Abstract
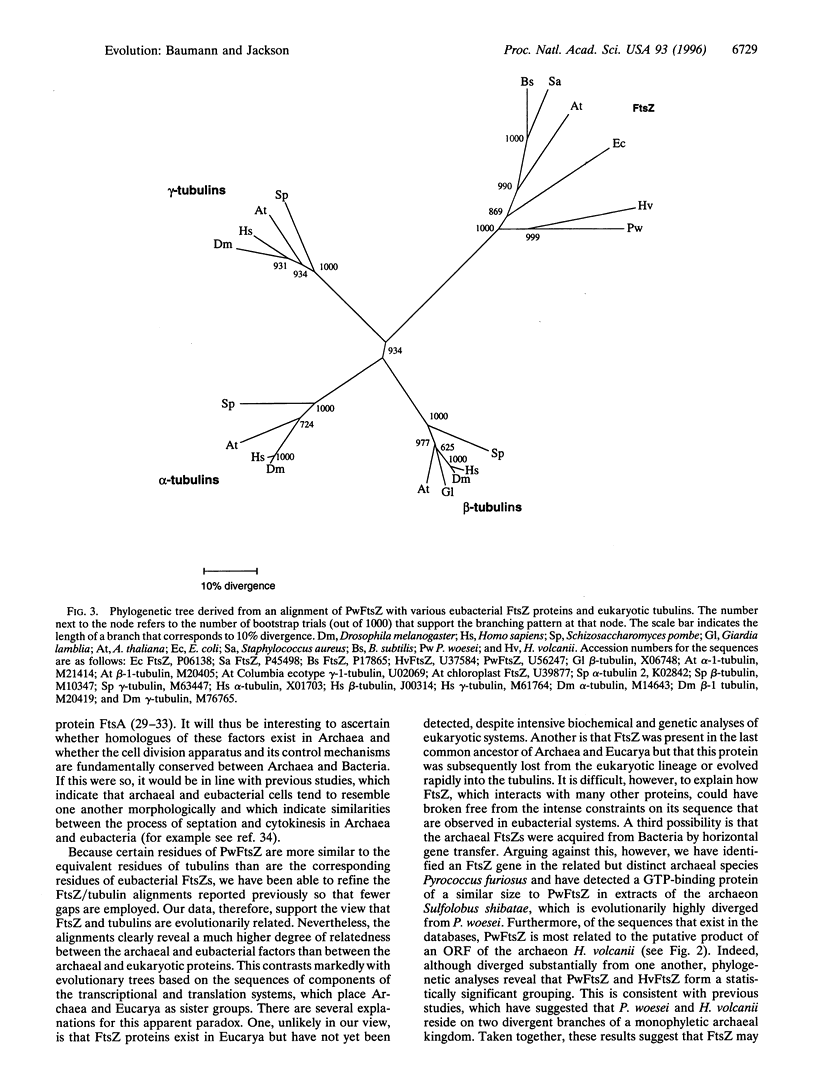
Life falls into three fundamental domains--Archaea, Bacteria, and Eucarya (formerly archaebacteria, eubacteria, and eukaryotes,. respectively). Though Archaea lack nuclei and share many morphological features with Bacteria, molecular analyses, principally of the transcription and translation machineries, have suggested that Archaea are more related to Eucarya than to Bacteria. Currently, little is known about the archaeal cell division apparatus. In Bacteria, a crucial component of the cell division machinery is FtsZ, a GTPase that localizes to a ring at the site of septation. Interestingly, FtsZ is distantly related in sequence to eukaryotic tubulins, which also interact with GTP and are components of the eukaryotic cell cytoskeleton. By screening for the ability to bind radiolabeled nucleotides, we have identified a protein of the hyperthermophilic archaeon Pyrococcus woesei that interacts tightly and specifically with GTP. Furthermore, through screening an expression library of P. woesei genomic DNA, we have cloned the gene encoding this protein. Sequence comparisons reveal that the P. woesei GTP-binding protein is strikingly related in sequence to eubacterial FtsZ and is marginally more similar to eukaryotic tubulins than are bacterial FtsZ proteins. Phylogenetic analyses reinforce the notion that there is an evolutionary linkage between FtsZ and tubulins. These findings suggest that the archaeal cell division apparatus may be fundamentally similar to that of Bacteria and lead us to consider the evolutionary relationships between Archaea, Bacteria, and Eucarya.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barns S. M., Fundyga R. E., Jeffries M. W., Pace N. R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Qureshi S. A., Jackson S. P. Transcription: new insights from studies on Archaea. Trends Genet. 1995 Jul;11(7):279–283. doi: 10.1016/s0168-9525(00)89075-7. [DOI] [PubMed] [Google Scholar]
- Bi E. F., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991 Nov 14;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993 Feb;175(4):1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Thompson C. M. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Doolittle W. F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarano P., Palm P., Creti R., Ceccarelli E., Sanangelantoni A. M., Tiboni O. Early evolutionary relationships among known life forms inferred from elongation factor EF-2/EF-G sequences: phylogenetic coherence and structure of the archaeal domain. J Mol Evol. 1992 May;34(5):396–405. doi: 10.1007/BF00162996. [DOI] [PubMed] [Google Scholar]
- Creti R., Londei P., Cammarano P. Complete nucleotide sequence of an archaeal (Pyrococcus woesei) gene encoding a homolog of eukaryotic transcription factor IIB (TFIIB). Nucleic Acids Res. 1993 Jun 25;21(12):2942–2942. doi: 10.1093/nar/21.12.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992 Oct;174(19):6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar S. J., Begg K. J., Donachie W. D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992 Oct;174(19):6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F. The origins and evolution of eukaryotic proteins. Philos Trans R Soc Lond B Biol Sci. 1995 Sep 29;349(1329):235–240. doi: 10.1098/rstb.1995.0107. [DOI] [PubMed] [Google Scholar]
