lines (Figure 6C , Table 3). In CK lines, globular, heart-shaped and cotyledonary embryos were clearly observed (Figure 6E–J ). However, somatic embryos of 35S:WUS lines exhibited several abnormal morphologies, such as leaf-like or multi-cotyledon embryos (Figure 6K–P ). Cotyledons are derived from the shoot apical meristem (SAM) region. SAM cells were arranged in an organized manner in CK somatic embryos (Figure 7B, D ), but the SAM cells of 35S:WUS lines were disorganized and rather unstructured (Figure 7A, C ).
Figure 6. AtWus overexpression results in abnormal development of somatic embryos.
A: Many abnormal somatic embryos were produced in 35S:WUS lines, and the somatic embryos were inflated and lacked cotyledons. B: Formation of normal somatic embryos in CK lines at different stages. Scanning electron microscopy: Holistic perspective of somatic embryos in 35S:WUS lines (C) and CK lines (D). E–J: Normal somatic embryos at different stages. E, F: globular embryo. G: heart-shape embryo. H–J: cotyledonary embryo. K–P: Abnormal somatic embryos having various appearance. O: leaf-like embryo. P: multiple-cotyledon embryo. Bar in A or B, 1 cm.
Table 3. Somatic embryo weight in CRI12.
| Replication | 35S:WUS (g) | CK (g) |
| 1 | 0.0138 | 0.00764 |
| 2 | 0.0124 | 0.00445 |
| 3 | 0.0181 | 0.00530 |
| Average | 0.0148 | 0.00580 |
p<0.05.
Figure 7. SAM structure of somatic embryos in transgenic lines observed with scanning electron microscopy.
A: Abnormal somatic embryo of 35S:WUS lines. B: Normal somatic embryo of CK lines. C: Enlarged image of SAM in A. D: Enlarged image of SAM in B.
Discussion
Wus is a key gene that controls renewal of stem cells in the apical meristem [29]. However, because AtWus promotes the vegetative-to-embryonic transition during SE in Arabidopsis, numerous studies have elucidated its functions during SE in several plant species [30]. During preparation of the current report, AtWus was shown to promote SE differentiation in G. hirsutum Coker cotton [31], a cultivar that easily undergoes SE. However, it is unknown if these results apply to other cotton genotypes. Furthermore, little work has been done on the regulatory role of Wus in the auxin signaling pathway during SE.
Here, we examined the role and molecular mechanism of Wus-promoted SE in G. hirsutum. Cultivar CRI12, a genotype that is difficult to regenerate. We constitutively overexpressed AtWus in CRI12 using an Agrobacterium-mediated transformation and found that AtWus can promote the formation of non-compact light green calli that induce EC easily. The EC differentiation rate was higher in 35S:WUS of CRI12 (47.75%) in comparison with <1% in CK. Hence, AtWus is a possible candidate that could promote the nonembryonic-to-embryonic transition during SE in cotton.
Transcript analysis has revealed that the auxin transport and signaling pathways may play a substantive role in EC induction [13]. Our QPCR results showed that GhPINs and GhSHY/GhARF may regulate the efficiency of callus differentiation into EC. PINs play a fundamental role in embryonic auxin distribution in plant embryos [16], and AtPIN1 is associated with the establishment of auxin gradients during SE in Arabidopsis. Previous studies revealed diverse changes in the endogenous auxin levels when EC was induced [13]. The antisense cDNA of AtWus suppresses PINs during SE in Arabidopsis [10]. When auxin levels are low, SHY (AUX/IAA) expression is induced and certain ARF activity is repressed, but at high auxin levels SHY is degraded by SCFTIR1/AFBs and the ARF inhibitory action is abolished [17], [19]. Our results revealed enhanced GhSHY2 expression and repressed GhARF3 expression in CRI12 calli overexpressing AtWus. However, AtWus is unable to modify the amount of auxin in cotton calli [31]. Hence, AtWus may play an important role in upregulating GhPINs to redistribute auxin gradients, which may alter expression patterns of SHY-ARF at low levels of auxin.
LEC1, LEC2, and FUS3 are crucial for SE. The capacity of SE is almost completely repressed in double and triple mutants of the three genes, indicating that LEC genes may function downstream of endogenous auxin-induced SE in Arabidopsis [21], [23]. In our present study, GhLEC1, GhLEC2, and GhFUS3 transcript levels were extremely low in callus that was unable to differentiate into EC, but levels were high in callus producing EC. Hence, AtWus positively regulated LECs and FUS3. In Arabidopsis, WUSCHEL and PGA37/MYB118 promote SE and activate the expression of LEC1, LEC2, and FUS3 [32]. In our study, GhLEC1, GhLEC2, and GhFUS3 were also upregulated in callus of 35S:WUS lines. These results suggest that AtWus may alter PIN expression, which leads to the establishment of new auxin gradients in the callus. Subsequently, a new auxin response was formed and stimulated GhLEC1, GhLEC2, and GhFUS3 in the callus of CRI12. AtWus may provoke the ability of differentiation in the callus by reactivating GhLECs and GhFUS3 expression through auxin transport and signaling mechanisms.
