Abstract
The entomopathogen Bacillus thuringiensis produces dense biofilms under various conditions. Here, we report that the transition phase regulators Spo0A, AbrB and SinR control biofilm formation and swimming motility in B. thuringiensis, just as they control biofilm formation and swarming motility in the closely related saprophyte species B. subtilis. However, microarray analysis indicated that in B. thuringiensis, in contrast to B. subtilis, SinR does not control an eps operon involved in exopolysaccharides production, but regulates genes involved in the biosynthesis of the lipopeptide kurstakin. This lipopeptide is required for biofilm formation and was previously shown to be important for survival in the host cadaver (necrotrophism). Microarray analysis also revealed that the SinR regulon contains genes coding for the Hbl enterotoxin. Transcriptional fusion assays, Western blots and hemolysis assays confirmed that SinR controls Hbl expression, together with PlcR, the main virulence regulator in B. thuringiensis. We show that Hbl is expressed in a sustained way in a small subpopulation of the biofilm, whereas almost all the planktonic population transiently expresses Hbl. The gene coding for SinI, an antagonist of SinR, is expressed in the same biofilm subpopulation as hbl, suggesting that hbl transcription heterogeneity is SinI-dependent. B. thuringiensis and B. cereus are enteric bacteria which possibly form biofilms lining the host intestinal epithelium. Toxins produced in biofilms could therefore be delivered directly to the target tissue.
Introduction
Bacillus subtilis and pathogenic bacteria of the Bacillus cereus group (B. cereus, B. thuringiensis and B. anthracis) are all Gram-positive, flagellated, sporulating, and aerobic bacteria clustering closely in the phylogenetic tree of the Bacillus genus ([1]; http://www.patricbrc.org/portal/portal/patric/Phylogeny?cType=taxon&cId=1386). They share a large number of transcriptional factors, including the sporulation regulator Spo0A, the stress response sigma factor σB, and the phase-transition regulators SinI, SinR, CodY and AbrB [2]. However, there are also important differences in the regulatory pathways between B. subtilis and B. cereus sensu lato. For example, the stress regulator σB is not activated in the same way in these species [3]; the two-component system DegU/DegS and the motility regulator SigD are absent from B. cereus sensu lato [2]; the virulence regulator PlcR, which promotes the transcription of numerous genes for extracellular enzymes and toxins and plays an important role in B. cereus and B. thuringiensis physiology [4]–[6], is absent from B. subtilis. These differences may well be the consequences of adaptation of these species to different ecosystems. B. subtilis is a saprophyte living on soil organic matter, whereas, B. thuringiensis is an entomopathogenic bacterium, genetically closely related to the human opportunistic pathogen B. cereus [7], [8], and to the human pathogen B. anthracis [9].
Both B. subtilis and B. thuringiensis, or B. cereus, can form biofilms at air-liquid interfaces. Biofilms are widely found structures in which microorganisms are protected against various stresses, allowing them to persist in adverse environmental conditions. The regulatory pathways in B. subtilis leading either to biofilm formation or to sporulation share the same initial steps. The transcriptional regulator Spo0A controls entry into sporulation [10], and is required for biofilm formation [11]. Spo0A represses abrB transcription [12] and promotes the transcription of sinI [13], the product of which is the SinR antagonist SinI. Both AbrB and SinR repress the two polycistronic operons tapA-sipW-tasA and epsA-O [14], [15]. The 15-gene epsA-O operon is involved in the biosynthesis of the exopolysaccharide component of the biofilm matrix [16] and the three-gene tapA-sipW-tasA operon is involved in the production of the protein component of the biofilm matrix [17], [18]. An inhibitor of flagellar motility is encoded by the epsE gene which is part of the epsA-O operon [19]. Therefore, deletion of sinR from B. subtilis results in an overproduction of biofilm and in impaired motility, whereas deletion of sinI results in the reverse phenotype. A paralogue of SinR, SlrR, is also involved in the control of biofilm formation and motility through its interaction with SinR [20], [21].
How biofilm formation is regulated in B. thuringiensis or in B. cereus is still unknown. In B. anthracis, SinR strongly represses the sipW-tasA operon [22], but the effect of sinR deletion on biofilm formation has not been studied. The quorum sensing molecule AI-2 is produced by B. cereus and inhibits biofilm formation when added exogenously [23], and the transcriptional regulators PlcR and CodY affect biofilm formation in the B. cereus reference strain ATCC 14579 [24]–[26]. PlcR is the main virulence regulator in B. cereus [6] and CodY, which represses the biosynthesis of branched amino-acids, might also be involved in the pathogenicity of B. cereus [26]–[29]. These findings suggest a connection between biofilm formation and virulence in this species. Here we report an investigation of the roles of Spo0A, AbrB and SinI/SinR in biofilm formation in the B. thuringiensis strain 407, which produces dense pellicles at the air-liquid interface. We found that SinI/SinR had a large effect on biofilm formation. We therefore analyzed the B. thuringiensis sinR regulon, which was found to include the sipW-tasA operon, but surprisingly no eps operon. SinR was also found to control the transcription of genes required for the production of lipopeptides previously shown to be involved in the bacterial survival in the host [30], and the transcription of enterotoxin genes.
Materials and Methods
Strains
Strains used in this study are listed in table S1. The acrystalliferous B. thuringiensis strain 407 Cry− (genome sequence at NCBI : NZ_CM000747) is genetically closely related to the B. cereus reference strain ATCC 14579 [31]; however, strain 407 forms thick biofilms, while ATCC 14579 is a poor biofilm producer. Locus tags listed below follow the annotations of the sequenced ATCC 14579 strain genome (NC_004622), and the corresponding locus tags in the sequenced 407 strain genome are given table S2.
Strain construction
The sinI-sinR locus in strain 407 was disrupted by insertion of a tetracycline resistance (TetR) cassette. A 937 bp HindIII-EcoRI fragment and a 758 bp XbaI–BamHI fragment, corresponding to the chromosomal DNA regions upstream and downstream of the sin genes locus, respectively, were generated by PCR using the primer pairs Sin1-Sin14 and Sin17-Sin18 (table 1). The TetR cassette was purified from pHTS2 [32] as a 1.5 kb XbaI-EcoRI fragment. The amplifed DNA fragments and the TetR cassette were inserted between the HindIII and BamHI sites of pRN5101 [33]. The resulting plasmid was used to transform the 407 wild type strain by electroporation, and the sin locus was deleted and replaced with the TetR cassette via allelic exchange by homologous recombination, as previously described [33]. The resulting mutant strain was designated sinI-sinR. The same procedure was used to disrupt sinI, sinR (BC1283 and BC1282, respectively) and abrB (BC0042): sinI and sinR were each disrupted with the tetracycline resistance cassette; and abrB with a kanamycin resistance cassette (a 1365 bp XbaI-PstI fragment of pDG783 [34]). The resulting mutant strains were designated sinI, sinR, and abrB. The primer pairs used for these disruptions are listed Table 1.
