Abstract
In the fission yeast Schizosaccharomyces pombe a chromosomal imprinting event controls the asymmetric pattern of mating-type switching. The orientation of DNA replication at the mating-type locus is instrumental in this process. However, the factors leading to imprinting are not fully identified and the mechanism is poorly understood. Here we show that the replication fork pause at the mat1 locus (MPS1), essential for imprint formation, depends on the lysine specific demethylase, Lsd1. We demonstrate that either Lsd1 or Lsd2 amine oxidase activity is required for these processes, working upstream of the imprinting factors Swi1 and Swi3 (homologs of mammalian Timeless and Tipin, respectively). We also show that the Lsd1/2 complex controls the replication fork terminators, within the rDNA repeats. These findings reveal a novel role for the Lsd1/2 demethylases in controlling polar replication fork progression, imprint formation and subsequent asymmetric cell divisions.
Keywords: Lsd1, DNA replication, fork barrier, imprint, asymmetric division, S. pombe
Introduction
Haploid S. pombe cells exist as two mating-types (MT), P (for plus) and M (for minus) that switch during cell divisions. A chromosomal imprinting event at the MT locus, mat1, controls the asymmetric pattern of MT switching in a cell lineage (Reviewed in Klar, 2007). It was shown that the polarity of DNA replication of mat1 is instrumental in the establishment of the imprint (Dalgaard and Klar, 1999) on one of the two sister chromatids and for MT switching during the following DNA replication (Arcangioli, 1998 and Figure 1A). One replication termination site (RTS1) located on the proximal side of the mat1 locus, together with the nearby origin of DNA replication ARS756, located a few Kbps distal to mat1, restrict the DNA replication of mat1 from its distal side (Dalgaard and Klar, 2001). In this configuration the second replication pause site (MPS1) located on the distal side of mat1, is optimized and competent for the establishment of imprinting (Klar, 1987; Dalgaard and Klar, 1999).
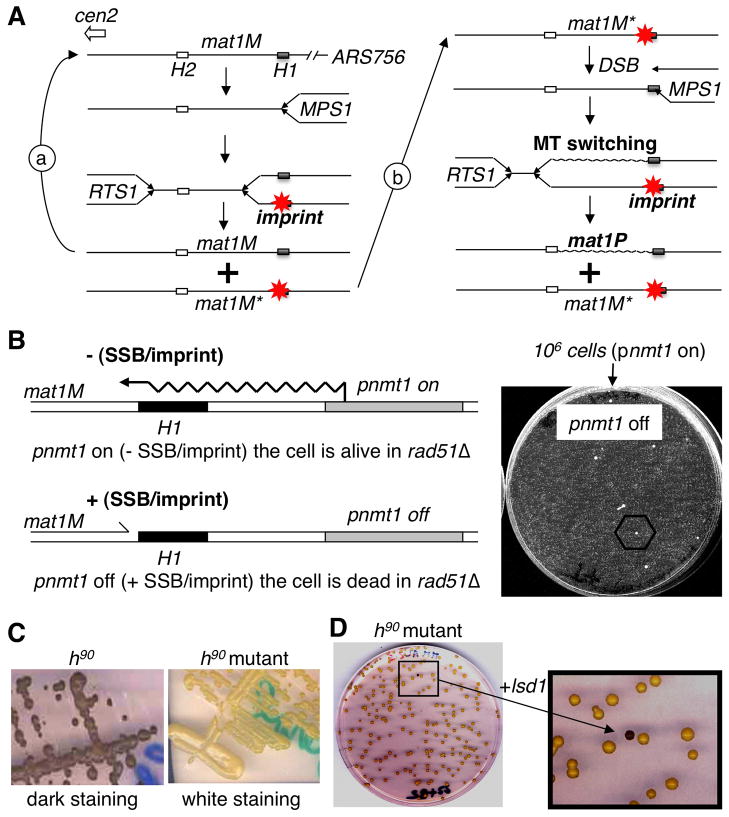
Figure 1. Selection and complementation of a new imprinting mutant.
(A) Schematic representation of two consecutive rounds of replication at mat1. Starting from a virgin mat1M locus (left). The close proximity of ARS756 with RTS1, constrains the polarity of mat1 replication to ensure optimal imprinting (*) at MPS1. Following DNA replication only one of the two mat1M loci is imprinted. During the following DNA replication: (a) replication of the virgin mat1M, (b) replication of the imprinted mat1M; the fork transforms the imprint or single-strand break into a polar one ended double-strand break (DSB) that promotes MT switching (mat1P) and the imprint is formed on the unswitched mat1M sister locus. The transformation of the imprint into a DSB requires homologous recombination (i.e. Rhp51) for cell survival.
(B) Selection of spontaneous survivors in the selection system. Left panel: By introducing the thiamine repressible promoter (pnmt1) upstream of mat1, we can force transcription through the imprinted region, erasing the imprint (-SSB), in a reversible manner (+SSB). Right panel: 106 cells grown under permissive conditions (pnmt1 on) are plated in the presence of thiamine (pnmt1 off) and only a few spontaneous mutants can form colonies.
(C) Iodine staining of mutants. By crossing the initial nbt-18 mutants with the wild-type h90 strain we introduced the mutation in the wild-type background and assayed by iodine staining. The left panel shows the dark staining of the wild-type h90 strain and the right panel shows the white staining of the h90strain carrying the mutant allele.
