Abstract
Background
Dengue fever and leptospirosis have partially overlapping geographic distributions, similar clinical presentations and potentially life-threatening complications but require different treatments. Distinguishing between these cosmopolitan emerging pathogens represents a diagnostic dilemma of global importance. We hypothesized that perturbations in host biomarkers can differentiate between individuals with dengue fever and leptospirosis during the acute phase of illness.
Methods
We randomly selected subjects from a prospective cohort study of acute febrile illness in Bucaramanga, Colombia and tested 19 serum biomarkers by ELISA in dengue fever (DF, n = 113) compared to subjects with leptospirosis (n = 47). Biomarkers were selected for further analysis if they had good discriminatory ability (area under the ROC curve (AUC) >0.80) and were beyond a reference range (assessed using local healthy controls).
Results
Nine biomarkers differed significantly between dengue fever and leptospirosis, with higher levels of Angptl3, IL-18BP, IP-10/CXCL10, Platelet Factor 4, sICAM-1, Factor D, sEng and sKDR in dengue and higher levels of sTie-2 in leptospirosis (p < 0.001 for all comparisons). Two biomarkers, sEng and IL18BP, showed excellent discriminatory ability (AUROC >0.90). When incorporated into multivariable models, sEng and IL18BP improved the diagnostic accuracy of clinical information alone.
Conclusions
These results suggest that host biomarkers may have utility in differentiating between dengue and leptospirosis, clinically similar conditions of different etiology.
Keywords: Dengue fever, Leptospirosis, Acute febrile illness, Host biomarkers, Clinical discrimination, Combinatorial models
Background
Dengue virus infection and leptospirosis represent important causes of acute febrile illness whose diagnosis and management in resource-poor settings remains challenging. Both diseases are potentially fatal, and represent important causes of morbidity and mortality globally. As emerging or re-emerging vector- and water-borne pathogens, respectively, dengue and leptospirosis are increasingly important considerations in patients with acute febrile illness, particularly in the context of decreasing malaria transmission in many areas of the world [1].
The burden of illness attributable to dengue viruses is estimated to be 390 million annually [2]. An estimated 2.5-4 billion people are at risk of infection, and transmission has increased in recent years, such that dengue is re-emerging as an international public health concern [3]. Dengue viruses are members of the flavivirus family and consist of four distinct serotypes. Transmission occurs in tropical and sub-tropical regions around the world through the bite of the Aedes mosquito vector. Clinical manifestations range from: (1) asymptomatic infection; to (2) a non-specific febrile illness (dengue fever); to (3) a life-threatening complication, severe dengue, often characterized by plasma leakage and coagulopathy. The pathogenesis of severe dengue is the result of a complex interaction of viral and host factors. The risk of severe dengue is increased 40–80 fold after a second infection with a different serotype [4,5], which may be explained by non-neutralizing heterotypic antibody-mediated enhancement of infection. Complement activation by virus-antibody complexes and T-cell mediated immunopathology have also been implicated in the progression of secondary dengue infection. Laboratory confirmation of infection relies on serologic methods, viral culture, and/or PCR and is not generally rapidly available to assist clinical decision-making, particularly in endemic resource-constrained areas. Specific anti-viral therapy is not available, but intensive supportive care may reduce mortality in severe dengue from 20% to less than 1% [6].
Leptospirosis accounts for a significant proportion of febrile illness among hospitalized patients in Asia and the Americas [7-9]. Pathogenic spirochetes of the genera Leptospira infect humans through contact of skin or mucous membranes with water or soil contaminated with the urine of host animals, who harbor the bacteria within their renal tubules. Outbreaks have been reported after monsoons and natural disasters [10], as well as among adventure travelers with prolonged water contact [11]. As in dengue, clinical manifestations vary widely across a spectrum of disease severity. The majority of infections produce asymptomatic seroconversion or undifferentiated fever; however, severe disease may develop in 5-10% of symptomatic patients, evidenced by renal and pulmonary involvement (Weil’s disease). The case-fatality rate may be as high as 5-15% when multisystem complications occur [12]. Serologic methods are used in clinical settings for a microbiologic diagnosis of leptospirosis, meaning that timely confirmation of infection is not widely available. The treatment involves specific antibiotic therapy, such that early diagnosis can assist treatment decisions and may arrest disease progression. Leptospirosis is often misdiagnosed as dengue fever, and is frequently under-recognized in endemic regions and during outbreaks [8,13].
Host immunopathology plays an important role in the progression of both dengue and leptospirosis [14,15]. Indeed, severe infection across a range of viral, bacteria and parasitic pathogens is characterized by engagement of shared host defense pathways, including inflammation, angiogenesis, coagulation and endothelial activation [16-18]. We hypothesized that peripheral blood biomarkers of these pathways may have clinical utility as diagnostic tools to differentiate between dengue and leptospirosis. We applied combinatorial approaches [17], using information provided by clinical features, standard clinical laboratory tests and multiple biomarkers from distinct pathobiological pathways, to examine novel strategies to distinguish dengue from leptospirosis.
Methods
Ethics statement
Ethical approval for this study was granted from the Medical Ethics Committee of the Universidad Industrial de Santander in Bucaramanga, Colombia. All subjects (or parents/guardians for minors) provided written informed consent to enter the study.
Study design
This study was a case–control study nested within a prospective cohort study of subjects with suspected dengue fever in Bucaramanga, Colombia. Cases and controls were randomly chosen using simple randomization from the cohort database following identification of subjects with available serum samples that had not been previously freeze-thawed. Cases were defined as individuals with dengue infection and controls were individuals with leptospirosis.
Study population
Participants greater than five years of age with an acute febrile syndrome less than 96 hours were enrolled in a prospective outpatient cohort study examining predictors of disease severity in dengue fever. Following enrollment, a physical examination was performed and a blood sample was collected to determine levels of hematocrit and albumin and to assess platelet and leukocyte counts. An additional serum sample was collected and stored at -80C for future biomarker assessment. Subjects were excluded based on the presence of the following conditions: history of concomitant diseases such as diabetes, acquired immunodeficiency syndrome, hematologic disorders, cancer, or cardiac disease; dengue hemorrhagic fever at baseline, major bleeding, hypoalbuminemia (<3 g/dL), effusions, or shock.
Study definitions
Dengue fever
Diagnosis of dengue infection was made based on either: viral isolation, a shift from a negative to a positive IgM test result, or a four-fold increase in existing dengue antibodies from admission to convalescence (7–15 days following symptom onset). Study subjects with negative convalescent IgM test were considered to have another cause for fever. We were not able to differentiate between primary and secondary dengue infections in this study.
Leptospirosis
In subjects negative for dengue fever a diagnosis of leptospirosis was made based on a shift from negative to positive IgM test result in admission and convalescence samples, or a four-fold increase in leptospirosis antibody titers.