Table 1. Primers used in this study.
| Primer | sequencea | restriction site |
| Sin1 | CCCAAGCTTTACCAGAAACTGTTAACC | HindIII |
| Sin2 | CCGGAATTCGGCAAGTTCAGTTAATGA | EcoRI |
| Sin3 | CGCTCAGATGCTGGTATCGCC | XbaI |
| Sin4 | CGGGATCCTGTATACGAAACAACTTTAGC | BamHI |
| Sin11 | CGCTCAGATGCTGGTATCGCC | XbaI |
| Sin12 | CCGGAATTCAAGTATTAAATCAATCCATTC | EcoRI |
| Sin14 | CCGGAATTCCCTCTATGGAAATTATAAATTG | EcoRI |
| Sin17 | GCTCTAGATGCAATGAACTCTGGTGTCTCC | XbaI |
| Sin18 | CGCGGATCCTAGGGGGAATTGATTGTGAGTC | BamHI |
| AbrB1FW | CCCAAGCTTGGGCTGCTAAATCTTCTAATCCCG | HindIII |
| AbrB1RV | GCTCTAGAGCCAATCATTTACATTTCCGTC | XbaI |
| AbrB2FW | AAACTGCAGCGACATGATAGATTTGATATACATC | PstI |
| AbrB2RV | CGCGGATCCAAAATATGTAGAGACCCACGAT | BamHI |
| Hbl_pHT304_FW | CCAAGCTTGATATTAGGATGTTTTGTGA | HindIII |
| Hbl_pHT304_RV | CGGGATCCTTTACCATTGTTTTTATAAC | BamHI |
| Yfp-F | GGGGTACCACATAAGGAGGAACTACTATGAGTAAAGGAGAAGAAC | KpnI |
| Yfp-R | CGGAATTCTTATTTGTATAGTTCATCCATGC | EcoRI |
| Apha3_pHT304_FW | CCAAGCTTGATAAACCCAGCGAACCATTTGAGG | HindIII |
| Apha3_pHT304_RV | CGGGATCCCCGGTGATATTCTCATTTTAGCC | BamHI |
| psinI-Rev BamHI | CGGGATCCCTAATTATCGGTCATAATTGC | BamHI |
| phbl-sinI-SOE-Fwd | aacatcctaatatTCTAGAatatttagtcatatcatg | none |
| phbl-sinI-SOE-Rev | atgactaaatatTCTAGAatattaggatgttttgtg | none |
| Hbl_pHT304_RV | CGGGATCCTTTACCATTGTTTTTATAAC | BamHI |
The primers were used to delete the sinI, sinR, sinI-sinR and abrB genes from the strain 407, or to create transcriptional fusions between the promoters of hbl or of sinI and lacZ, yfp or mcherry on the pHT304-18 plasmid.
: underlined sequences indicate the location of restriction sites, and lower case letters indicate overlapping sequences complementary to Phbl (not underlined) or to PsinI (underlined).
The antibiotic concentrations used for bacterial selection were as follows: 100 µg.ml−1 of ampicillin for E. coli; and 10 µg.ml−1 of erythromycin, 200 µg.ml−1 of kanamycin and 10 µg.ml−1 tetracycline for B. thuringiensis.
Transcriptional fusions and beta-galactosidase assays
Expression of the hbl operon in the 407 wild type and mutant strains was monitored using a transcriptional fusion between the hbl promoter region and lacZ. The DNA sequence containing the hbl promoter was amplified using primers Hbl_pHT304_FW and Hbl_pHT304_RV (table 1) and inserted into pHT304-18Z [35], to give pHT304-18ΩPhbl'-lacZ. The same DNA sequence was inserted into pHT304-18YFP, resulting in pHT304-18ΩPhbl'-yfp. The DNA sequence containing the apha3 promoter was amplified from pDG783 [34] with primers Apha3_pHT304_FW and Apha3_pHT304_RV (table 1) and inserted into pHT304-18YFP, resulting in pHT304-18ΩPapha3'-yfp. The plasmid pHT304-18YFP was constructed by insertion, between the sites EcoRI and KpnI of pHT304-18 [36], of the yfp gene amplified from pKL183 [37] using the primer pair Yfp-F and Yfp-R (table 1). The plasmid pHT304-18ΩPhbl'yfp-PsinI'mCherry used to monitor simultaneously, in the same cell, hbl and sinI expressions, was constructed as follows. DNA fragments containing the promoter of sinI and hbl were amplified by PCR using the primers pairs phbl-sinI-SOE-Fwd/psinI-Rev BamHI and phbl-sinI-SOE-Rev/Hbl_pHT304_RV, respectively (table 1). These fragments were annealed to each other through complementary overlapping sequences introduced in primers phbl-sinI-SOE-Fwd and phbl-sinI-SOE-Rev. A single DNA fragment containing the promoter elements of sinI and hbl in opposite directions was then generated by PCR amplification with the primers psinI-Rev BamHI and Hbl_pHT304_RV. The resulting 1225 bp fragment was digested with BamHI and cloned in the promoter free pHT304-18Ωyfp-mCherry digested with the same enzyme.
Electroporation was used to transfer pHT304-18ΩPhbl'-lacZ, pHT304-18ΩPhbl'-yfp, pHT304-18ΩPapha3'-yfp and pHT304-18ΩPhbl'yfp-PsinI'mCherry into 407 wild type or into 407 sinI strains. Beta-galactosidase specific activities were measured as described previously, and are expressed in units of beta-galactosidase per milligram of protein [38]. Beta-galactosidase was extracted from cells in biofilm obtained in glass tube assays (see below), and from planktonic cultures grown in LB medium at 30°C and 175 rpm. Three replicates were performed for each assay.
Biofilm assays
The ability of 407 wild type and mutant strains to form biofilms in PVC (polyvinylchloride) microtiter plates was measured as described previously [23]. The method used to obtain biofilm in glass tubes was similar to that used in microtiter plates, with the following differences. Sterilized 6 ml glass tubes were inoculated with 2 ml of the cultures diluted to an OD600 of 0.01, and incubated for 48 h. The 2 ml culture medium was then removed using a Pasteur pipette and the OD600 of the ring and of the pellicle, thoroughly vortexed in 1 ml PBS, was measured. The microtiter plate and glass tube assays measure different parts of the biofilm: in the microtiter plate assay, the pellicle is lost during the staining procedure and the biofilm mass determined corresponds to the ring adhering at the air-liquid-solid interface. In the glass tube assay, the entire biofilm is recovered. Biofilms were formed in glass tubes for binocular microscopy observation, for beta-galactosidase assays, for fluorescence microscopy observation or for flow cytometry experiments.
Swimming assays
The swimming ability of the 407 mutant strains was determined on LB 0.3% agar plates. Strains were grown in LB medium at 37°C with shaking at 175 rpm until the culture reached an OD600 of 1. A 5 µl drop of each culture was spotted on an agar plate and incubated at 30°C for 12 hours. Each experiment was repeated four times.
Microarray analysis
Cells used for microarray analysis were grown in bactopeptone medium at 30°C and 250 rpm. Samples were collected 2 hours after entry into stationary phase (t2). Entry into stationary phase (t0) was determined as the breakpoint of the growth curve, i.e. the time point when the slope of the growth curve starts to decrease, which usually occurs in the 407 strain around OD 2.5 in these culture conditions. RNA isolation and cDNA synthesis, labeling and purification were performed as described previously [6]. Microarray slides were printed at the microarray core facility of the Norwegian University of Science and Technology (NTNU). Design, printing, prehybridization, hybridization and scanning of the slides and analysis of the data was as previously described [6]. The microarray experiments were based on four slides, all being true biological replicates. Genes with false discovery rate corrected p-values<0.05, and for which differential expression between the sinR-negative and sinR-positive strains was at least two-fold, were considered to be repressed (fold change FC>2.0) or induced (FC<0.5) by the deleted gene.