- Erickson H. P. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995 Feb 10;80(3):367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- Hausner W., Thomm M. The translation product of the presumptive Thermococcus celer TATA-binding protein sequence is a transcription factor related in structure and function to Methanococcus transcription factor B. J Biol Chem. 1995 Jul 28;270(30):17649–17651. doi: 10.1074/jbc.270.30.17649. [DOI] [PubMed] [Google Scholar]
- Hixon W. G., Searcy D. G. Cytoskeleton in the archaebacterium Thermoplasma acidophilum? Viscosity increase in soluble extracts. Biosystems. 1993;29(2-3):151–160. doi: 10.1016/0303-2647(93)90091-p. [DOI] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J., Charlebois R. L., Doolittle W. F. Archaebacterial genomes: eubacterial form and eukaryotic content. Curr Opin Genet Dev. 1994 Dec;4(6):816–822. doi: 10.1016/0959-437x(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Langer D., Hain J., Thuriaux P., Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci U S A. 1995 Jun 20;92(13):5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh T. L., Reich C. I., Whitelock R. B., Olsen G. J. Transcription factor IID in the Archaea: sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Dai K., Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994 May;176(9):2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K. W., Vierling E. Conserved cell and organelle division. Nature. 1995 Aug 10;376(6540):473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992 Oct 16;71(2):189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S. A., Baumann P., Rowlands T., Khoo B., Jackson S. P. Cloning and functional analysis of the TATA binding protein from Sulfolobus shibatae. Nucleic Acids Res. 1995 May 25;23(10):1775–1781. doi: 10.1093/nar/23.10.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S. A., Khoo B., Baumann P., Jackson S. P. Molecular cloning of the transcription factor TFIIB homolog from Sulfolobus shibatae. Proc Natl Acad Sci U S A. 1995 Jun 20;92(13):6077–6081. doi: 10.1073/pnas.92.13.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D., Park J. T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992 Sep 17;359(6392):251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Rowlands T., Baumann P., Jackson S. P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994 May 27;264(5163):1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- Searcy D. G., Hixon W. G. Cytoskeletal origins in sulfur-metabolizing archaebacteria. Biosystems. 1991;25(1-2):1–11. doi: 10.1016/0303-2647(91)90008-9. [DOI] [PubMed] [Google Scholar]
- Sogin M. L. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991 Dec;1(4):457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- Sánchez M., Valencia A., Ferrándiz M. J., Sander C., Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994 Oct 17;13(20):4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Rothfield L. I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P., Crossley R., Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992 Sep 17;359(6392):254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]