Healthy controls
Serum samples were collected from 15 healthy adults from Bucaramanga to derive a population-based normal range. Samples were not tested for past dengue virus or leptospirosis infection, but all controls were asymptomatic at the time of blood sampling and were unlikely to have an acute infection.
Biomarker assessment
Nineteen biomarkers were selected because they represent markers of novel pathways implicated in infectious disease pathobiology, including inflammation, coagulation, and endothelial activation [19-26]. These proteins provide information about a broad range of innate immune responses and serve to characterize the host response to different pathogens. Serum concentrations of biomarkers were measured using ELISA DuoSets from R&D Systems (Minneapolis, MN). Biomarkers measured were: angiopoietin (Ang)-1, Ang-2, soluble Tie-1 (sTie-1), soluble Tie-2 (sTie-2), vascular endothelial growth factor (VEGF), soluble VEGFR-1/Flt-1(sFlt-1), soluble VEGR-2/KDR (sKDR), soluble Endoglin (sEng), C-reactive protein (CRP), interleukin-10 (IL-10), interleukin-18 binding protein (IL-18BP), 10 kDa-interferon induced protein (IP-10, CXCL10), soluble ICAM-1, chitinase 3-like 1 (CHI3L1), complement factor 5a (C5a), complement factor D (Factor D), angiopoietin-like 3 (Angptl3), and angiopoietin-like 4 (Angptl4). All ELISA kits were validated prior to use and appropriate samples dilutions were obtained for each biomarker by testing a dilution curve of serum obtained from febrile subjects. ELISAs were performed as previously described [16] and biomarker concentrations were extrapolated using a 4-parameter logistic slope curve (GraphPad Prism v5.0).
Statistical analysis
GraphPad Prism v5, SPSS v16 and MedCalc software were used for statistical analysis. Comparisons of continuous variables were performed using the Mann–Whitney U test. Bonferonni adjustment was used to account for multiple testing. Receiver operating characteristic (ROC) curves were used to assess the discriminatory ability of the biomarkers. The area under the ROC curves (AUC) or c-indices (ROC generated from the predicted probabilities of logistic regression models) were compared using the Delong-Delong Clarke Pearson method [27]. Biomarker cut-points were established by using the Youden index.
Comparisons of proportions were performed using Pearson chi-square test or Fisher’s exact test, as appropriate. Adjusted odds ratios were calculated using logistic regression. Variables were selected for inclusion in logistic regression models using forward stepwise logistic regression. All logistic regression models were validated by ensuring the Hosmer-Lemeshow goodness-of-fit test was not significant (p > 0.05). Complete model validation is provided as an Additional file 1: Table S1 and Table S2.
Results
Study population
160 subjects between the ages of 5 and 81 years with an acute febrile illness were included in the study. The median age for individuals with dengue fever and leptospirosis was 25 and 27 years respectively. There was an equal distribution of dengue and leptospirosis in both men and women. The demographic and clinical characteristics and laboratory findings at presentation are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of population
| Dengue (n = 113) | Leptospirosis (n = 47) | P value | |
|---|---|---|---|
|
Demographic characteristics |
|
|
|
| Age, years |
25.0 (16.0–41.0) |
27.0 (20.0–38.0) |
0.370 |
| Sex, number (% M) |
52 (46.0) |
26 (55.3) |
0.284 |
| Height, cm |
163 (152–170) |
164 (157–174) |
0.050 |
| Weight, kg |
61.0 (50.5–74.5) |
62.0 (51.5–71.0) |
0.821 |
| Body mass index (kg/m2) |
23.2 (19.7–26.9) |
21.7 (19.6–24.5) |
0.213 |
| Duration of fever, hours |
77.0 (64.0–88.8) |
70.5 (55.0–79.7) |
0.042 |
|
Laboratory findings |
|
|
|
| Axillary temperature,ºC |
36.5 (36.0–37.4) |
36.0 (35.6–36.5) |
0.003 |
| Number (%) >38ºC |
12 (10.6) |
3 (6.5) |
0.557 |
| Platelet count (x103) |
126 (83–180) |
202 (175–240) |
<0.001 |
| Number (%) <100 x103/uL |
33 (30) |
2 (5.7) |
<0.001 |
| Leukocyte count |
2900 (2200–3950) |
4900 (3600–6300) |
<0.001 |
| Number (%) <5000/uL |
99 (90.0) |
21 (48.8) |
<0.001 |
| Hematocrit |
38.7 (35.9–42.2) |
41.1 (36.7–44.7) |
0.062 |
| Positive tourniquet test |
75 (66.4) |
23 (48.9) |
0.039 |
|
Signs and symptoms, number (%) |
|
|
|
| Headache |
107 (94.7) |
45 (95.7) |
0.780 |
| Retro-orbital pain |
79 (69.9) |
31 (66.0) |
0.623 |
| Asthenia |
78 (69.0) |
37 (78.7) |
0.214 |
| Muscle pain |
102 (91.1) |
43 (91.5) |
0.932 |
| Joint pain |
90 (79.6) |
35 (74.5) |
0.576 |
| Chills |
107 (94.7) |
44 (93.6) |
0.788 |
| Cough |
41 (37.6) |
22 (46.8) |
0.283 |
| Nasal congestion |
38 (34.9) |
22 (46.8) |
0.159 |
| Sore throat |
36 (31.9) |
26 (55.3) |
0.006 |
| Rash |
48 (42.5) |
7 (14.9) |
<0.001 |
| Facial erythema |
57 (50.4) |
7 (14.9) |
<0.001 |
| Pruritis |
35 (32.1) |
11 (23.4) |
0.274 |
| Nausea |
86 (76.1) |
37 (78.7) |
0.721 |
| Vomiting |
38 (33.6) |
17 (36.2) |
0.758 |
| Diarrhea |
42 (37.2) |
11 (23.4) |
0.092 |
| Abdominal pain |
61 (54.5) |
25 (53.2) |
0.883 |
| Blurred Vision |
46 (41.1) |
17 (36.2) |
0.564 |
| Dizziness |
75 (67.0) |
40 (85.1) |
0.020 |
| Drowsiness |
68 (60.7) |
28 (59.6) |
0.893 |
| Dehydrated |
66 (60.6) |
32 (69.6) |
0.288 |
| Conjunctival injection |
48 (42.5) |
11 (23.4) |
0.023 |
| Orthostatic hypotension |
17 (15.0) |
2 (4.3) |
0.055 |
| Hepatomegaly | 1 (0.9) | 5 (10.6) | 0.003 |
Data are presented as median (interquartile range) unless otherwise indicated.
Group comparison by Chi-square or Fisher’s exact test (where <5 cases in any specified cell) for dichotomous variables or Mann–Whitney for continuous variables.