The microarrays used contain 70-mer oligonucleotide probes designed to detect open reading frames (ORFs) in B. anthracis strain Ames, B. anthracis strain A2012, B. cereus strain ATCC 14579, and B. cereus strain ATCC 10987 [39]. Only probes with 93% identity or greater to a transcript/feature sequence of 407 were included in the analysis. Of the predicted genes of the 407 genome, 1719 did not have corresponding probes on the array. However, among these genes, 1165 were annotated as hypotheticals (68%), and 761 were on contigs 00213 and 00060, which have later been shown to be plasmid-borne [40]. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-1806.
Hemolysis assays
The various strains were grown in LB medium at 37°C until they reached an OD600 of 1. The hemolytic activity of the strains was assayed by loading a 5 µl drop of each culture on sheep blood agar plates. The agar plates were incubated overnight at 30°C and then scanned: the area of hemolysis and the colony size were determined from the scans with ImageJ software [41]. To minimize differences due to variations in growth rates, a hemolytic index was calculated using the following formula:
were Hi is the hemolytic index, Ha the hemolysis area and Cs the colony size at the end of the incubation time. Results are presented as means of four independent experiments.
Western blot analysis
Cultures were grown in bactopeptone medium at 30°C and 250 rpm, and culture supernatants were collected by centrifugation at t2. SDS-PAGE was carried out using 12% acrylamide gels, and silver stained according to Blum et al. [42]. Proteins were blotted onto Immun-Blot PVDF membranes (Bio-Rad), and nonspecific binding was blocked with 5% nonfat milk. The HblB (binding) component was detected using monoclonal antibody 2A3 diluted 1∶15 [43], kindly provided by Dr Erwin Märtlbauer (Ludwig-Maximilians-Universität, Munich, Germany). Peroxidase-conjugated AffiniPure Goat-anti-mouse IgG (Jackson Immuno Research Laboratories Inc) were used at 0.8 µg/ml as secondary antibody, and bands were detected using the SuperSignal West Femto Substrate (Pierce) and quantified using ImageJ [41]. After subtraction of the background, the mean gray value for each band was normalized to the intensity of the band in the 407 wild type sample, arbitrarily defined as 1.
Flow cytometry experiments
Biofilms recovered from glass tubes assays were homogenized by aspirating/pushing ten times through a 26-gauge needle. Planktonic cultures, or homogenized biofilms, were mixed with an equal volume of ice-cold, 0.2 µm-filtered PBS containing 8% formaldehyde, washed with ice-cold PBS and resuspended in TEG (Tris 20 mM EDTA 10 mM glucose 0.5M pH 7.2). Fluorescence was recorded on a Cyflow SL flow cytometer (Partec GmbH, Münster, Germany). YFP fluorescence was measured by using a solid blue-laser emitting at 488 nm, a 620-nm Long Pass Dichroic Mirror and a 590-nm band pass filter (565–615). mCherry fluorescence was measured with a solid yellow-laser emitting at 561 nm combined to a 585-nm band-pass filter. Gating on FSC/SSC was used to discriminate bacteria from the background. For each sample, at least 40,000 gated events were measured. Data were collected with the FlowMax software (Partec GmbH, Münster, Germany) and analysed with the Weasel 2.0 software (WEHI, USA).
Results
Spo0A, AbrB and SinI/SinR control biofilm formation and swimming motility
Deletion of abrB, sinI or sinI-sinR did not result in significant changes in growth (figure S1a and S1b). In contrast, the spo0A mutant strain grew more slowly (figure S1a), and the sinR mutant strain grew poorly (figure S1b). This is not true in B. anthracis strain Sterne, where it has previously been described that deletion of sinR do not impair bacterial growth [22].
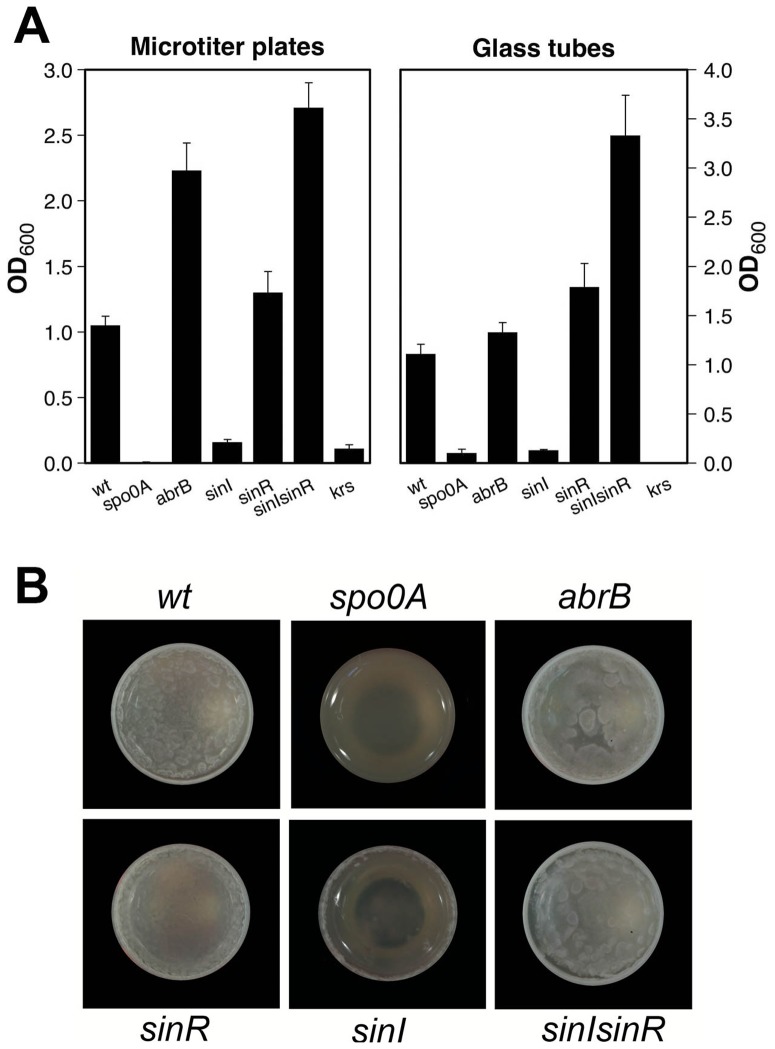
The effect of spo0A, abrB, sinI and sinR on biofilm formation and on motility was similar to B. subtilis as previously reported. In microtiter plates and in glass tubes, Spo0A promoted biofilm formation while AbrB repressed this phenotype (figure 1A). Both mutants had no effect on swimming motility on 0.3% LB agar plates (figure 2). The sinI mutant was highly motile but unable to form biofilms, the sinR mutant was non-motile and overproduced bofilms and the sinI-sinR mutant was highly motile and overproduced biofilms. The architecture of the wild type strain biofilm, viewed from above in glass tubes, appeared as a thick ring sticking to the tube wall, surrounding a floating pellicle on which protrusions could be seen (figure 1B). This architecture was similar in the abrB and the sinI-sinR mutants, whereas the sinR mutant produced a thick ring and a flat pellicle (figure 1B). In contrast, the spo0A mutant produced no ring and no pellicle, and the sinI mutant displayed a small ring but no pellicle.