(D) Functional complementation. The h90 strain carrying the mutation (white) is transformed with a genomic library and colonies are assayed for coloration by iodine vapors in order to reveal the complemented cells (black colony in the enlarged view).
The imprint is made on the newly synthesized lagging strand during the resumption of DNA synthesis at MPS1 (Dalgaard and Klar, 1999; Holmes et al., 2005), and has been mapped at the nucleotide level (Nielsen and Egel, 1989). Several cis-acting elements and trans-acting factors are required for imprint formation (Egel et al., 1984; Arcangioli and Klar, 1991; Kaykov et al., 2004; Sayrac et al., 2011). Four gene products, Swi1, Swi3, Swi7 and Sap1, are necessary for imprinting (Egel et al., 1984; Arcangioli et al., 1994). Swi1 and Swi3 promote imprinting by pausing the replication fork at MPS1 (Dalgaard and Klar, 2000) and accumulate during S-phase at MPS1 (Kaykov et al., 2004; Holmes et al., 2005). The Swi1/3 complex stabilizes replication fork blocks at three locations, MPS1 and RTS1 at the mat1 locus and at RFBs at the rDNA loci. The stabilized fork at mat1 promotes gene conversion and subsequent mating-type switching (Klar, 2007), whereas, the stabilized fork at the rDNA maintains the number of copies (Sommariva et al., 2005). It was also shown that RTS1 is not recombinogenic at its endogenous locus, but when placed ectopically, it generates replication instability (Lambert et al., 2005; Ahn et al., 2005). Thus, Swi1/3 functions differentially to control the recombinogenic potential of site-specific replication fork barriers (Pryce et al., 2009). The swi7 gene encodes for the essential large catalytic polymerase α subunit (Singh and Klar, 1993) and Sap1 is an essential gene product important for replication termination (Arcangioli et al., 1994; Mejia-Ramirez et al., 2005; Krings and Bastia, 2005; Zaratiegui et al., 2011).
LSD1/KDM1B is a FAD-dependent lysine specific demethylase enzyme that represses transcription by demethylating histone H3 (H3K4me1 and H3K4me2) (Shi et al., 2004) and activates transcription by demethylating histone H3 (H3K9me1 and H3K9me2) (Metzger et al., 2005). LSD1 specificity, and therefore downstream function, is dictated by the associated DNA-binding transcription factors. LSD1 also demethylates non-histone proteins, such as the tumor suppressor protein p53 (Huang et al., 2007) and DNA methyltransferase 1 (Dnmt1) (Wang et al., 2009). A second protein related to LSD1 named KDM1B is thought to contribute to the resetting of epigenetic marks in germ cells (Katz et al., 2009; Ciccone et al., 2009). In fission yeast, two histone demethylases, Lsd1 and Lsd2, have been identified within a complex (Nicolas et al., 2006). Lsd1 is required for optimal cell growth, whereas Lsd2 is essential for viability. Lsd1 has been co-purified along with Lsd2 and two PHD finger proteins, Phf1 and Phf2, also essential for viability (Nicolas et al., 2006; Gordon et al., 2007; Opel et al., 2007 and Lan et al., 2007). Lsd1 exhibits a weak in vitro H3K9 demethylation activity consistent with a slight in vivo global increase in H3K9 methylation of the lsd1 catalytically-dead mutant. Furthermore, the double catalytically inactive lsd1-ao and lsd2-ao strain is viable, hence does not fully phenocopy the deletion of lsd1 (or lsd2), strongly indicating a nonenzymatic role for these two proteins (Opel et al., 2007; Gordon et al., 2007 and Lan et al., 2007). Finally, Lsd1 acts at boundary elements between euchromatin and heterochromatin in fission yeast and Drosophila (Lan et al., 2007; Gordon et al., 2007; Rudolph et al., 2007; Li et al., 2008).
Almost thirty years ago, a genetic screen in S. pombe identified switching (swi) mutants (Egel et al., 1984). In this study, we used an inducible MT switching system (Holmes et al., 2005 and Roseaulin et al., 2008) to identify new imprinting mutants. Among the imprinting candidates discovered, we found Lsd1. Combining genetic and molecular approaches, we examined which step of the MT switching process, Lsd1 intervenes. Our findings are predicted to have important implications for cellular differentiation and development in eukaryotes.
Results
Lsd1 - a new MT switching mutant
We recently showed that the homologous recombination machinery is essential for viability when the replication fork collides with the imprint at mat1 (Roseaulin et al, 2008). This allowed us to convert the inducible imprinting strain (Holmes et al., 2005) into a conditional strain, by deleting the rhp51 gene, a critical protein involved in the process of homologous recombination. We reasoned that imprinting mutants, arising spontaneously during permissive growth conditions, would allow the rhp51Δ strain to survive, when plated in non-permissive growth conditions (Figure 1B). Using this approach, 100 spontaneous mutants were isolated from 100 independent cultures.