Clinical and laboratory factors that discriminate between dengue fever and leptospirosis
To identify clinical signs and laboratory parameters that could aid in differentiating dengue fever from leptospirosis, we generated adjusted odds ratios for variables with p < 0.10 by bivariate analysis. Following adjustment for age, sex, height, and duration of illness, thrombocytopenia and leukopenia were independently associated as risk factors for dengue fever with adjusted odds ratios (aOR) of 10.0 (95% CI, 2.2-45.8), p = 0.003 and 9.3 (95% CI, 3.6-24.0), p < 0.001 respectively. In addition, the presence of rash (aOR (95% CI), p-value: 5.8 (2.1-15.7), p = 0.001) facial erythema (7.3 (2.8-19.3), p < 0.001) and conjunctival injection (3.0 (1.3-7.1), p = 0.012) were more common in subjects with dengue fever. Dizziness (aOR (95% CI), p-value: 0.4 (0.2-1.0), p = 0.047) and sore throat (0.3 (0.1-0.7), p = 0.004) were more common in leptospirosis compared to dengue fever.
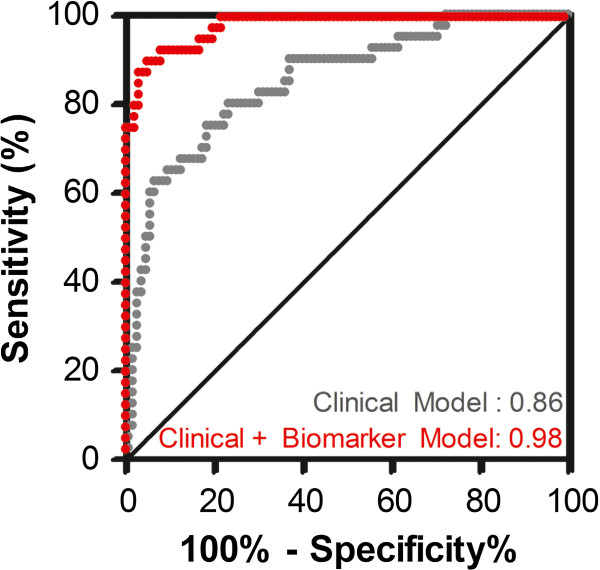
To further explore how clinical and laboratory parameters could be incorporated into a model to differentiate between dengue fever and leptospirosis, we used logistic regression and forward step-wise selection to identify variables with the best discriminatory ability. All factors with significant adjusted odds ratios from Table 2 were included into logistic regression models and leukopenia, rash and dizziness were identified as the best three discriminatory measures. A clinical model including age, sex, height, duration of illness, leukopenia, rash and dizziness had a c-index (equivalent to the AUC) of 0.86 indicating that these parameters have good discriminatory ability (Figure 1, Additional file 1: Table S1).
Table 2.
Clinical and laboratory factors associated with dengue fever, relative to leptospirosis
| Odds Ratio (95% CI) | P-Value | Adjusted OR (95% CI)* | P-value* | |
|---|---|---|---|---|
| Positive tourniquet test |
1.4 (0.9–2.3) |
0.156 |
1.4 (0.7–2.9) |
0.354 |
| Thrombocytopenia |
6.2 (1.6–24.3) |
0.001 |
10.0 (2.2–45.8) |
0.003 |
| Leukopenia |
3.8 (2.4–6.0) |
<0.001 |
9.4 (3.7–23.8) |
<0.001 |
| Orthostatic hypotension |
3.0 (0.8–11.5) |
0.055 |
3.4 (0.7–15.9) |
0.117 |
| Rash |
3.1 (1.5–6.5) |
<0.001 |
5.8 (2.1–15.7) |
0.001 |
| Facial erythema |
3.8 (1.8–8.0) |
<0.001 |
7.3 (2.8–19.3) |
<0.001 |
| Conjunctival injection |
1.9 (1.1–3.4) |
0.023 |
3.0 (1.3–7.1) |
0.012 |
| Diarrhea |
1.6 (0.9–2.9) |
0.092 |
1.9 (0.8–4.4) |
0.125 |
| Dizziness |
0.5 (0.2–0.9) |
0.020 |
0.4 (0.2–1.0) |
0.047 |
| Hepatomegaly |
0.3 (0.2–0.5) |
0.003 |
0.1 (0.01–1.4) |
0.093 |
| Sore throat | 0.5 (0.3–0.8) | 0.006 | 0.3 (0.1–0.7) | 0.004 |
*Adjustment for: age, sex, height, duration of illness (logistic regression analysis). Model fit was assessed using the Hosmer-lemeshow test and ensuring the term was insignificant (p > 0.05).
Figure 1.
Integrating Clinical and Laboratory Data with Biomarker Data Improves Discrimination of Dengue Fever and Leptospirosis. Logistic regression analysis was used to generate two models to discriminate between dengue fever and leptospirosis and the predicted probabilities from those models were plotted using ROC curve analysis. The first model used clinical and laboratory data (clinical model: age, sex, height, duration of illness, leukopenia, rash, dizziness) and had good discriminatory performance with a c-index (equivalent to the AUC) of 0.86 (95% CI: 0.79-0.91). By adding in the biomarker data, we generated a model with excellent discriminatory ability and a c-index of 0.979 (95% CI: 0.94-0.996). The biomarker model (clinical model with IL-18BP, sEng) had a c-index that was statistically higher than that of the clinical model (p = 0.0003).
Taking another more intuitive approach we used the clinical parameters identified by logistic regression to generate a clinical score that could easily be implemented and interpreted to identify subjects with dengue fever. For each dichotomous measure, we assigned a value of 0 (not present), -1 (more common in leptospirosis) or +1 (more common in dengue fever) for each variable (Figure 2). Thus, using a clinical score ranging from -1 to 2, leukopenia, rash and dizziness were able to differentiate between dengue fever and leptospirosis with an AUC (95% CI) of 0.81(0.73-0.87), p < 0.001. Using a cut-point of ≥1, this model had a sensitivity of 61%, a specificity of 88% and a positive and negative likelihood ratio of 5.2 and 0.4 respectively to correctly diagnose dengue fever.
Figure 2.
Integrating Biomarker Data into a Clinical Score Improves Diagnosis of Dengue Fever. A clinical score (from -1 to 2) was created for each study participant by assigning a value of 0 (not present), -1 (more common in leptospirosis) or +1 (more common in dengue) for leukopenia, rash and dizziness. The score was used to create an area under the ROC curve (AUC) of 0.81 (95% CI, 0.73-0.86), p < 0.001. Biomarker data were then integrated into the clinical score (from -1 to 4) by assigning a value of +1 if IL-18BP (>24.5 ng/mL) and sEng (> 9.12 ng/mL) levels were higher than the assigned cut-offs to generate an AUC of 0.96 (95% CI, 0.91-0.98), p < 0.001.