Although AtWus improved EC induction, the observed abnormal somatic embryos were an unexpected consequence of AtWus overexpression and prevented seedling generation. AtWus expression is limited to the SAM because auxin accumulates in the cotyledon primordial cells during somatic embryo development in Arabidopsis [10]. Therefore, ectopic expression of Wus could cause loss of expression specificity in somatic embryos, leading to asymmetric growth. In Arabidopsis, LEC1 and FUS3 may control multiple aspects of seed development [33]. Constitutive expression of LEC1 leads to occasional formation of somatic embryo like structures [34]. Therefore, constitutive expression of AtWus in CRI12 may have led to constitutive expression of GhLEC1, GhLEC2 and GhFUS3 in embryos, and then the expression of GhLECs and GhFUS3 may have resulted in the observed abnormal embryos and failure of seedling regeneration. Using inducible promoters such as the estradiol-inducible promoter [35] rather than the 35S promoter during SE in cotton may avoid abnormal embryo formation. Estradiol could be added into the CIM and EIM for EC induction with subsequent transfer of ECs onto SIM without estradiol for normal embryo development. This promoter has been successfully applied for SE in several species [30].
Cotton is an important source of textile fiber and edible oil, but cotton yield is adversely affected by abiotic or biotic stresses [36]. Therefore, efforts to improve cotton resistance against such stresses by genetic modification may play a vital role in efforts to increase production. Agrobacterium-mediated transformation via SE has been the most popular transgenic technology in cotton. Most genotypes cannot undergo EC induction or have low rates of differentiation, although many of those recalcitrant genotypes have certain positive agronomic characters [37]. Thus, the difficulty of EC induction always restricts the application of transgenic breeding and in vitro regeneration in additional cultivars. For example, CRI12 used to be an important elite cultivar widely cultured in China for its disease resistance, high yeild and superior fiber quality. However, SE production in this cultivar is not easy, making it difficult to improve traits using transgenic technology. In our study, the introduction of AtWus into the recalcitrant cotton cultivars enhanced their somatic embryogenesis. With this foundation established, we may now construct a vector to overexpress AtWus with an estradiol-inducible promoter and transfer it into CRI12 or other cultivars. This will require a simple addition of estradiol to CIM and EIM to ensure EC induction with high frequency and production of transgenic seedlings. This will then enable us to improve the rate of cotton transformation for many foreign genes or cotton genes. Such transgenic plants can be used directly as germplasm for cotton breeding. This protocol will achieve our goals of creating more germplasm resources that facilitate SE and expand the scope of transgenic breeding in more cultivars.
Supporting Information
Characterization of GhLEC1, GhLEC2 and GhFUS3.
(TIF)
Specific primers for cloning GhLEC1, GhLEC2, GhFUS3 and AtWuschel.
(XLSX)
Primers used for quantitative real time PCR.
(DOCX)
Acknowledgments
We are grateful to Lihua Ma and Tianping Suo (Cotton Research Institute, Chinese Academy of Agricultural Sciences) for their technical assistance in scanning electronic microscopy. We thank Dr. Jianru Zuo (State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for helpful suggestions and discussions.