Each mutant was backcrossed with the wild type (h90, MT switching proficient) strain in order to introduce the mutated allele into the wild-type background for assaying MT switching phenotypes. Our first assay relied on the staining of starch, produced during sporulation with iodine staining (Figure 1C). Using this assay, we observed that all of the mutants exhibited white or streaky iodine staining patterns, indicating a drastic reduction of MT switching and sporulation. As Swi1 and Swi3 were previously characterized as imprinting genes, all of the mutants were individually transformed with plasmids expressing Swi1 or Swi3 (more than 90% of the mutants). The mutant nbt-18 was neither complemented by swi1 nor swi3 and was therefore transformed with a bank of plasmid-containing genomic DNA. Among more than 40,000 white colonies, a single black colony was isolated (Figure 1D) and the partial sequence of the corresponding plasmid-inserted genomic DNA identified the lysine specific demethylase gene, lsd1/swirm1 (SPBC146.09c) (Nicolas et al., 2006).
The HMG domain of Lsd1 is essential for replication fork pausing at MPS1
Sequence analysis of the lsd1 gene, from the nbt-18 strain, revealed a nonsense mutation (GAA into TAA) at position E918 (called lsd1-E918) within the high mobility group (HMG/B) domain (Figure 2A). To determine whether the HMG domain or the C-terminal end of Lsd1 is required for function at mat1, two mutants were constructed. The lsd1-W891A mutation converts tryptophan at position 891 into an alanine and the lsd1-P946 mutation converts proline at position 946 into a stop codon (Figure 2A and 2B). Subsequently, we replaced the endogenous wild-type lsd1 gene with these mutated alleles. We found that the lsd1Δ null mutant strain confers white iodine staining along with severe growth defects, whereas lsd1-E918 and lsd1-W891A confer white staining and mildly reduced growth and lsd1-P946 cells behave like the wild type strain. This indicates that the C-ter region is dispensable, whereas the HMG domain is necessary for wild type functions.
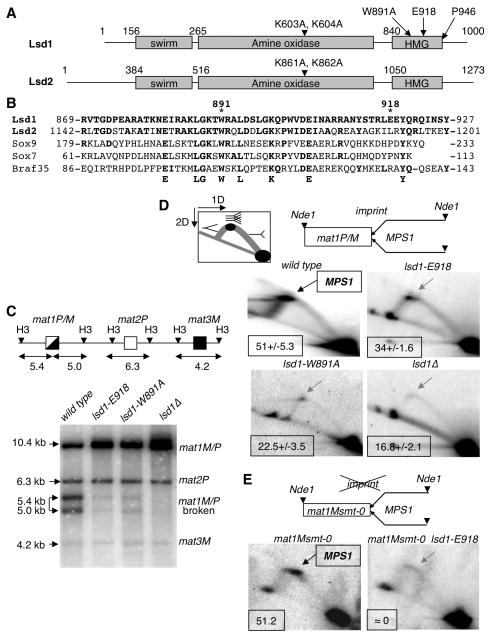
Figure 2. The Lsd1 HMG domain is required for DSB and MPS1 activity at mat1.
(A) Schematic representation of Lsd1 and Lsd2. The protein domains and the mutations in lsd1 and lsd2 are indicated.
(B) Alignment of the Lsd1 HMG domain with HMG domains from Lsd2, Sox9, Sox7 and Braf35 from mouse. The conserved residues are indicated.
(C) lsd1 mutants affect the level of the break at mat1. Top panel: Schematic representation of the mat region, showing mat1P/M, mat2P and mat3M cassettes. The sizes of the HindIII fragments (H3) are indicated (Kbp). Lower panel, Southern blot of HindIII digested DNA from the lsd1, lsd1-E918, lsd1-W891A and lsd1Δ strains, hybridized with the labeled mat1P HindIII-containing fragment. The mat1P probe also hybridizes with the mat2P and mat3M cassettes. The sizes and identities of the DNA fragments are indicated.
(D) Lsd1 is required for MPS1 activity at mat1P/M. Top panel: diagram of the migration pattern of the replication intermediates detected in 2D gel electrophoresis. The position of the imprint, MPS1 pause site and the polarity of DNA replication are indicated. DNA replication intermediates at mat1 from the wild type, lsd1-E918, lsd1-W891A and lsd1Δ strains are analyzed (lower panel). In the wild type panel the DNA replication intermediates accumulating at MPS1 are indicated by an arrow. The percentage of pause is shown for each strain from 2 to 3 independent 2D gels.
(E) Lsd1 is required for MPS1 activity in mat1Msmt-0 (deletion of 263 bp distal to mat1). DNA replication intermediates from the Msmt-0 and Msmt-0 lsd1-E918 strains were analyzed and show the decrease of MPS1 activity in the lsd1-E918 background.
To determine when Lsd1 is required for the MT switching process, we examined the imprint at mat1. The imprint (SSB or ribonucleotides) behaves as a fragile site and is transformed into a DSB (breaking the HindIII restriction fragment of 10.4 kb containing mat1 into two fragments of 5.4 kb and 5.0 kb, Figure 2C), when the genomic DNA is extracted by the conventional DNA purification procedure (Beach, 1983; Arcangioli 1998; Dalgaard and Klar, 1999). Southern blot analysis shows that the level of DSB formation at mat1 is drastically reduced in the lsd1-E918, lsd1-W891A and lsd1Δ strains compared to the wild type (Figure 2C). We then examined the same mutants for their role in promoting replication fork pausing at MPS1 by native 2D gel electrophoresis. In contrast to the prominent pause site induced by MPS1 in wild-type cells, MPS1 activity is significantly reduced in lsd1-E918, lsd1-W891A and in lsd1Δ strains (Figure 2D). We also analyzed the activity of MPS1 in the mat1-Msmt-0 strain, which contains a 263 bp deletion of the cis-acting elements necessary for imprint formation/maintenance but not for MPS1 activity. We found that MPS1 activity in this context was also Lsd1-dependent (Figure 2E). Taken together, these results demonstrate that the HMG domain of Lsd1 is required for replication pausing at MPS1, a pre-requisite and early step of imprinting.