Host biomarkers differentiate between dengue fever and leptospirosis
We hypothesized that host biomarkers derived from pathways of disease pathogenesis in dengue may further improve discrimination between these clinically non-specific acute febrile syndromes. We examined 19 different serum biomarkers from different pathways implicated in dengue pathogenesis focusing on: endothelial activation and angiogenesis (Ang-1, Ang-2, sTie-2, sTie-1, VEGF, sFlt-1, sFlk-2, sEng, angiopoietin-like 4), inflammation (CRP, IL-10, IL-18BP, IP-10/CXCL10, sICAM-1, CHI3L1), complement activation and coagulation (C5a, Factor D, PF4) and the regulation of lipids (angiopoietin-like 3). Data are summarized in Table 3.
Table 3.
Biomarkers levels measured at time of presentation in dengue fever and leptospirosis
| Healthy control (n = 15) | Dengue (n = 113) | Leptospirosis (n = 47) |
P value |
|
|---|---|---|---|---|
| Dengue vs. Leptospirosis | ||||
| Angiopoietin-1 |
44.9 (32.9-53.6) |
32.3 (24.1-45.0) |
35.8 (29.0-46.7) |
0.236 |
| Angiopoietin-2 |
1.4 (1.0-2.6) |
1.8 (1.3-2.5) |
2.0 (1.3-3.0) |
0.474 |
| sTie-1 |
8.3 (6.8-12.7) |
10.1 (8.1-13.5) |
9.0 (7.6-11.6) |
0.120 |
| VEGF |
0.21 (0.15-0.28) |
0.13 (0.05-0.30) |
0.24 (0.08-0.49) |
0.017 |
| sFlt-1 |
0.04 (0.04-0.34) |
0.04 (0.04-0.88) |
0.15 (0.04-0.70) |
0.864 |
| C5a |
18.5 (13.8-22.1) |
53.8 (34.3-63.8) |
41.2 (30.2-59.6) |
0.103 |
| CRP a |
1.8 (1.2-6.1) |
19.7 (9.1-41.4) |
13.3 (4.1-39.8) |
0.125 |
| IL-10 |
0.02 (0.02-0.16) |
0.09 (0.02-0.37) |
0.08 (0.02-0.20) |
0.688 |
| CHI3L1 |
44.5 (35.8-61.3) |
58.1 (43.0-78.6) |
52.1 (36.5-74.1) |
0.114 |
| Angiopoietin-like 4 |
44.7 (30.2-90.9) |
40.4 (30.6-57.1) |
49.8 (35.9-67.8) |
0.094 |
| Angiopoietin-like 3 |
108 (83–118) |
165 (141–195) |
122 (96–148) |
<0.001* |
| IL-18BP |
5.8 (4.4-8.3) |
67.8 (41.5-90.9) |
11.4 (7.4 (21.5) |
<0.001* |
| CXCL10 |
0.2 (0.2-0.4) |
3.2 (1.2-4.4) |
0.5 (0.2-1.4) |
<0.001* |
| Platelet Factor 4 a |
18.5 (15.6-27.1) |
25.2 (16.2-39.1) |
19.1 (14.0-24.6) |
<0.001* |
| sICAM-1 |
169 (136–187) |
352 (300–488) |
228 (159–275) |
<0.001* |
| Factor D |
1143 (1018–1265) |
1376 (1172–1615) |
970 (837–1128) |
<0.001* |
| sEng |
6.6 (5.7-8.4) |
11.7 (9.6-13.9) |
7.6 (7.2-8.2) |
<0.001* |
| sKDR |
5.3 (4.5-6.1) |
6.1 (5.5-7.1) |
5.2 (4.6-6.1) |
<0.001* |
| sTie-2 | 8.7 (6.4-9.7) | 7.7 (5.9-9.3) | 10.0 (7.8-13.0) | <0.001* |
Biomarker concentrations are presented as median (IQR) in ng/mL unless otherwise indicated, aμg/mL.
*p < 0.05 following adjustment for 19 pair-wise comparisons.
Of the 19 biomarkers measured, 9 biomarkers differed significantly between dengue fever and leptospirosis following Bonferonni correction for multiple testing. Median levels of Angptl3, IL-18BP, CXCL10, Platelet Factor 4, sICAM-1, Factor D, sEng and sKDR were higher in dengue fever compared to leptospirosis (p < 0.001 for all). sTie-2 was the only biomarker with median levels higher in subjects with leptospirosis compared to dengue fever. Data for these markers are shown in Figure 3, superimposed on population-derived normal range (median, 5-9%). Receiver operating characteristic (ROC) curves were generated to assess the discriminatory ability of the biomarkers (Figure 3) and the performance characteristics of the ROC curves are reviewed in Table 4.
Figure 3.
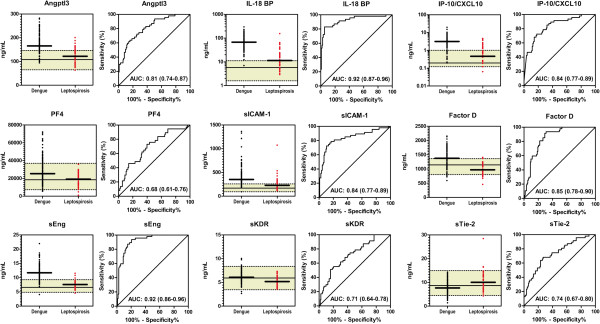
Biomarkers Discriminate Between Dengue Fever and Leptospirosis. Aligned dot plots and median of serum biomarker levels in dengue fever (n = 113) and leptospirosis (n = 47) measured at time of presentation during the acute phase of febrile illness. A population derived healthy range for adults in Bucaramanga, Colombia (n = 15) is represented by the shaded area with the median and 5-95% shown by the horizontal limits. All biomarkers were significantly different between cases with dengue fever and controls with leptospirosis (p < 0.001) following Bonferonni adjustment for multiple comparisons (19 pair-wise comparisons).
Table 4.