Figure 1. Biofilm formation by mutant strains.
Biofilm production by the wild type strain 407 (wt) and by the 407 mutants spo0A, abrB, sinI, sinR, sinI-sinR and krsABC, was assessed in microtiter plates and in glass tubes. A: Bars represent the means of six (microtiter plates) to 12 (glass tubes) replicates from three independent experiments, and error bars represent the standard error of the mean. B: Pictures of the floating pellicles obtained in glass tubes for wild type and mutant strains.
Figure 2. Swimming assays with the mutant strains.

Swimming assays were performed on LB agar plates incubated overnight at 30°C, with the wild type strain (wt) and the spo0A, abrB, sinI, sinR and sinIsinR mutant strains. Each swimming assay was repeated four times in independent experiments, and the results obtained were fairly reproducible. Only one replicate is shown here.
Microarray analysis of the sinI, sinR and sinIsinR mutants reveals the SinR regulon
We used microarray analysis of strain 407 to identify genes mediating the effects of sinI and sinR on biofilm formation. A comparative microarray analysis of the sinR mutant and of the wild type strain 407, in the early stationary growth phase, revealed 421 repressed genes in the mutant, many of which are associated with the translation machinery (data not shown). The large decrease in the growth rate of strain 407 upon deletion of sinR is consistent with this result. To overcome this problem, the transcriptome in the presence and absence of sinR was analyzed in a sinI-deletion background. Indeed, both sinI and sinI-sinR mutant strains grew similarly to the wild type strain (figure S1b).
From this analysis, 32 genes appeared to be repressed by SinR two hours after the onset of stationary phase (t2) (Table 2). The B. cereus homologue of B. subtilis sipW and two tasA homologues (BC1278, BC1279 and BC1281), as well as the Hbl enterotoxin genes BC3102 (hblA encoding the binding component HblB) and BC3104 (hblC encoding the lytic component HblL2) were highly repressed by SinR (Table 2). The krsEABC operon (BC2450–BC2453), recently shown to be involved in the biosynthesis and export of a non-ribosomal lipopeptide [30], was also found to be SinR-regulated. Deletion of the krs locus abolished the capability to form biofilms either in microtiter plates or in glass tubes (figure 1). The SinR regulon also included as many as nine genes coding for proteins of unknown functions, some of them being strongly repressed by the sinR deletion. In addition, two motility genes, encoding the chemotaxis protein CheA (BC1628) and the flagellar basal body rod protein FlgC (BC1642), were slightly repressed by SinR (signal ratio 2.5 and 2, respectively), but this moderate repression probably does not explain the large opposite effect of SinR on motility. Genes possibly involved in detoxification processes (BC2230, BC3076, BC3078, BC4272), in sugar metabolism (BC2960, BC2854, BC3759), in DNA recombination (BC2556) or degradation (BC1072), in peptidoglycan turnover (BC5234), and in energy production (BC3142) were also identified as being regulated by SinR in strain 407 at t2 (table 2). Finally, SinR repressed a gene (BC2410) encoding a PlcR-controlled transcriptional regulator [6].
Table 2. Microarray results for the 407 sinRsinI mutant compared to the 407 sinI mutant.
| Locus taga | Gene Productb | Bac | SRd |
| BC1278 | Signal peptidase I, SipW | GBAA1287 | 36.8 |
| BC1279 | TasA homologue | GBAA1288 | 26.0 |
| BC1281 | Camelysin CalY | GBAA1290 | 88.6 |
| BC1072 | endonuclease/exonuclease/phosphatase family protein | GBAA1075 | 3.6 |
| BC2556 | DNA integration/recombination/invertion protein | NH | 2.4 |
| BC3102 | Enterotoxin binding component precursor Hbl | NH | 8.9 |
| BC3104 | Enterotoxin lytic component HblL2 | NH | 3.9 |
| BC0418 | hypothetical protein | NH | 9.8 |
| BC1280 | hypothetical protein | NH | 5.4 |
| BC2409 | hypothetical protein | NH | 3.9 |
| BC2875 | hypothetical protein | NH | 4.2 |
| BC3283 | hypothetical protein | NH | 2.7 |
| BC3290 | hypothetical protein | NH | 2.2 |
| BC3697 | hypothetical protein | NH | 3.2 |
| BC4216 | hypothetical protein | NH | 8.1 |
| BC4259 | hypothetical protein | NH | 3.1 |
| BC1628 | chemotaxis protein CheA | NH | 2.5 |
| BC1642 | flagellar basal body rod protein FlgC | NH | 2.1 |
| BC2230 | macrolide-efflux protein MFS-1 family | NH | 2.4 |
| BC3076 | acetyltransferase | NH | 4.4 |
| BC3078 | aminoglycoside 3′-phosphotransferase | NH | 5.4 |
| BC4272 | superoxide dismutase | NH | 2.7 |
| BC2854 | aldo-keto-oxidoreductase | NH | 6.2 |
| BC2960 | Glycosyl transferase | NH | 2.9 |
| BC3759 | 6-phospho-beta-glucosidase | NH | 3.0 |
| BC2410 | TetR family transcriptional regulator | NH | 2.2 |
| BC3142 | NADPH-dependent oxidoreductase | NH | 2.2 |
| BC5234 | N-acetylmuramoyl-L-alanine amidase | NH | 2.2 |
| BC2450 | macrolide-efflux protein MFS-1 family | NH | 4.1 |
| BC2451 | peptide synthetase | NH | 1.3 |
| BC2452 | peptide synthetase | NH | 1.1 |
| BC2453 | peptide synthetase | NH | 2.2 |
Genes are grouped into functional families.
: locus tag and gene product, respectively, according to the annotation of the ATCC14579 genome.
: homologues in the SinR regulon of B. anthracis (Pflughoeft et al., 2011); NH: no homologues.