Funding Statement
This study was supported by National Science Fund for Distinguished Young Scholars (Grant no. 31125020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schmidt ED, Guzzo F, Toonen MA, de Vries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049–2062. [DOI] [PubMed] [Google Scholar]
- 2. Thomas TL (1993) Gene expression during plant embryogenesis and germination: an overview. Plant Cell 5: 1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis . Development 122: 87–96. [DOI] [PubMed] [Google Scholar]
- 4. Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M (2003) Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J 35: 429–441. [DOI] [PubMed] [Google Scholar]
- 5. Yadav RK, Reddy GV (2011) WUSCHEL-mediated cellular feedback network imparts robustness to stem cell homeostasis. Plant Signal Behav 6: 544–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, et al. (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yadav RK, Tavakkoli M, Reddy GV (2010) WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137: 3581–3589. [DOI] [PubMed] [Google Scholar]
- 8. Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814. [DOI] [PubMed] [Google Scholar]
- 9. Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis . Plant J 30: 349–359. [DOI] [PubMed] [Google Scholar]
- 10. Su YH, Zhao XY, Liu YB, Zhang CL, O’Neill SD, et al. (2009) Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis . The Plant Journal 59: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arroyo-Herrera A, Gonzalez AK, Moo RC, Quiroz-Figueroa FR, Loyola-Vargas VM, et al. (2008) Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue and Organ Culture 94: 171–180. [Google Scholar]
- 12. Ikeda-Iwai M, Satoh S, Kamada H (2002) Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J Exp Bot 53: 1575–1580. [DOI] [PubMed] [Google Scholar]
- 13. Yang X, Zhang X, Yuan D, Jin F, Zhang Y, et al. (2012) Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol 12: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, et al. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- 15. Zazimalova E, Krecek P, Skupa P, Hoyerova K, Petrasek J (2007) Polar transport of the plant hormone auxin - the role of PIN-FORMED (PIN) proteins. Cell Mol Life Sci 64: 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weijers D, Sauer M, Meurette O, Friml J, Ljung K, et al. (2005) Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis . Plant Cell 17: 2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H (2012) Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci 367: 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sauer M, Balla J, Luschnig C, Wisniewska J, Reinohl V, et al. (2006) Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20: 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, et al. (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fambrini M, Durante C, Cionini G, Geri C, Giorgetti L, et al. (2006) Characterization of LEAFY COTYLEDON1-LIKE gene in Helianthus annuus and its relationship with zygotic and somatic embryogenesis. Development genes and evolution 216: 253–264. [DOI] [PubMed] [Google Scholar]
- 21. Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis . Planta 222: 977–988. [DOI] [PubMed] [Google Scholar]
- 22. Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, et al. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proceedings of the National Academy of Sciences 105: 3151–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, et al. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences 98: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu L, Yang X, Yuan D, Zeng F, Zhang X (2011) GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton. Plant Biotechnol J 9: 1038–1048. [DOI] [PubMed] [Google Scholar]
- 25. Jin S, Zhang X, Nie Y, Guo X, Huang C (2005) Factors affecting transformation efficiency of embryogenic callus of upland cotton (Gossypium hirsutum) with Agrobacterium tumefaciens . Plant cell, tissue and organ culture 81: 229–237. [Google Scholar]
- 26. Wan C-Y, Wilkins TA (1994) A Modified Hot Borate Method Significantly Enhances the Yield of High-Quality RNA from Cotton (Gossypium hirsutum L.). Analytical biochemistry 223: 7–12. [DOI] [PubMed] [Google Scholar]
- 27. Wang K, Wang Z, Li F, Ye W, Wang J, et al. (2012) The draft genome of a diploid cotton Gossypium raimondii . Nat Genet 44: 1098–1103. [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, Zhang C, Zhang X, Liu C, Wu Z, et al.. (2013) Transcriptome Profiling Reveals Auxin and Cytokinin Regulating Somatic Embryogenesis in Different Sister Lines of Cotton Cultivar CCRI24. J Integr Plant Biol. [DOI] [PubMed]
- 29. Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, et al. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- 30. Solís-Ramos LY, González-Estrada T, Nahuath-Dzib S, Zapata-Rodriguez LC, Castaño E (2009) Overexpression of WUSCHEL in C. chinense causes ectopic morphogenesis. Plant Cell, Tissue and Organ Culture (PCTOC) 96: 279–287. [Google Scholar]
- 31. Bouchabke-Coussa O, Obellianne M, Linderme D, Montes E, Maia-Grondard A, et al. (2013) Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep 32: 675–686. [DOI] [PubMed] [Google Scholar]
- 32. Wang X, Niu QW, Teng C, Li C, Mu J, et al. (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis . Cell Res 19: 224–235. [DOI] [PubMed] [Google Scholar]
- 33. Parcy F, Valon C, Kohara A, Miséra S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. The Plant Cell Online 9: 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lotan T, Ohto M-a, Yee KM, West MA, Lo R, et al. (1998) Arabidopsis LEAFY COTYLEDON1 Is Sufficient to Induce Embryo Development in Vegetative Cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- 35. Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273. [DOI] [PubMed] [Google Scholar]
- 36. Leelavathi S, Sunnichan V, Kumria R, Vijaykanth G, Bhatnagar R, et al. (2004) A simple and rapid Agrobacterium-mediated transformation protocol for cotton (Gossypium hirsutum L.): embryogenic calli as a source to generate large numbers of transgenic plants. Plant Cell Rep 22: 465–470. [DOI] [PubMed] [Google Scholar]
- 37. Wu J, Zhang X, Nie Y, Jin S, Liang S (2004) Factors affecting somatic embryogenesis and plant regeneration from a range of recalcitrant genotypes of Chinese cottons (Gossypium hirsutum L.). In Vitro Cellular & Developmental Biology-Plant 40: 371–375. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of GhLEC1, GhLEC2 and GhFUS3.
(TIF)
Specific primers for cloning GhLEC1, GhLEC2, GhFUS3 and AtWuschel.
(XLSX)
Primers used for quantitative real time PCR.
(DOCX)