Performance characteristics of biomarkers significantly different in dengue fever following adjustment for multiple comparisons
| AUC (95% CI) | Cut-off* | Sensitivity (95% CI) | Specificity (95% CI) | Positive LR (95% CI) | Negative LR (95% CI) | |
|---|---|---|---|---|---|---|
| Angiopoietin-like 3 |
0.81 (0.74–0.87) |
>135.75 |
84.1 (76.0–90.3) |
63.8 (48.5–77.3) |
2.3 (1.8–2.9) |
0.3 (0.1–0.5) |
| IL-18BP |
0.92 (0.87–0.96) |
>24.52 |
94.7 (88.8–98.0) |
83.0 (69.2–92.4) |
5.6 (4.9–6.4) |
0.06 (0.02–0.2) |
| CXCL10 |
0.84 (0.77–0.89) |
>0.96 |
82.3 (74.0–88.8) |
72.3 (57.4–84.4) |
2.9 (2.4–3.6) |
0.2 (0.1–0.5) |
| Platelet Factor 4 a |
0.69 (0.61–0.76) |
>29.98 a |
39.8 (30.7–49.5) |
97.9 (88.7–99.9) |
18.7 (14.9–23.6) |
0.6 (0.1–4.3) |
| sICAM-1 |
0.84 (0.77–0.89) |
>285.9 |
83.2 (75.0–89.6) |
78.7 (64.3–89.3) |
3.9 (3.3–4.6) |
0.2 (0.1–0.4) |
| Factor D |
0.85 (0.79–0.90) |
>1248.1 |
69.0 (59.6–77.4) |
93.6 (82.5–98.7) |
10.8 (9.4–12.5) |
0.3 (0.1–1.1) |
| sEng |
0.92 (0.86–0.96) |
>9.12 |
79.7 (71.0–86.6) |
93.6 (82.5–98.7) |
12.5 (11.1–14.1) |
0.2 (0.07–0.7) |
| sKDR |
0.71 (0.64–0.78) |
>5.18 |
84.1 (76.0–90.3) |
51.1 (36.1–65.9) |
1.7 (1.3–2.3) |
0.3 (0.2–0.5) |
| sTie-2 | 0.73 (0.65–0.80) | ≤ 9.18 | 72.6 (63.4–80.5) | 68.1 (52.9–80.9) | 2.3 (1.8–2.9) | 0.4 (0.2–0.7) |
*Concentration in ng/mL unless otherwise indicated, aμg/mL.
ROC curve analysis- non-parametric analysis by method of Delong et al. with the confidence interval estimated using binomial.
Although several biomarkers were able to discriminate between dengue fever and leptospirosis, the median biomarker levels in dengue fever or leptospirosis did not extend beyond the population derived normal range, suggesting that these biomarkers may not have clinical utility. There were five biomarkers where the median levels were outside the population-derived normal range in dengue fever, but not leptospirosis: Angptl3, IL-18BP, CXCL10, sICAM-1, and sEng. Two biomarkers had excellent discriminatory ability (AUC >0.90) and using the Youden index to dichotomize the biomarkers one had high sensitivity, IL-18BP (sensitivity: 94.7%) while the other had high specificity, sEng (specificity: 93.6%).
Combining clinical and biomarker data improve discrimination
Since we had identified biomarkers with excellent discriminatory ability on their own, we wanted to explore whether these biomarkers could be integrated into the clinical models to improve clinical prediction. Intuitively, we assumed the two biomarkers with the best individual performance would be the best candidates to integrate into the clinical models. To test this assumption we used forward stepwise logistic regression and confirmed that IL-18BP and sEng were the best discriminatory biomarkers.
By adding the biomarkers into the clinical logistic regression model (clinical + biomarker model), we achieved a c-index (95% CI) of 0.98 (0.94-1.0), which was significantly better than the clinical model alone (p = 0.0003 comparing the AUCs by the method of Delong et al.) (Figure 1). Next, we added the biomarkers into the clinical score to generate a clinical + biomarker score that ranged from -1 to 4 (Figure 2). The AUC for the clinical + biomarker model was 0.96 (95% CI, 0.91-0.98) and at a cut-point ≥2 the model had a sensitivity of 87.6% (95% CI, 79.8-93.2) and specificity of 90.7 (77.9-97.4). This model had an AUC that was significantly higher than the AUC of the clinical model alone (0.96 vs. 0.81, p < 0.0001).
Discussion
In areas where both dengue and leptospirosis co-circulate, misdiagnosis of these febrile illnesses is common, given the considerable overlap in clinical signs and symptoms [8,9,13,28]. The lack of affordable, timely and practical diagnostic tests for both dengue and leptospirosis in many settings contributes to the diagnostic dilemma. Previous studies have examined strategies to distinguish between dengue and leptospirosis on the basis of clinical and traditional laboratory parameters, including petechial counts on a standardized tourniquet test, total leukocyte count, aspartate transaminase, and albumin level [9,13]. Our study adds to this limited diagnostic toolbox, describing a panel of novel, clinically informative biomarkers to distinguish dengue from leptospirosis. Activation of specific host defense pathways appears to differ between the two infections, as indicated by differences in circulating concentrations of key regulatory proteins. Of note, this observation may be leveraged to generate clinically useful diagnostic tests based on host proteins that could complement or replace pathogen-based microbiologic diagnosis. To this end, we have taken preliminary measures to assess the clinical utility of these host biomarkers, demonstrating that, alone and in combination, biomarkers improve the ability to discriminate between dengue and leptospirosis above and beyond clinical information alone. Elevated levels of sEng and IL18BP point strongly to a diagnosis of dengue virus infection, rather than leptospirosis. Additional studies will be required to validate these findings in independent populations.
The primary objective of this work was to identify clinical and classical laboratory features, as well as novel biomarkers that can be used by clinicians faced with the diagnostic dilemma of distinguishing between dengue and leptospirosis. Thrombocytopenia, leukopenia, rash, facial erythema, conjunctival injection and a positive tourniquet test were more common among patients with dengue whereas dizziness and sore throat were reported more commonly by patients with leptospirosis. Similarly, previous comparative studies have documented lower total leukocyte counts and higher petechial counts on tourniquet test in dengue infection relative to leptospirosis [9,13]. Nonetheless, these features are variably present in both infections, such that clinical criteria alone, even when formalized into multivariate models and clinical scores, remained suboptimal for distinguishing between dengue and leptospirosis in our prospective sample. Information derived from circulating levels of novel biomarkers improved diagnostic accuracy, with sEng and IL18BP showing the best performance individually and in multivariable models to distinguish between dengue and leptospirosis. Furthermore, both of these biomarkers were markedly elevated relative to normal controls, suggesting they could be integrated into clinical practice as a diagnostic tool for dengue virus infection.
Host biomarkers sEng and IL-18BP compare favourably with another recently commercialized point-of-care diagnostic modality for dengue based on detection of the virus NS1 antigen. In our study, the sensitivity and specificity of elevated sEng for the diagnosis of dengue (relative to patients with leptospirosis) were 80% and 94%, and for IL-18BP were 95% and 83%, respectively, using optimal thresholds for the study population. In comparison, sensitivity of the NS1 antigen was lower (62% [29] and 69% [30] in previous reports). The specificity, however, of direct viral antigen detection was marginally higher (100% [29] and 96% [30] in these reports). Of note, our approach using host biomarker signatures for diagnosis contrasts with the more conventional approach of direct viral antigen detection.