: SR, microarray signal ratio, computed as the signal for the sinIsinR mutant divided by the signal for the sinI mutant.
hbl expression is controlled by SinR
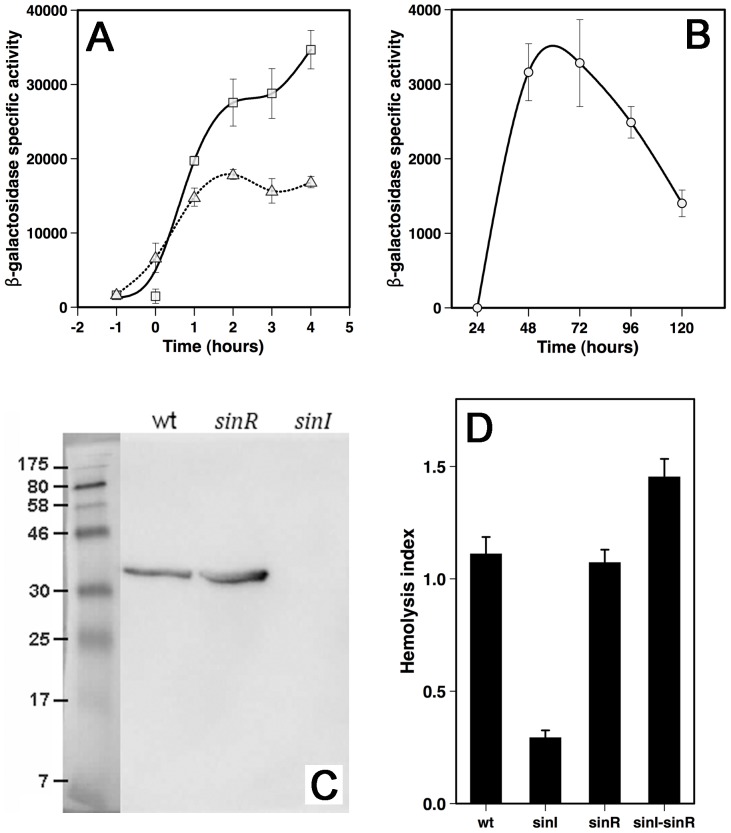
Microarray analysis suggested that expression of the Hbl enterotoxin may be controlled by SinR. The Hbl enterotoxin is already known to be controlled by the virulence transcriptional regulator PlcR [44], [45]. To confirm that the hbl genes were also under SinR regulation, we used a transcriptional fusion between the hblC (BC3104; the first gene of the hblCDA operon) promoter region and the lacZ gene. As already described [46], expression of hblC in the wild type strain increased sharply after t0 and reached a plateau at t2 (figure 3A). Deletion of sinI did not abolish hblC expression, but reduced it greatly, resulting in a ratio of expression of 2.3 at t4 (figure 3A). The effect of SinR on extracellular HblB (encoded by hblA; the third gene of the operon) was then assessed by Western immunoblotting (figure 3C). The amount of extracellular HblB component produced by the sinR mutant strain was higher than that of the wild type, whereas no band could be detected in the sinI mutant. Hence, SinR repressed HblB production while SinI had the reverse effect. Hbl enterotoxins have hemolytic activity, and these toxins are the major hemolysins acting on sheep blood [47]. Therefore, we studied sinI, sinR and sinI-sinR deletion mutants by hemolysis assays on sheep blood agar plates (figure 3D). The hemolytic activity of the sinI mutant strain was much lower than that of the wild type strain. In contrast, deletion of sinR had no effect on hemolytic activity, and deletion of both sinI and sinR resulted in higher hemolytic activity.
Figure 3. hbl expression and hemolytic activity.
a and b: transcriptional activity of the hbl promoter region (time on the x-axis is relative to entry into stationary phase, and bars represent standard errors of the mean); a: the wt strain (squares) was compared to the sinI strain (triangles) in planktonic cultures; b: biofilm glass tube assay. c: Western blot of supernatants from cultures of mutant strains ; band intensities, relative to that for the wild type strain, were: wt: 1.00; sinR: 1.64 ; sinI : 0.01; d: hemolytic properties of the 407 mutant strains.
hbl is expressed in the biofilm produced by strain 407
The co-regulation of biofilm formation and hbl expression by SinR suggests that hbl could be expressed by strain 407 in biofilm. To test this, we produced biofilms in glass tubes with the wild type strain 407 transformed with pHT304-18ΩPhbl'-lacZ, the plasmid carrying the Phbl'-lacZ transcriptional fusion, and we followed hbl expression in the floating pellicle and the planktonic bacterial population right underneath (figure 3B): hbl was expressed in the biofilm, peaking after 48 to 72 hours of culture, when the biofilm has reached its maximal development in glass tubes. This expression, despite decreasing from the peak at 48 h, was sustained and lasted for more than 120 hours, whereas hbl expression in planktonic cultures was shut down after t8 (data not shown). Therefore, B. cereus biofilms are persistent structures in which the bacteria produce significant amounts of hbl over long periods of time.
hbl is expressed by a small subpopulation of biofilm cells
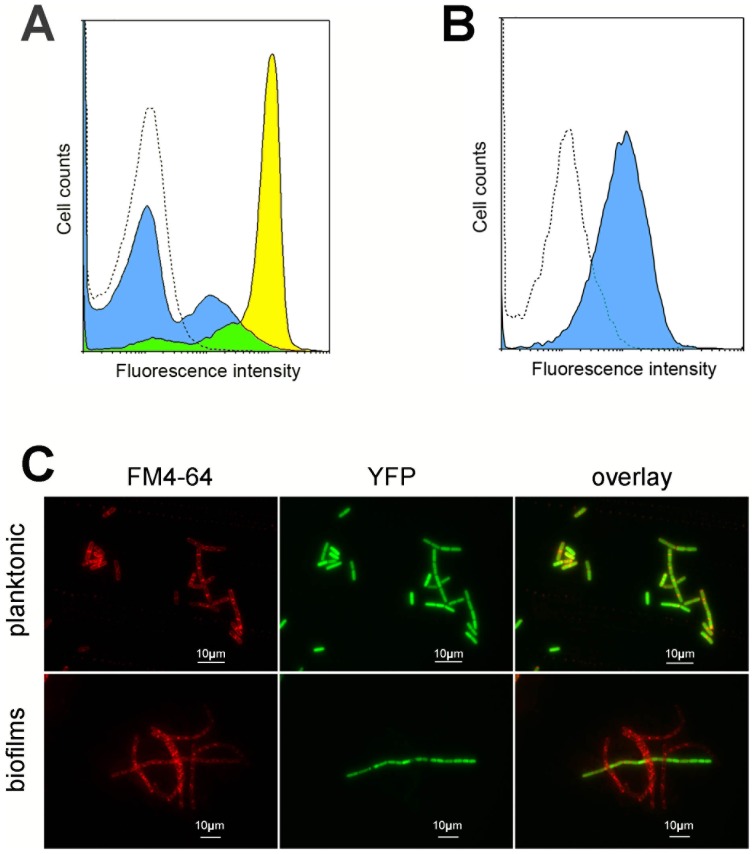
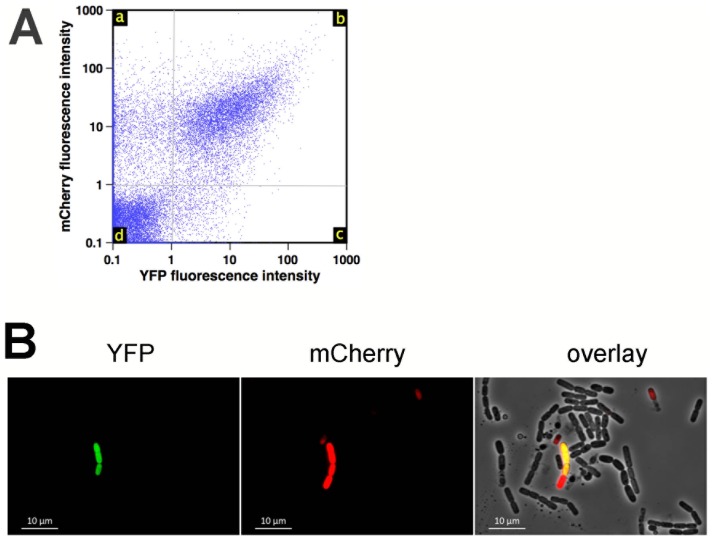
To determine the proportion of B. cereus cells expressing hbl enterotoxin genes, we used a transcriptional fusion between the promoter region of hbl and the yellow fluorescence protein gene yfp. Strain 407 expressing yfp under the control of the hbl promoter was grown in planktonic cultures and in biofilms, and samples were harvested when hbl expression reached a plateau or was maximal as determined with lacZ fusions (figure 3A and 3B: t2 for planktonic cultures and 48 h for biofilms). As shown by flow cytometry, the hbl promoter was active in 90% of bacteria in planktonic cultures in early stationary phase and in 33% of bacteria in homogenized 48 h-old biofilms (figure 4A). In contrast, the apha3 constitutive promoter was active in 88% of bacteria in 48 h-old biofilms (figure 4B), showing that the heterogeneity of hbl expression in biofilms was not consecutive to non-viable bacterial cells or to plasmid loss in this culture condition. In addition, colonies recovered from 48 h-old biofilms formed with the 407 strain carrying the pHT304-18ΩPhbl'-yfp, and transfered to LB- and erythromycin-LB plates, were 100%±0.0 resistant to erythromycin, the pHT304 resistance marker (3 independent experiments). Flow cytometry also revealed that hbl was on average transcribed at a lower level in the biofilm than in planktonic cultures (figure 4A). These results are supported by epifluorescence microscopy, which showed that almost all bacteria in planktonic cultures expressed hbl whereas only a few expressed it in biofilms (figure 4C). By using a plasmid carrying both the Phbl'-yfp and the PsinI'-mcherry transcriptional fusions, we have monitored the expression of hbl and sinI in the same cells in 48 h-old biofilms. We found that 16% of the bacteria expressed hbl (figure 5A), which is in the same range as our previous results. Furthermore, flow cytometry and microscopy observation revealed that almost all bacteria expressing hbl also expressed sinI (figure 5A and 5B). In addition, 12% of the bacteria expressed sinI but not hbl.
