Abstract
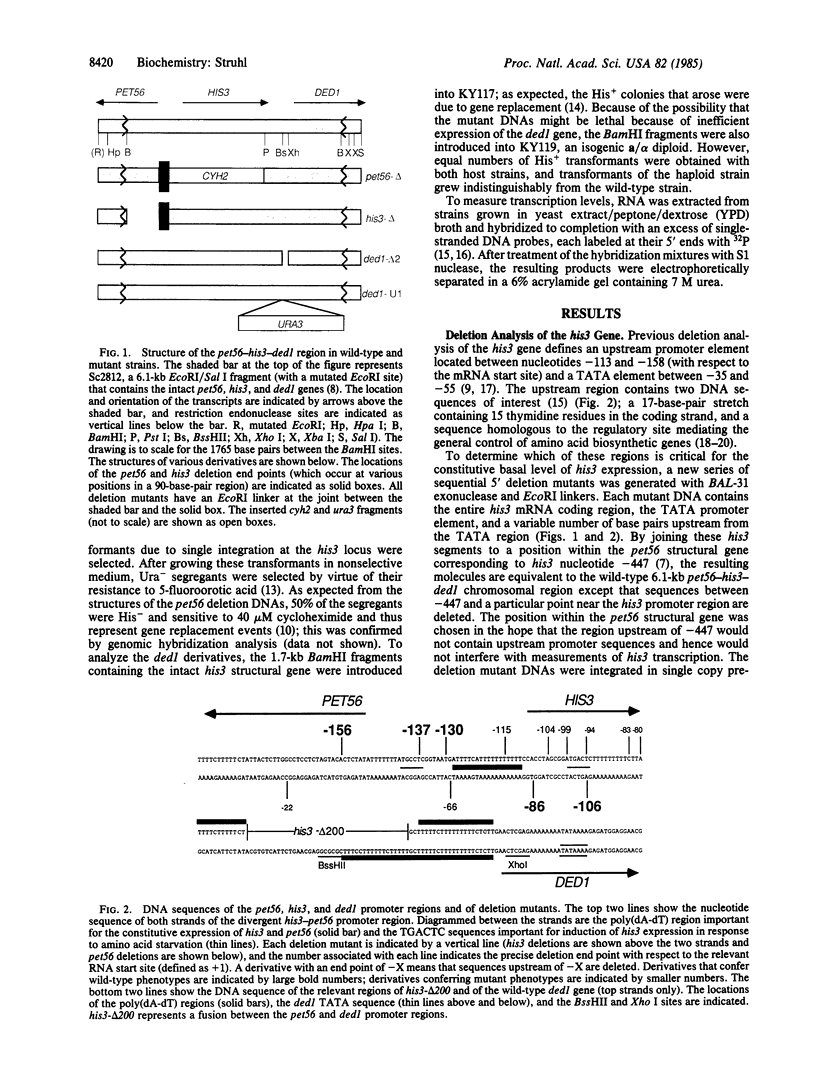
pet56, his3, and ded1 are adjacent but unrelated genes located on chromosome XV of the yeast Saccharomyces cerevisiae. his3 and pet56 are transcribed in opposite directions from initiation sites separated by approximately equal to 200 base pairs. Under normal growth conditions, both genes are transcribed at a similar basal level. Deletion analysis of the his3 gene indicates that the upstream promoter element for constitutive expression is defined by a 17-base-pair region that contains 15 thymidine residues in the coding strand. Sequential deletions of the pet56 gene indicate that this same region is required for wild-type transcription levels. Thus, this poly(dA-dT) sequence acts bidirectionally to activate transcription of two unrelated genes. Transcription of the ded1 gene is initiated approximately equal to 300 base pairs downstream from the his3 gene, and it occurs at a 5-fold higher level. This gene contains a 34-base-pair region containing 28 thymidine residues in the coding strand located upstream from the ded1 TATA box. Deletion of this dA-dT stretch significantly reduces transcription below the wild-type level. Thus, for at least three different yeast genes, naturally occurring stretches of poly(dA-dT) serve as upstream promoter elements for constitutive expression. In addition, it appears that longer stretches of poly(dA-dT) are more effective upstream promoter elements. These transcriptional effects may be due to exclusion of nucleosomes from poly(dA-dT) regions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach M. L., Lacroute F., Botstein D. Evidence for transcriptional regulation of orotidine-5'-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Kaback D. B., Angerer L. M., Davidson N. Improved methods for the formation and stabilization of R-loops. Nucleic Acids Res. 1979 Jun 11;6(7):2499–2317. doi: 10.1093/nar/6.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. A bent helix in kinetoplast DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):279–283. doi: 10.1101/sqb.1983.047.01.033. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Oettinger M. A., Struhl K. Suppressors of Saccharomyces cerevisiae his3 promoter mutations lacking the upstream element. Mol Cell Biol. 1985 Aug;5(8):1901–1909. doi: 10.1128/mcb.5.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Cox D., Williamson V. M., Young E. T. DNA sequences of two yeast promoter-up mutants. Nature. 1983 Aug 18;304(5927):652–654. doi: 10.1038/304652a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. Isolation of galactose-inducible DNA sequences from Saccharomyces cerevisiae by differential plaque filter hybridization. Cell. 1979 Feb;16(2):443–452. doi: 10.1016/0092-8674(79)90020-5. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Promotor mutants of the yeast his3 gene. J Mol Biol. 1981 Nov 5;152(3):553–568. doi: 10.1016/0022-2836(81)90268-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Transcription of the his3 gene region in Saccharomyces cerevisiae. J Mol Biol. 1981 Nov 5;152(3):535–552. doi: 10.1016/0022-2836(81)90267-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Direct selection for gene replacement events in yeast. Gene. 1983 Dec;26(2-3):231–241. doi: 10.1016/0378-1119(83)90193-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]