Endoglin, a component of the tumor necrosis factor-beta (TGF-β) receptor complex, participates in angiogenic and inflammatory signaling pathways. Endoglin is abundantly expressed on endothelial cells and is upregulated during inflammation [31]. The soluble form of the receptor (sEng) is shed from the endothelial surface into the circulation in the setting of critical illness by cleavage through matrix metalloproteinases [31]. By indirect inhibition of TGF-β signaling, sEng has antiangiogenic effects, produces endothelial dysfunction with vascular leak, and abrogates anti-inflammatory effects of TGF-β1 [32,33]. Elevated levels of sEng have been found in patients with pre-eclampsia [32], as well as severe and placental malaria [34,35]. We examined sEng in patients with dengue or leptospirosis and found significantly elevated serum levels in the majority of patients with dengue. Upregulation and shedding of sEng into the peripheral circulation due to endothelial activation during dengue virus infection may account for this observation, and may be a relatively specific indicator of dengue infection, since levels were not elevated in leptospirosis compared to healthy controls. This finding suggests that sEng may have clinical utility as a diagnostic biomarker of dengue in populations where dengue and leptospirosis co-circulate. Mechanistically, high sEng levels may attenuate TGF-β1 mediated anti-inflammatory responses, which may contribute to the disease manifestations of severe dengue. Further studies are warranted to investigate a putative pathogenic role of sEng in dengue virus infection.
IL-18 is a proinflammatory cytokine that stimulates natural killer cell activity and interferon gamma production in T-helper type I cells. IL-18 binding protein (IL18BP) is a constitutively expressed and secreted endogenous antagonist of IL-18. It is produced by mononuclear cells and can be upregulated by IFN-gamma, presumably as part of a feedback mechanism to downregulate IL-18 activity. Elevated levels of IL-18 and IL-18BP have been observed in patients with idiopathic thrombocytopenia purpura [25], suggesting a possible link to reduced platelet number and/or function, as in severe dengue and leptospirosis. IL18BP may be a determinant of immune response to viral infections including hepatitis C [36], and bacterial infections such as brucellosis [37]; however, no prior studies have examined IL-18BP in the context of dengue and leptospirosis. In our cohort, nearly all dengue patients and approximately half of leptospirosis patients had elevated levels of IL-18BP relative to healthy norms and IL-18BP was further elevated in dengue compared to leptospirosis. This finding may be explained by the increased levels of IFN-gamma observed in both dengue and leptospirosis [22,38], which may serve to stimulate IL-18BP production. IL-18BP may play an immunomodulatory role in these inflammatory conditions, although further studies are needed to elucidate a causal role for IL-18BP in limiting disease progression. Quantitative IL-18BP levels were higher in dengue than leptospirosis and could accurately be used to differentiate the two infections, both as an individual quantitative biomarker and in combination with other clinical and biochemical features.
In addition to identifying serum proteins that distinguish between dengue and leptospirosis, our findings provide noteworthy insights into numerous host pathways that are engaged during infection with dengue and leptospirosis. We examined several categories of host response to infection, including endothelial activation or quiescence, inflammation, coagulation, and the complement system. These factors have previously been studied in the context of malaria [16,17,20], sepsis [18,39], hemolytic-uremic syndrome [40], HIV/AIDS [19,41]. We studied regulatory proteins involved in these pathways, as well as endothelial cell surface receptors that are abnormally shed into the circulation during endothelial activation (sTie-1, sTie-2, sFlt-1, sKDR, sEng, and sICAM-1).
Dysregulation of the vascular endothelium with plasma leakage plays a defining role in dengue shock syndrome [42]. Leptospirosis is also associated with vascular injury and endothelial pathology that may contribute to the frank hemorrhages that characterize Weil syndrome (severe leptospirosis) [43]. The angiopoietins (Angs) and their tyrosine kinase receptors (Tie) are regulators endothelial activation or quiescence in mature vascular beds [44,45], and vascular permeability. Previous investigators have found derangements in Ang-1 and Ang-2 in patients with severe dengue infection that are associated with plasma leakage [46]. In our study, Ang-1 levels were lower in dengue patients than healthy controls (p = 0.017) but were not different between individuals with leptospirosis and healthy controls (p = 0.107). Low levels of Ang-1 may therefore contribute to endothelial activation and vascular leak in dengue infection.
Angiopoeitin-like-3 (Angptl3) and Angiopoietin-like-4 (Angptl4) are secreted glycoproteins which share sequence homology with the angiopoietins. Unlike the angiopoietins, Angptls do not bind Tie-1 or Tie-2 and their cognate receptors are unknown. Circulating levels of Angptl3 and Angptl4 have not previously been studied in dengue or leptospirosis. In our study, we found elevated levels of Angptl3 in patients with dengue fever relative to healthy controls and leptospirosis, whereas Angptl4 levels were similar in the two patient groups and did not differ from healthy controls. Angptl3 plays a role in lipid metabolism [47], hematopoietic stem cell activity [48], angiogenesis [49], and endothelial permeability in the glomerulus [50]. Our findings suggest that Angptl3 may participate in pathologic processes in dengue virus infection, with a possible role in modulating endothelial permeability, a hallmark of severe dengue.
We examined circulating levels of the angiopoietin tyrosine kinase receptors Tie-1 and Tie-2 and found statistically significant, albeit quantitatively modest differences in sTie-2 between dengue and leptosirosis patients, whereas sTie-1 levels were similar. Elevated levels of sTie-2 have been detected in the peripheral circulation of patients with sepsis [51], severe malaria [20,52], malignancy [53,54], and inflammatory bowel disease [55]. Soluble Tie-2 is released from the endothelial cell surface by proteolytic cleavage of the extracellular domain of the Tie-2 receptor by matrix metalloproteases [51]. In our study, levels of sTie-2 were elevated in leptospirosis relative to dengue. However, levels of sTie-2 were generally within the range of healthy controls in both conditions, suggesting that leptospirosis infection produces subtle derangements in sTie-2 levels.
Another critical regulatory pathway of endothelial activation is vascular endothelial growth factor (VEGF) and its tyrosine kinase receptors, Flt-1 (VEGFR-1) and KDR (VEGFR-2). KDR expressed on the endothelial cell surface, mediates most of the endothelial growth and survival signals, whereas Flt-1 acts as a negative regulator. We found that sKDR levels were higher in dengue fever than in leptospirosis. This suggests that sKDR may be useful as a biomarker to discriminate between leptospirosis and dengue.
Endothelial adhesion molecules, including ICAM-1, appear to be involved in host response to both leptospirosis and dengue. Recombinant leptospira antigens increase ICAM-1 expression on human umbilical vein endothelial cells in vitro[56,57]. ICAM-1 is shed from endothelial cells after exposure to the pro-inflammatory cytokines TNF and IL-1. The soluble form of ICAM-1 (sICAM-1) is increased in sepsis, severe malaria, during the febrile stage of dengue infection [34,58], and in leptospirosis [59]. In our study, sICAM-1 was elevated relative to healthy controls in both dengue and leptospirosis, as in previous studies. Our study extends these observations, demonstrating that quantitative levels of sICAM-1 are more markedly elevated in dengue relative to leptospirosis, suggesting that sICAM-1 may have diagnostic utility in differentiating the two infections.