Figure 4. Heterogeneity of hbl expression in planktonic cultures and in biofilms.
A: Expression from the hbl promoter was monitored in planktonic cultures and in biofilms by epifluorescence microscopy through a transcriptional fusion to yfp. Cell limits are shown by the membrane stain FM4-64 (red). B: Flow cytometry analysis of bacteria expressing Phbl'-yfp in planktonic cultures or in biofilms, shown as histogram plot. The blue-filled curve shows biofilm data, the yellow-filled curve shows planktonic cultures data and the unfilled dashed curve shows data from bacteria lacking yfp. C: Flow cytometry analysis of bacteria expressing Papha3'-yfp in biofilms (blue-filled curve) compared to bacteria lacking yfp (unfilled dashed curve), shown as histogram plot.
Figure 5. Expression of hbl and of sinI in biofilms.
A: Observation by epifluorescence microscopy of bacteria expressing Phbl'-yfp (left, in green) and PsinI'-mcherry (center, in red) in 48 h-old biofilms. An overlay of YFP fluorescence (hbl expression), mCherry fluorescence (sinI expression) and phase contrast microscopy is shown on the right. B: Flow cytometry analysis of bacteria expressing Phbl'-yfp and PsinI'-mcherry in 48 h-old biofilms, shown as dot-plot. While 72% of the bacteria do not express hbl nor sinI (quadrant d), 15% of the cells which express hbl also express sinI (quadrant b), and 12% of the bacteria express sinI but not hbl (quadrant a).
Discussion
We deleted from B. thuringiensis the genes encoding Spo0A, AbrB and SinR, which are regulators of the transition phase of growth. These regulators were previously shown to control biofilm formation and swarming motility in B. subtilis. We report here that, in B. thuringiensis, SinR represses biofilm formation and is required for swimming motility, whereas SinI has the reverse effect. Consequently, the SinI/SinR antirepressor/repressor pair is likely to act as a switch between biofilm formation and swimming motility, as it does in B. subtilis between biofilm formation and swarming motility [48]. In addition, Spo0A is required in B. thuringiensis for biofilm formation and AbrB represses this phenotype, and neither of these regulators affects motility. These findings suggest that the regulation of biofilm formation and of motility by Spo0A, AbrB, and SinI/SinR show similarities in B. cereus and in B. subtilis. Similarities between the two species for control of biofilm formation is supported by the presence of the sipW-tasA operon in their respective SinR regulons. The sipW-tasA operon was shown to be required for the production of the proteic component of the biofilm matrix in B. subtilis, and to be directly controlled by SinR in B. anthracis [22].
However, within the 32 genes included in the B. thuringiensis SinR regulon, only sipW and tasA are shared with the B. subtilis SinR regulon reported previously [15]. B. thuringiensis and B. cereus display a chromosomal conserved locus (genes BC5267 to BC5278 in B. cereus strain ATCC14579) similar to the epsAO locus which in B. subtilis is involved in the biosynthesis of the exopolysaccharide component of the biofilm matrix. A 120 bp antitermination RNA element named EAR is found, in B. subtilis, exclusively only in the epsAO locus [49], and a corresponding element is predicted to be present in the BC5267–BC5278 locus, consistent with these loci being homologous. But while the B. subtilis epsAO genes are repressed by SinR, the B. thuringiensis BC5267–BC5278 orthologs are not.
Conversely, the B. thuringiensis - but not the B. subtilis - SinR regulon includes genes required for the production of a lipopeptide. This lipopeptide, kurstakin, is required for biofilm formation. In B. subtilis, production of the lipopeptide surfactin, also important for biofilm formation [16], is controlled by the two-component system ComA-ComP [50]. This two-component system is not present in bacteria of the B. cereus group, but it was recently shown that kurstakin production is activated by the NprR cell-cell communication system during stationary phase [30]. In addition to its role in biofilm formation, kurstakin was shown to be required for bacterial survival in the host cadaver [50].
Our transcriptional analysis also revealed that SinR repressed the hbl enterotoxin genes. The transcription of hbl genes is promoted by the virulence regulator PlcR [44], [45]. The products of these genes are active against the rabbit intestine [51] and may be responsible for the diarrheal symptoms associated with B. cereus-dependent gastroenteritis [7]. Transcriptional fusion experiments confirmed that SinI/SinR controls hbl transcription, and Western blotting and hemolysis assays also showed that SinI/SinR regulates the production of Hbl enterotoxin components. The SinR regulon includes another gene controlled by PlcR: BC2410. The product of BC2410 is a transcriptional regulator, the targets of which are still unknown, but which might be involved in bacterial pathogenesis [6]. Therefore, SinR in B. cereus controls virulence factors that are part of the PlcR regulon, in addition to biofilm formation and motility. These findings suggest that inactivation of SinR by SinI both serve to trigger biofilm formation and to enhance enterotoxin production.