MPS1 activity requires either Lsd1 or Lsd2 amine oxidase activities
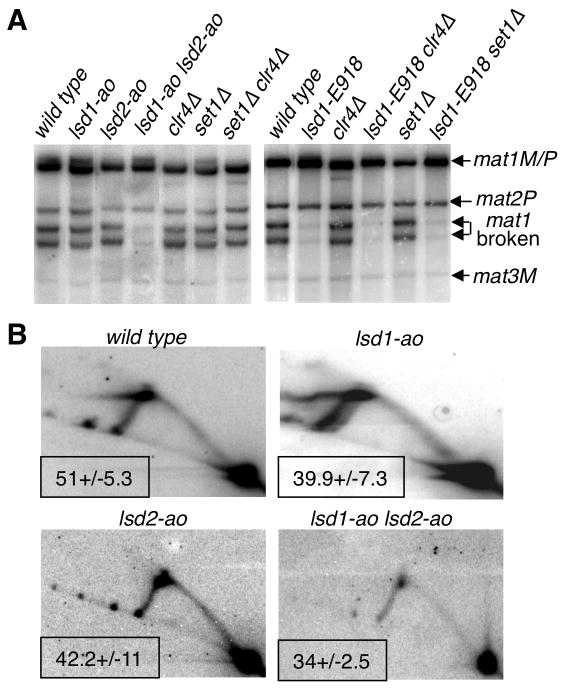
Because a non-enzymatic role for Lsd1 has been proposed (Lan et al., 2007, Gordon et al., 2007), we examined mutant strains, harboring the catalytically dead, lsd1-ao or lsd2-ao, in which K603 and K604 or K861 and K862 are substituted with alanine residues, respectively (Shi et al., 2004; Lan et al., 2007, Gordon et al., 2007 and Figure 2A). We found that the two catalytically dead mutants exhibit DSB and MPS1 activities similar to the wild-type strain (Figure 3A and 3B), whereas the double mutant exhibits a strong reduction in DSB levels and a more subtle reduction of the MPS1 levels (Figure 3A and 3B). Taken together, these findings indicate that either Lsd1 or Lsd2 amine oxidase activities are required to control replication fork progression at MPS1 and imprint formation/maintenance.
Figure 3. Lsd1/2 amine-oxidase activities are required for replication fork pausing at MPS1 and imprinting.
(A) Analysis of the DSB at mat1. The genomic DNA is analyzed by Southern blot, as in Figure 2. The relevant genotypes of the strains, the sizes and identities of the DNA fragments are indicated.
(B) MPS1 activity in demethylase mutants. 2D gel analysis of the replication fork intermediates at mat1, showing that the accumulating replication material at the apex of the Y-arc is not significantly reduced in single lsd1-ao and lsd2-ao mutants but is diminished in the double mutant strain. The gels are treated as in Figure 2 and the relevant genotypes are indicated. The percentage of pause is indicated for each strain from 2 independent 2D gels.
Contrary to higher eukaryotes, the fission yeast Set1 and Clr4 methyltransferases are the dominant (if not the only) methyltransferases for H3K4 and H3K9, respectively (Noma and Grewal, 2002; Nakayama et al., 2001; Cam et al., 2005). Therefore, we analyzed the methyltransferase and demethylase single and double mutant strains for imprinting by southern blot analysis (Figure 3A). We found that single set1Δ, clr4Δ and double set1Δ clr4Δ mutants exhibit DSB levels similar to the wild type (Figure 3A), indicating that H3K4 and H3K9 demethylation by Lsd1 and Lsd2, is not involved in imprint formation. Moreover, we found that lsd1-E918 set1Δ and lsd1-E918 clr4Δ double mutants exhibit similarly low DSB (Figure 3A) and MPS1 pause levels (data not shown) at mat1 as compared to the single lsd1-E918 mutants, indicating that the lysine methylation status at H3K4 and H3K9 is not involved in DSB or imprinting formation.