Abnormal hemostasis is a defining feature of dengue hemorrhagic fever and Weil’s syndrome (severe leptospirosis that can be associated withpulmonary hemorrhage). Alterations in platelet number and function contribute to the coagulopathy in both diseases. Dengue infection is associated with thrombocytopenia, a positive tourniquet test [9,13], and release of platelet contents, including platelet factor 4 (PF4), into the circulation [60]. Leptospira proteins bind fibrinogen and block platelet aggregation [61]. In our study, the platelet count was reduced in both dengue and leptospirosis, with more profound thrombocytopenia in dengue fever. A positive tourniquet test was found in 75% of laboratory-confirmed dengue cases, versus 23% of leptospirosis cases, as has been described in other studies comparing the clinical features of dengue and leptospirosis [9,13]. Finally, the platelet alpha granule factor PF4 was increased in dengue fever relative to healthy controls, but not in leptospirosis. This suggests that levated levels of PF4 (>40,000 ng/mL) may be useful for positively identifying patients with dengue infection. Of note, another phylogenetically ancient role for PF4 is as a CXC chemokine that participates in innate host defenses, forming immunogenic neoantigens by binding polyanions including components of foreign pathogens [24]. Our finding of elevated levels of PF4 in dengue, but not leptospirosis, suggests a role of PF4 as a mediator of innate immunity or pathogenesis in dengue virus infection. Further studies are warranted to test this hypothesis.
The complement system appears to play an important role in innate immune control of both dengue virus and leptospires. Dysregulation of the alternative complement pathway is associated with severe dengue infection [62]. Furthermore, the mannose binding lectin (MBL) pathway of the complement system has been implicated in controlling dengue virus infections and modulating disease manifestations [63]. The alternative pathway of the complement system also plays a central role in innate defense against leptospirosis, as illustrated by the sensitivity of non-pathogenic and resistance of pathogenic Leptospira species to the cidal activity of human serum [64,65]. We evaluated components of the complement system, including the anaphylatoxin C5a and Factor D, a trypsin peptidase involved in the alternative pathway of complement system activation. Factor D was significantly elevated in patients with dengue, relative to leptospirosis and relative to healthy controls. Factor D has previous been shown to be elevated in patients with DHF compared to uncomplicated dengue fever, suggesting an important role for Factor D in the immunopathology of DHF, perhaps through the amplification of downstream complement factors and inflammatory mediators. Our study lends support to these findings and additionally suggests that elevated Factor D levels in dengue infection may serve to distinguish it from leptospirosis.
Our investigation into diagnostic biomarkers for dengue and leptospirosis is subject to several limitations. Because patients were sampled from a single site (Bucaramanga, Columbia), and biomarker levels may vary according to genetic background, age, presence of co-infections, nutritional status, pre-existing immunity, or other environmental factors, results should be extrapolated with caution to different geographic areas or different demographic groups. Whereas our study used well-defined patient samples with dengue or leptospirosis, other pathogens may confound the clinical diagnosis in different settings (e.g., malaria or scrub typhus). It would be important to validate our findings in different areas and patient populations, and across a range of pathogens with a similar clinical presentation to examine the robustness as well as the specificity and predictive value of diagnostic host biomarkers. Ideally, biomarker levels should also be tested across the full spectrum of disease manifestations, including severe dengue and Weil syndrome. Finally, a larger sample of healthy control patients would yield more precise estimates of the upper and lower normal limits of the serum level of these novel proteins.
Despite similarities in their clinical presentation, dengue (an intracellular virus) and leptospirosis (an extracellular bacterium) differ microbiologically, such that divergent host responses to these pathogens might be exploited to develop tools to discriminate between the infections in clinical practice. Our work examined a broad and diverse panel of host proteins, demonstrating that sEng and IL18BP in particular were differentially upregulated in dengue relative to leptospirosis and healthy controls. Detection of elevated levels of these biomarkers strongly points to dengue virus at the likely etiology in patients with the typical, overlapping clinical presentation.
Conclusion
Following validation in independent patient populations, these data may accelerate the development of simple clinical instruments, such as point-of-care lateral flow immunochromatographic rapid tests that could be widely implemented in resource-limited settings where dengue and leptospirosis co-circulate. Our findings also produced noteworthy insights into the activation of diverse host response programs, generating novel hypotheses into the pathogenesis of dengue and leptospirosis.
Abbreviations
Ang-1: Angiopoietin-1; Ang-2: Angiopoietin-2; sTie-2: Soluble tie-2; VEGF: Vascular endothelial growth factor; sVEGFR-1/sFlt-1: Soluble VEGF receptor 1; sVEGFR-2/sKDR: Soluble VEGF receptor 2; sEng: Soluble endoglin; CRP: C-reactive protein; IL-10: Interleukin-10; IL-18BP: Interleukin-18 binding protein; IP-10/CXCL10: 10 kDa-interferon induced protein; sICAM-1: Soluble intercellular adhesion molecule 1; CHI3L1: Chitinase 3-like 1; C5a: Complement factor 5a; Factor D: Complement factor D; Angptl3: Angiopoietein-like 3; Angptl4: Angiopoietin-like 4; PF4: Platelet factor 4; ROC: Receiver operating characteristic; AUC: Area under the curve; CI: Confidence interval.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ALC, MG, MH, NR, WCL, LAVC and KCK participated in the design of the study. ALC, MG and NR carried out the immunoassays. ALC and MH performed the statistical analysis. WCL, LAVC and KCK contributed reagents/materials/analysis tools. ALC, MH and KCK wrote the initial draft of the manuscript. MG, NR, WCL and LAVC critically revised the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1 and Table S2 accompany the manuscript. They provide statistical validation for the two logistic regression models presented in Figure 1.
Contributor Information
Andrea L Conroy, Email: andrea.conroy@utoronto.ca.
Margarita Gélvez, Email: margarita.gelvez@hotmail.com.
Michael Hawkes, Email: michael.hawkes@utoronto.ca.
Nimerta Rajwans, Email: nrajwans@uhnresearch.ca.
W Conrad Liles, Email: WCLiles@medicine.washington.edu.
Luis Angel Villar-Centeno, Email: luisangelvillarc@gmail.com.
Kevin C Kain, Email: kevin.kain@uhn.on.ca.
Acknowledgments
The authors wish to express their appreciation to all the study participants and study staff.