We assessed the expression of hbl in biofilms using lacZ fusions. We found that this expression was sustained and lasted for more than 48 h but was moderate as compared to the strong expression of hbl in planktonic cultures. We determined hbl expression at the cell level using yfp fusions: most of the bacteria in planktonic cultures expressed hbl, while only a small subpopulation of cells expressed it in biofilms. This heterogeneity in the expression of hbl in biofilms is likely to be a consequence of the heterogeneity in the expression of sinI, since we found that bacteria expressing hbl also expressed sinI. SinI-dependent heterogeneity of genes expression in biofilms has been described in B. subtilis, where the sipW-tasA operon is expressed in the same subpopulation as sinI, whereas sinR is expressed in the whole biofilm bacterial population [52].
The B. thuringiensis SinR regulon shares only 4 genes with the B. anthracis SinR regulon as determined previously [22] : the homologue of B. subtilis sipW and two homologues of tasA, and a gene encoding an endonuclease. The 28 other genes of the B. thuringiensis SinR regulon have not been identified as components of the B. anthracis SinR regulon, indicating possible differences between these two species for the role of SinR. More specifically, the inhA1 gene, encoding a metalloprotease, has previously been reported to be SinR-dependent in B. anthracis [22]. This gene is also present in B. thuringiensis and B. cereus, were it is likely to play an important role in the bacterial pathogenesis [53], [54]. Although inhA1 was previously reported to be controlled by Spo0A, AbrB and SinR in the 407 strain [55], we do not confirm, by microarray analysis, the role of SinR in the regulation of inhA1 transcription. However, the previous study used an overexpression of sinI or of sinR on high copy plasmids to investigate the role of SinR in the control of inhA1, which might have introduced bias in that study.
The sinI-sinR mutant produced more biofilm and was more hemolytic than the sinR mutant, which was unexpected. If the function of SinI is only antagonizes SinR, then sinI-sinR and sinR mutants should have similar phenotypes, and give similar results. The observed difference could suggest that SinI also acts on another regulator, different from SinR. In B. subtilis, biofilm formation is stimulated by SlrR, a paralogue of SinR [21], [56], and SlrR interacts with SlrA, a paralogue of SinI [57]. Possibly, SinI in B. thuringiensis inactivates both SinR and a putative paralogue of SinR.
In the present work, we have shown that B. thuringiensis and B. subtilis have similarities and differences in the control of biofilm formation. The SinR regulon of the two species have only two genes in common under the conditions studied. In B. thuringiensis, the SinR regulon includes the hbl gene and the krsEABC locus, which are involved in the interaction of the bacterium with its host. The Hbl toxin complex is cytotoxic and causes damage to the intestinal tract [7], and kurstakin is required for biofilm formation and for bacterial survival in the host cadaver. Therefore, SinR co-regulates both biofilm formation and part of the infectious process in B. thuringiensis, which makes sense if we consider that this pathogen can settle in heterologous biofilms [58], and therefore potentially integrate into the biofilm microbiota lining the host intestinal epithelium. In this biofilm, toxins could be delivered directly to their target tissue.
Supporting Information
Growth curves. The various strains were grown in LB medium at 37°C and 175 rpm. The OD was measured at 600 nm and plotted against time. a: wild-type strain (black circle), spo0A mutant (white circle), abrB mutant (white triangle). b: wild-type strain (black circle), sinR mutant (white circle), sinI mutant (white triangle), sinI—sinR mutant (white inverted triangle).
(DOC)
Strains used in this study.
(DOC)
Correspondence between the locus tags in the ATCC14579 and in the 407 strains. No ortholog annotated in B. thuringiensis 407 and no DNA with similarity to the indicated B. cereus ATCC 14579 ORF present in the B. thuringiensis 407 genome. Could potentially be a result of missing data in the draft B. thuringiensis 407 sequence.
(DOC)
Acknowledgments
The flow cytometer was acquired with the financial support of the DIM Astrea program, and bioinformatics analyses were performed using computing services available from the Norwegian EMBnet node.
Funding Statement
TD was funded by the ‘Direction Générale à L'Armement’, France and IB was funded by the ‘Région Ile de France’, France. AF, OAØ and ABK were funded by a project grant from the Norwegian Research Council through the FUGE II Programme (Channel 3 grant ; project nr. 183421). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Priest FG (1993) Systematics and Ecology of Bacillus In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria. Washington: American Society for Microbiology. pp. 3–16. [Google Scholar]
- 2. Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, et al. (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis . Nature 423: 87–91. [DOI] [PubMed] [Google Scholar]
- 3. Van Schaik W, Abee T (2005) The role of sigmaB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr Opin Biotechnol 16: 218–224. [DOI] [PubMed] [Google Scholar]
- 4. Lereclus D, Agaisse H, Gominet M, Salamitou S, Sanchis V (1996) Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J Bacteriol 178: 2749–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gohar M, Okstad OA, Gilois N, Sanchis V, Kolsto AB, et al. (2002) Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2: 784–791. [DOI] [PubMed] [Google Scholar]
- 6. Gohar M, Faegri K, Perchat S, Ravnum S, Okstad OA, et al. (2008) The PlcR virulence regulon of Bacillus cereus . PLoS One 3: e2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stenfors Arnesen LP, Fagerlund A, Granum PE (2008) From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev 32: 579–606. [DOI] [PubMed] [Google Scholar]
- 8. Bottone EJ (2010) Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev 23: 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, et al. (2000) Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis - One species on the basis of genetic evidence. Appl Environ Microbiol 66: 2627–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piggot PJ, Hilbert DW (2004) Sporulation of Bacillus subtilis . Curr Opin Microbiol 7: 579–586. [DOI] [PubMed] [Google Scholar]
- 11. Hamon MA, Lazazzera BA (2001) The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis . Mol Microbiol 42: 1199–1210. [DOI] [PubMed] [Google Scholar]
- 12. Strauch M, Webb V, Spiegelman G, Hoch JA (1990) The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA 87: 1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shafikhani SH, Mandic-Mulec I, Strauch MA, Smith I, Leighton T (2002) Postexponential regulation of sin operon expression in Bacillus subtilis . J Bacteriol 184: 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA (2004) Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis . Mol Microbiol 52: 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu F, Kearns DB, Branda SS, Kolter R, Losick R (2006) Targets of the master regulator of biofilm formation in Bacillus subtilis . Mol Microbiol 59: 1216–1228. [DOI] [PubMed] [Google Scholar]
- 16. Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, et al. (2004) Genes involved in formation of structured multicellular communities by Bacillus subtilis . J Bacteriol 186: 3970–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Branda SS, Chu F, Kearns DB, Losick R, Kolter R (2006) A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59: 1229–1238. [DOI] [PubMed] [Google Scholar]
- 18. Romero D, Vlamakis H, Losick R, Kolter R (2011) An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol 80: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB (2008) A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320: 1636–1638. [DOI] [PubMed] [Google Scholar]