Lsd1 is directly controlling MPS1 within the mat1 locus
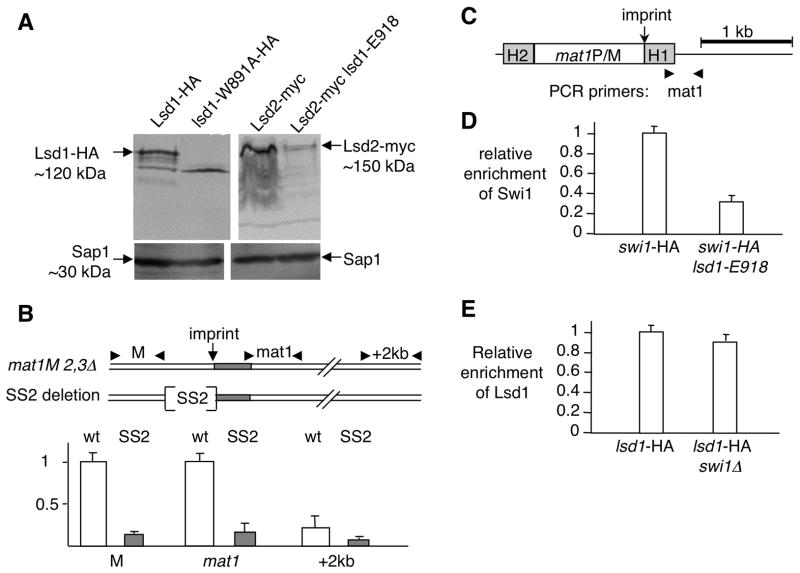
We confirmed that Lsd1-HA and Lsd2-MYC, tagged proteins are greatly enriched at the imprinted site at mat1 (Nicolas et al., 2006; Lan et al., 2007; Gordon et al., 2007) but are lost in the lsd1-HMG mutant background (data not shown). We reasoned that the dramatic decrease of Lsd1 and Lsd2 enrichment in the context of the lsd1-HMG mutations might result from protein instability rather than problems of direct recruitment at mat1. Western blot experiments showed that the lsd1-W891A-HA protein is unstable. Similarly, the Lsd2-myc protein in the presence of the lsd1-E918 mutated allele is strongly destabilized (Figure 4A). Knowing that Lsd2 is essential for cell viability, we infer that the remaining level of Lsd2, in the lsd1-E918 mutant (Figure 4A), is sufficient to ensure cell viability, but not imprinting at mat1. It was previously shown that lsd1-ao and lsd2-ao single and double mutant strains exhibit wild type levels of both proteins (Gordon et al., 2007 and this work, data not shown), indicating that the catalytic activity of both proteins are not necessary for protein stability and that the HMG domain of Lsd1 contributes to the non-enzymatic role of the Lsd1/2 complex.
Figure 4. Lsd1 functions at mat1 to promote replication fork pausing at MPS1.
(A) The HMG domain of Lsd1 stabilizes the Lsd1/2 complex. Western blot of Lsd1-HA and Lsd2-myc tagged proteins in wt and HMG mutant strains. Sap1 protein is used as a loading control.
(B) Lsd1 binds to the SS2 cis-acting element proximal to the imprint site at mat1. Schematic representation of the wild type (wt) and deletion (SS2: 110 bp) region and primers used for the ChIP experiment. The two sets of primers are localized ~200 bp to the left (M) and ~100 bp to the right of the imprint (mat1). Quantitative PCR analysis of the ChIP for both wt lsd1-myc and SS2 lsd1-myc strains are shown.
(C) Schematic representation of the mat1 region and primers used for the ChIP experiment. The PCR primer pair (mat1) overlaps the imprinting regulatory region from mat1.
(D) Lsd1 recruitment is Swi1-independent. Quantitative PCR analysis of the ChIP of Lsd1-HA in the swi1+ and swi1 Δ backgrounds. Lsd1- HA swi1+ was set to 1.
(E) Swi1 recruitment is Lsd1-dependent. Quantitative PCR analysis of the ChIP of Swi-HA in the lsd1+ and lsd1-E918 backgrounds. Swi1-HA lsd1+ was set to 1.
Another way to directly implicate Lsd1 function at mat1 is to assay the enrichment of Lsd1 by ChIP on strains containing deletions of cis-acting elements required for imprinting. Recently, a small deletion (called SS2) within the mat1-M locus has been reported to be important for both the replication fork pause (MPS1) and the imprint at mat1 (Sayrac et al., 2011). Thus we compared the enrichment of Lsd1 in the wild type and SS2 mutant backgrounds by ChIP (Figure 4B). Both strains are deleted for the silent donors, mat2-P and mat3-M cassettes, to avoid removal of the deletions at mat1 during MT switching and interference during PCR amplification. The mat2-mat3 deletion strain retains wild type pausing and imprinting functions (Klar and Miglio, 1986; Dalgaard and Klar, 1999; Roseaulin et al., 2008). The data (Figure 4B) shows that Lsd1 enrichment depends on SS2, indicating that the Lsd1/2 complex is directly controlling replication fork pausing at MPS1, hence imprinting through this cis-acting element.
Lsd1 functions upstream of Swi1 to control MPS1 activity
Consistent with the association of Swi1 with the replication fork, Swi1 accumulates at MPS1 during S-phase and is released during early G2 of the cell cycle (Holmes et al., 2005). Using the inducible MT switching strain, we found that Lsd1-HA is present at mat1 before S phase, i.e. before Swi1 accumulation, and remains stably associated with mat1 during the entire length of the cell cycle (Figure S1). To further investigate the relative order of action of Lsd1 and Swi1 in this process, we analyzed by ChIP-qPCR (Figure 4C) the presence of Lsd1-HA at MPS1 in the swi1Δ mutant background (Figure 4D) and conversely the presence of Swi1-HA at MPS1 in the lsd1-E918 mutant background (Figure 4E). We found that Swi1 is not required for the enrichment of Lsd1-HA at MPS1, whereas Lsd1 is required for the accumulation of Swi1-HA at MPS1, consistent with Lsd1 controlling MPS1 upstream of Swi1/3 activity.
Swi1 and Swi3 not only stabilize replication forks at MPS1, but also are required to preserve genomic integrity when the cells are treated with drugs affecting DNA replication or destabilizing microtubules (Noguchi et al., 2004). We observed (Figure S2) little or no effect for the cells mutated for Lsd1, indicating that Lsd1/2 activity does not simply overlap with all Swi1/3 functions.
Lsd1 and Lsd2 control the replication fork barriers at RFB within the rDNA repeats
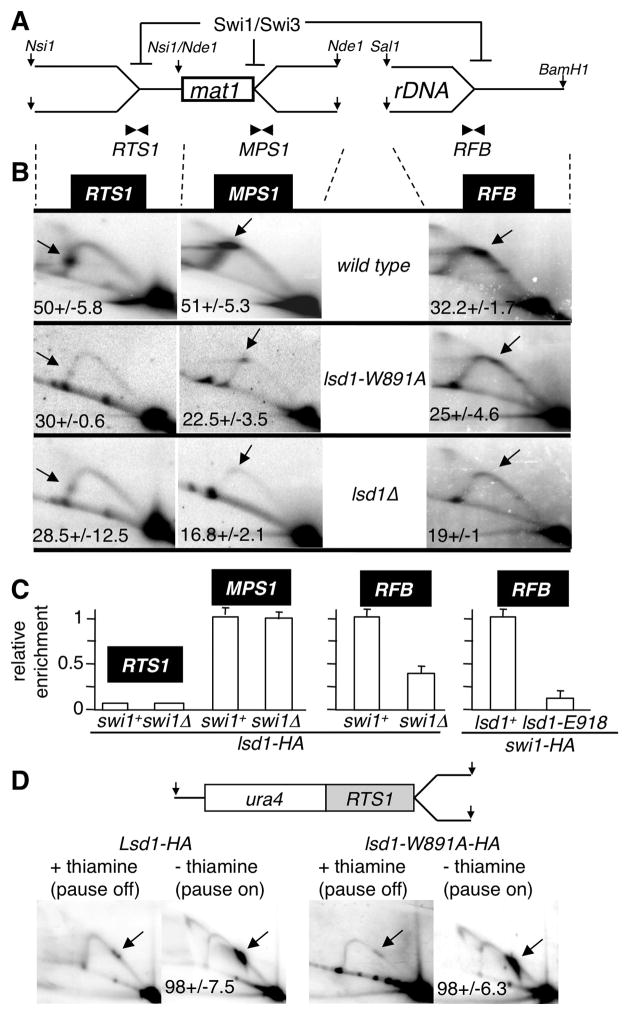
The Swi1/3 complex controls two other known site-specific replication fork barriers in fission yeast, RTS1 located 700 bp proximal to mat1 and RFB, within the rDNA locus (Dalgaard and Klar, 2000; Sanchez et al., 1998; Noguchi et al., 2003). The lsd1-W891A and lsd1Δ mutants exhibit a significant reduction of the replication fork arrest at RTS1 and RFB as observed by 2D gel electrophoresis analysis (Figure 5B). These results show that Lsd1 controls several site-specific replication pauses, revealing a novel epigenetic program controlling DNA replication fork progression. We found that Lsd1-HA is significantly enriched at RFB but only weakly at RTS1 by ChIP-qPCR analysis (Figure 5C). As observed at MPS1, Swi1 enrichment at RFB is severely reduced (≈7 fold) in the lsd1-E918 background. The result for RTS1 is intriguing and suggests an indirect role of Lsd1 at this site. To further assess the role of Lsd1 at RTS1, we used the ectopic RTS1 system on chromosome 3, known to induce fork pausing and homologous recombination (Lambert et al., 2005; Ahn et al., 2005). RTS1 activity is controlled by the expression of the rtf1 ORF under control of the thiamine repressible promoter nmt41 (Lambert et al., 2005; Eydmann et al., 2008). We found that in cells harboring the lsd1-W891A mutant, ectopic RTS1 activity is similar to wild-type cells (Figure 5D), ruling out a direct action of Lsd1 at RTS1. A simple model is that the lack of MPS1 activity in the absence of Lsd1 allows the replication fork to replicate through RTS1, in a non-active orientation, prior to the arrival of the converging fork. In this context, the polar terminating RTS1 activity will appear Lsd1-dependent, but indirectly through MPS1 inactivation.
Figure 5. Lsd1 controls directly RFB and indirectly RTS1 replication fork barriers.
(A) Schematic representation of the Swi1/Swi3-dependent DNA replication fork pauses and arrests on the proximal (RTS1) and distal (MPS1) sides of mat1 and at the rDNA (RFB) loci. The restriction enzymes used for genomic digestions are indicated.
(B) 2D gel autoradiograms of RTS1 (left), MPS1 (middle) and RFB (right) in the wild type (top), lsd1-W891A (middle) and lsd1Δ (bottom) strains. The restriction enzymes used are indicated. Arrows show the accumulating DNA replication material at pause/arrest sites.
(C) ChIP analysis of Lsd1-HA at RTS1, MPS1 and RFB. Quantitative PCR analysis of the ChIP of lsd1-HA in swi1+ and swi1Δ backgrounds using primer pairs close to the pause/arrest sites (RTS1, MPS1 and RFB). On the left panel, Lsd1-HA swi1+ was set to 1 at MPS1 (as a single copy locus) and on the right panel, Lsd1-HA swi1+ was set to 1 at RFB, independently due to the high rDNA copy number.