- 20. Kobayashi K (2008) SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis . Mol Microbiol 69: 1399–1410. [DOI] [PubMed] [Google Scholar]
- 21. Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, et al. (2008) A novel regulatory protein governing biofilm formation in Bacillus subtilis . Mol Microbiol 68: 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pflughoeft KJ, Sumby P, Koehler TM (2011) Bacillus anthracis sin locus and regulation of secreted proteases. J Bacteriol 193: 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auger S, Krin E, Aymerich S, Gohar M (2006) Autoinducer 2 affects biofilm formation by Bacillus cereus . Appl Environ Microbiol 72: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsueh YH, Somers EB, Lereclus D, Wong AC (2006) Biofilm Formation by Bacillus cereus Is Influenced by PlcR, a Pleiotropic Regulator. Appl Environ Microbiol 72: 5089–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsueh YH, Somers EB, Wong AC (2008) Characterization of the codY gene and its influence on biofilm formation in Bacillus cereus . Arch Microbiol 189: 557–568. [DOI] [PubMed] [Google Scholar]
- 26. Lindback T, Mols M, Basset C, Granum PE, Kuipers OP, et al. (2012) CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus . Environ Microbiol 14: 13. [DOI] [PubMed] [Google Scholar]
- 27. Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, et al. (2003) Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 185: 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mader U, Hennig S, Hecker M, Homuth G (2004) Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. J Bacteriol 186: 2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frenzel E, Doll V, Pauthner M, Lucking G, Scherer S, et al. (2012) CodY orchestrates the expression of virulence determinants in emetic Bacillus cereus by impacting key regulatory circuits. Mol Microbiol 85: 67–88. [DOI] [PubMed] [Google Scholar]
- 30. Dubois T, Faegri K, Perchat S, Lemy C, Buisson C, et al. (2012) Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis . PLoS Pathog 8: e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tourasse NJ, Helgason E, Okstad OA, Hegna IK, Kolsto AB (2006) The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol 101: 579–593. [DOI] [PubMed] [Google Scholar]
- 32. Sanchis V, Agaisse H, Chaufaux J, Lereclus D (1996) Construction of new insecticidal Bacillus thuringiensis, recombinant strains by using the sporulation non-dependent expression system of cryIIIA and a site specific recombination vector. J Biotechnol 48: 81–96. [DOI] [PubMed] [Google Scholar]
- 33. Lereclus D, Vallade M, Chaufaux J, Arantes O, Rambaud S (1992) Expansion of insecticidal host range of Bacillus thuringiensis by in vivo genetic recombination. Biotechnology (N Y) 10: 418–421. [DOI] [PubMed] [Google Scholar]
- 34. Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P (1995) Antibiotic-resistance cassettes for Bacillus subtilis . Gene 167: 335–336. [DOI] [PubMed] [Google Scholar]
- 35. Agaisse H, Lereclus D (1994) Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis . Mol Microbiol 13: 97–107. [DOI] [PubMed] [Google Scholar]
- 36. Arantes O, Lereclus D (1991) Construction of cloning vectors for Bacillus thuringiensis . Gene 108: 115–119. [DOI] [PubMed] [Google Scholar]
- 37. Lemon KP, Grossman AD (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282: 1516–1519. [DOI] [PubMed] [Google Scholar]
- 38. Perchat S, Dubois T, Zouhir S, Gominet M, Poncet S, et al. (2011) A cell-cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol Microbiol 82: 619–633. [DOI] [PubMed] [Google Scholar]
- 39. Kristoffersen SM, Ravnum S, Tourasse NJ, Okstad OA, Kolsto AB, et al. (2007) Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579. J Bacteriol 189: 5302–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheppard AE, Poehlein A, Rosenstiel P, Liesegang H, Schulenburg H (2013) Complete Genome Sequence of Bacillus thuringiensis Strain 407 Cry-. Genome announcements 1: e00158–00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- 42. Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99. [Google Scholar]
- 43. Dietrich R, Fella C, Strich S, Martlbauer E (1999) Production and characterization of monoclonal antibodies against the hemolysin BL enterotoxin complex produced by Bacillus cereus . Appl Environ Microbiol 65: 4470–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Agaisse H, Gominet M, Okstad OA, Kolsto AB, Lereclus D (1999) PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis . Mol Microbiol 32: 1043–1053. [DOI] [PubMed] [Google Scholar]
- 45. Okstad OA, Gominet M, Purnelle B, Rose M, Lereclus D, et al. (1999) Sequence analysis of three Bacillus cereus loci carrying PIcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology 145: 3129–3138. [DOI] [PubMed] [Google Scholar]
- 46. Gilois N, Ramarao N, Bouillaut L, Perchat S, Aymerich S, et al. (2007) Growth-related variations in the Bacillus cereus secretome. Proteomics 7: 1719–1728. [DOI] [PubMed] [Google Scholar]
- 47. Lindback T, Okstad OA, Rishovd AL, Kolsto AB (1999) Insertional inactivation of hblC encoding the L2 component of Bacillus cereus ATCC 14579 haemolysin BL strongly reduces enterotoxigenic activity, but not the haemolytic activity against human erythrocytes. Microbiology 145 (Pt 11) 3139–3146. [DOI] [PubMed] [Google Scholar]
- 48. Kearns DB, Chu F, Branda SS, Kolter R, Losick R (2005) A master regulator for biofilm formation by Bacillus subtilis . Mol Microbiol 55: 739–749. [DOI] [PubMed] [Google Scholar]
- 49. Irnov I, Winkler WC (2010) A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales . Mol Microbiol 76: 559–575. [DOI] [PubMed] [Google Scholar]
- 50. Msadek T (1999) When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis . Trends Microbiol 7: 201–207. [DOI] [PubMed] [Google Scholar]
- 51. Beecher DJ, Schoeni JL, Wong AC (1995) Enterotoxic activity of hemolysin BL from Bacillus cereus . Infect Immun 63: 4423–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chai Y, Chu F, Kolter R, Losick R (2008) Bistability and biofilm formation in Bacillus subtilis . Mol Microbiol 67: 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramarao N, Lereclus D (2005) The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell Microbiol 7: 1357–1364. [DOI] [PubMed] [Google Scholar]
- 54. Dalhammar G, Steiner H (1984) Characterization of inhibitor A, a protease from Bacillus thuringiensis which degrades attacins and cecropins, two classes of antibacterial proteins in insects. Eur J Biochem 139: 247–252. [DOI] [PubMed] [Google Scholar]
- 55. Grandvalet C, Gominet M, Lereclus D (2001) Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147: 1805–1813. [DOI] [PubMed] [Google Scholar]
- 56. Kobayashi K (2007) Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol 189: 4920–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chai Y, Kolter R, Losick R (2009) Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis . Mol Microbiol 74: 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Houry A, Gohar M, Deschamps J, Tischenko E, Aymerich S, et al. (2012) Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc Natl Acad Sci USA 109: 13088–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curves. The various strains were grown in LB medium at 37°C and 175 rpm. The OD was measured at 600 nm and plotted against time. a: wild-type strain (black circle), spo0A mutant (white circle), abrB mutant (white triangle). b: wild-type strain (black circle), sinR mutant (white circle), sinI mutant (white triangle), sinI—sinR mutant (white inverted triangle).
(DOC)
Strains used in this study.
(DOC)
Correspondence between the locus tags in the ATCC14579 and in the 407 strains. No ortholog annotated in B. thuringiensis 407 and no DNA with similarity to the indicated B. cereus ATCC 14579 ORF present in the B. thuringiensis 407 genome. Could potentially be a result of missing data in the draft B. thuringiensis 407 sequence.
(DOC)