(D) RTS1 activity is lsd1-independent. Schematic representation of the Ase1 restriction fragment containing the RTS1 pause at the ura4 locus on chromosome 3 and 2D gel autoradiograms. RTS1 activity is “off” by repressing (+ thiamine) and “on” by inducing (- thiamine) expression of the barrier protein Rtf1, as indicated. 2D gels were probed using the ura4 fragment.
Discussion
In this work we have identified the lysine-specific demethylase 1 (lsd1) gene as a new mating-type switching player. We found that Lsd1 and Lsd2 work in a redundant manner, upstream of the Swi1/3 complex to promote replication fork pausing at MPS1 and imprinting at mat1. We further showed that Lsd1 interacts in vivo with the cis-acting element, SS2, within mat1 (Figure 4B), demonstrating its role in the initial pausing step, prior to imprinting. Furthermore, the well-conserved replication fork block (RFB) at the rDNA loci was also found to be under the control of Lsd1/2 in S. pombe. Altogether, these results lead to the idea that the histone H3K4 or H3K9 methylation status might control replication fork progression. Intriguingly, the dedicated methyltransferases (Set1 and Clr4) were not involved in this regulation, indicating a novel mechanism to regulate replication fork progression in eukaryotes. Thus, Lsd1 and Lsd2 control MPS1 pausing activity, the epigenetic initiator event marking the newly replicated lagging strand at the mat1 locus to ensure asymmetric cell division (Klar, 1987).
In mammals, the LSD1 protein is not fused to an HMG domain, as in S. pombe, but instead the LSD1 complex contains an HMG-containing protein called BRAF35 (Hakimi et al., 2002 and Figure 2B). We found that mutation of the HMG domain of Lsd1 significantly decreases the level of Lsd1 and Lsd2 proteins. Similarly, in mammals, it was shown that CoREST recruits LSD1 to chromatin and protects LSD1 from proteasomal degradation in vivo, suggesting that in fission yeast and mammals, chromatin-free Lsd1 is unstable (Shi et al., 2005, Perillo et al., 2008).
In addition to Set1 and Clr4 methyltransferases we further showed that Set6 (unknown target) and Set9 (H4K20) (Sanders et al., 2004) play no significant role in imprinting formation (Figure 3 and S3). Furthermore, we found that a mutation in the JmjC domain of Lid2 (lid2-j) does not affect MT switching and Lsd1 recruitment at MPS1, indicating that Lid2, a trimethyl H3K4 demethylase (Li et al., 2008), is not involved in the imprinting process at mat1 (Figure S4). Therefore, the well-characterized methylated lysine targets within histone H3 and H4 do not appear to be required for imprinting at mat1.
In S. pombe, the imprint requires MPS1 pausing activity and marks the lagging strand after replication restart. This implies that Lsd1 functions within the chromatin prior to or upon the arrival of the replication fork machinery. Interestingly, it was recently proposed in C. elegans that short truncations within the C-terminal domain of histone H3 prevent efficient nucleosomal assembly on newly replicated DNA in a chromatin assembly factor-1 (CAF-1) dependent manner (Nakano et al., 2011). This supports the notion that Lsd1 in S. pombe could target the C-terminus of histone H3 to generate different sister genomes, although any non-histone protein involved in replication fork pause, progression or stabilization at MPS1, are also potential targets. Primary candidates could be barrier proteins, replicative DNA helicases, the Swi1/3 complex or CAF-1 assembly factor.
Experimental Procedures
Fission Yeast strains and Genetic Procedures
The S. pombe strains used in this study are listed in Table S1. Standard molecular genetic protocols for fission yeast were previously described (Moreno et al., 1991). A C-terminal HA tag of lsd1 was constructed using the PCR-based gene tagging system (Bähler et al, 1998).
Drop Assays
Cells are grown at mid-exponential phase, diluted and lawn on YES plate. The incubation was performed three days at 32°C.
ChIP Assays
Exponentional cultures were fixed with 1% formaldehyde for 15 min. Spheroplastes were sonicated and the chromatin (0.2–0.5 Kbp) was immunoprecipitated by antibodies (anti-HA affinity matrix from Roche and anti-MYC coupled to protein G-sepharose 4B from Sigma). Recovered DNA was analyzed by PCR, the amplified products separated on 1.5% agarose gels and stained with ethidium bromide. Relative enrichments were obtained by first normalizing the ChIP DNA value by the total input DNA value (WCE). The normalized ChIP DNA value of the wild type background was set to 1 and compared to the normalized ChIP DNA value of the mutants. The ChIP experiments were repeated at least twice, with independent cultures and provided similar results. The sequence of the oligonucleotides is available upon request.
Southern Blot Analysis
Extracted genomic DNA (Moreno et al., 1991) was digested with the restriction enzyme HindIII, separated by 0.8% agarose gel electrophoresis and blotted onto Hybond-N+ nylon membranes. The probes were labeled with alpha-32P and the blot visualized with a phosphoImager.
2D gel Analysis
2D gel analysis of replication intermediates was carried out as described (Brewer and Fangman, 1988). DNA was prepared and digested, with the indicated restriction enzymes, in agarose plugs (Kaykov et al., 2004). Enriched fractions for replication intermediates were obtained using BND cellulose columns. Gels were blotted on Hybond-N+ nylon membrane. The probes were labeled with alpha-32P and the blot visualized with phosphoImager and quantified with ImageQuant software (pause/Y arc + pause in percentage).
Supplementary Material
Highlights.
The Lysine specific demethylase, Lsd1, is a new mating-type switching mutant
Lsd1 is essential for replication fork pausing at MPS1
Lsd1 is essential for replication fork arrest at RFB
Lsd1 and Lsd2 act in a redundant manner for replication fork pausing
Acknowledgments
We thank the scientific community for yeast strains, in particular J.Z. Dalgaard for SS2, F. Lan and Y. Shi for lsd1-ao and lsd2-ao and K. Ekwall; X. Sun and J. Pelloux for their help and J. Cooper for critical reading of the manuscript. This work was supported by Grants from the ARC and conseil regional de Martinique to L.R. and ANR-06-BLAN-0271 to S.L. and B.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JS, Osman F, Whitby MC. Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J. 2005;24:2011–2023. doi: 10.1038/sj.emboj.7600670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B, Klar AJ. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 1991;10:3025–3032. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B, Copeland TD, Klar AJ. Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:2058–2065. doi: 10.1128/mcb.14.3.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 1998;17:4503–4510. doi: 10.1093/emboj/17.15.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;10:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Beach DH. Cell type switching by DNA transposition in fission yeast. Nature. 1983;305:682–687. [Google Scholar]
- Brewer BJ, Fangman WL. A replication fork barrier at the 3′end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. Orientation of DNA replication establishes mating-type switching patterns in S. pombe. Nature. 1999;400:181–184. doi: 10.1038/22139. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. A DNA replication-arrest site in RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 2001;15:2060–2068. doi: 10.1101/gad.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R, Beach DH, Klar AJ. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci USA. 1984;81:3481–3485. doi: 10.1073/pnas.81.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eydmann T, Sommariva E, Inagawa T, Mian S, Klar AJ, Dalgaard JZ. Rtf1-mediated eukaryotic site-specific replication termination. Genetics. 2008;180:27–39. doi: 10.1534/genetics.108.089243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M, Holt DG, Panigrahi A, Wilhelm BT, et al. Genome-wide dynamics of SAPHIRE, an essential complex for gene activation and chromatin boundaries. Mol Cell Biol. 2007;27:4058–4069. doi: 10.1128/MCB.02044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AM, Kaykov A, Arcangioli B. Molecular and cellular dissection of mating-type switching steps in Schizosaccharomyces pombe. Mol Cell Biol. 2005;25:303–311. doi: 10.1128/MCB.25.1.303-311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Kaykov A, Holmes AM, Arcangioli B. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 2004;23:930–938. doi: 10.1038/sj.emboj.7600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987;326:466–470. doi: 10.1038/326466a0. [DOI] [PubMed] [Google Scholar]
- Klar AJ. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Miglio LM. Initiation of meiotic recombination by double-strand breaks in S. pombe. Cell. 1986;46:725–731. doi: 10.1016/0092-8674(86)90348-x. [DOI] [PubMed] [Google Scholar]
- Krings G, Bastia D. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem. 2005;280:39135–39142. doi: 10.1074/jbc.M508996200. [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, et al. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Lan F, Zaratiegui M, Villén J, Vaughn MW, et al. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Li F, Huarte M, Zaratiegui M, Vaughn MW, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Ramírez E, Sánchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernández P. The mating type switch-activating protein Sap1 is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol. 2005;25:8755–8761. doi: 10.1128/MCB.25.19.8755-8761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nakano S, Stillman B, Horvitz HR. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell. 2011;147:1525–1536. doi: 10.1016/j.cell.2011.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, et al. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Lee MG, Hakimi MA, Cam HP, et al. Fission yeast homologs of human histone H3 lysine 4 demethylase regulate a common set of genes with diverse functions. J Biol Chem. 2006;281:35983–35988. doi: 10.1074/jbc.M606349200. [DOI] [PubMed] [Google Scholar]
- Nielsen O, Egel R. Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J. 1989;8:269–276. doi: 10.1002/j.1460-2075.1989.tb03373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, et al. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci U S A. 2002;99:16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel M, Lando D, Bonilla C, Trewick SC, et al. Genome-wide studies of histone demethylation catalysed by the fission yeast homologues of mammalian LSD1. PLoS One. 2007;2:e386. doi: 10.1371/journal.pone.0000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Pryce DW, Ramayah S, Jaendling A, McFarlane RJ. Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swi1. Proc Natl Acad Sci U S A. 2009;106:4770–4775. doi: 10.1073/pnas.0807739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, et al. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Sanchez JA, Kim SM, Huberman JA. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitments of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sayrac S, Vengrova S, Godfrey EL, Dalgaard JZ. Identification of a novel type of spacer element required for imprinting in fission yeast. PLoS ONE. 2011;7:e1001328. doi: 10.1371/journal.pgen.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Singh J, Klar AJ. DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature. 1993;361:271–273. doi: 10.1038/361271a0. [DOI] [PubMed] [Google Scholar]
- Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Schizosaccharomyces pombe Swi1, Swi3 and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Vaughn MW, Irvine DV, Goto D, et al. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.