Funding
This work was supported by Colciencias [1102-459-21561 to LAVC] and the Canadian Institutes of Health Research [MOP-115160 and MOP-13721 to KCK; Canada Research Chairs to KCK and WCL; Clinician Scientist Training Award to MH; Fellowship award to ALC]. The funders had no role is study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, Fullman N, Naghavi M, Lozano R, Lopez AD. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;14(9814):413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O. et al. The global distribution and burden of dengue. Nature. 2013;14(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Dengue and dengue hemorrhagic fever. 2012. Fact sheet 117.
- Thomas L, Verlaeten O, Cabie A, Kaidomar S, Moravie V, Martial J, Najioullah F, Plumelle Y, Fonteau C, Dussart P. et al. Influence of the dengue serotype, previous dengue infection, and plasma viral load on clinical presentation and outcome during a dengue-2 and dengue-4 co-epidemic. The American journal of tropical medicine and hygiene. 2008;14(6):990–998. [PubMed] [Google Scholar]
- Halstead SB. Dengue. Curr Opin Infect Dis. 2002;14(5):471–476. doi: 10.1097/00001432-200210000-00003. [DOI] [PubMed] [Google Scholar]
- Ranjit S, Kissoon N, Jayakumar I. Aggressive management of dengue shock syndrome may decrease mortality rate: a suggested protocol. Pediatr Crit Care Med. 2005;14(4):412–419. doi: 10.1097/01.PCC.0000163676.75693.BF. [DOI] [PubMed] [Google Scholar]
- Daher EF, Lima RS, Silva Junior GB, Silva EC, Karbage NN, Kataoka RS, Carvalho Junior PC, Magalhaes MM, Mota RM, Liborio AB. Clinical presentation of leptospirosis: a retrospective study of 201 patients in a metropolitan city of Brazil. The Brazilian journal of infectious diseases : an official publication of the Brazilian Society of Infectious Diseases. 2010;14(1):3–10. [PubMed] [Google Scholar]
- Ellis T, Imrie A, Katz AR, Effler PV. Underrecognition of leptospirosis during a dengue fever outbreak in Hawaii, 2001–2002. Vector Borne Zoonotic Dis. 2008;14(4):541–547. doi: 10.1089/vbz.2007.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libraty DH, Myint KS, Murray CK, Gibbons RV, Mammen MP, Endy TP, Li W, Vaughn DW, Nisalak A, Kalayanarooj S. et al. A comparative study of leptospirosis and dengue in Thai children. PLoS Negl Trop Dis. 2007;14(3):e111. doi: 10.1371/journal.pntd.0000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JT, Gayer M, Connolly MA. Epidemics after natural disasters. Emerg Infect Dis. 2007;14(1):1–5. doi: 10.3201/eid1301.060779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J, Bornstein SL, Karpati AM, Bruce M, Bolin CA, Austin CC, Woods CW, Lingappa J, Langkop C, Davis B. et al. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin Infect Dis. 2002;14(12):1593–1599. doi: 10.1086/340615. [DOI] [PubMed] [Google Scholar]
- Farr RW. Leptospirosis. Clin Infect Dis. 1995;14(1):1–6. doi: 10.1093/clinids/21.1.1. quiz 7–8. [DOI] [PubMed] [Google Scholar]
- LaRocque RC, Breiman RF, Ari MD, Morey RE, Janan FA, Hayes JM, Hossain MA, Brooks WA, Levett PN. Leptospirosis during dengue outbreak, Bangladesh. Emerg Infect Dis. 2005;14(5):766–769. doi: 10.3201/eid1105.041212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajiki H, Salomao R. Association of plasma levels of tumor necrosis factor alpha with severity of disease and mortality among patients with leptospirosis. Clin Infect Dis. 1996;14(5):1177–1178. doi: 10.1093/clinids/23.5.1177. [DOI] [PubMed] [Google Scholar]
- Wagenaar JF, Gasem MH, Goris MG, Leeflang M, Hartskeerl RA, van der Poll T, van’t Veer C, Van Gorp EC. Soluble ST2 levels are associated with bleeding in patients with severe leptospirosis. PLoS Negl Trop Dis. 2009;14(6):e453. doi: 10.1371/journal.pntd.0000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy AL, Lafferty EI, Lovegrove FE, Krudsood S, Tangpukdee N, Liles WC, Kain KC. Whole blood angiopoietin-1 and -2 levels discriminate cerebral and severe (non-cerebral) malaria from uncomplicated malaria. Malar J. 2009;14(1):295. doi: 10.1186/1475-2875-8-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman LK, Dhabangi A, Musoke C, Conroy AL, Hawkes M, Higgins S, Rajwans N, Wolofsky KT, Streiner DL, Liles WC. et al. Combinations of host biomarkers predict mortality among Ugandan children with severe malaria: a retrospective case–control study. PloS one. 2011;14(2):e17440. doi: 10.1371/journal.pone.0017440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciuto DR, dos Santos CC, Hawkes M, Toltl LJ, Conroy AL, Rajwans N, Lafferty EI, Cook DJ, Fox-Robichaud A, Kahnamoui K. et al. Angiopoietin-1 and angiopoietin-2 as clinically informative prognostic biomarkers of morbidity and mortality in severe sepsis. Critical care medicine. 2011;14(4):702–710. doi: 10.1097/CCM.0b013e318206d285. [DOI] [PubMed] [Google Scholar]
- Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D. et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;14(10):e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy AL, Phiri H, Hawkes M, Glover S, Mallewa M, Seydel KB, Taylor TE, Molyneux ME, Kain KC. Endothelium-based biomarkers are associated with cerebral malaria in Malawian children: a retrospective case–control study. PLoS One. 2010;14(12):e15291. doi: 10.1371/journal.pone.0015291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankhambo LA, Banda DL, Jeffers G, White SA, Balmer P, Nkhoma S, Phiri H, Molyneux EM, Hart CA, Heyderman RS, Carol ED. The role of angiogenic factors in predicting clinical outcome in severe bacterial infection in Malawian children. Crit Care. 2010;14(3):R91. doi: 10.1186/cc9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butthep P, Chunhakan S, Yoksan S, Tangnararatchakit K, Chuansumrit A. Alteration of cytokines and chemokines during febrile episodes associated with endothelial cell damage and plasma leakage in dengue hemorrhagic fever. The Pediatric infectious disease journal. 2012;14(12):e232–e238. doi: 10.1097/INF.0b013e31826fd456. [DOI] [PubMed] [Google Scholar]
- Khongphatthanayothin A, Phumaphuti P, Thongchaiprasit K, Poovorawan Y. Serum levels of sICAM-1 and sE-selectin in patients with dengue virus infection. Jpn J Infect Dis. 2006;14(3):186–188. [PubMed] [Google Scholar]
- Krauel K, Weber C, Brandt S, Zahringer U, Mamat U, Greinacher A, Hammerschmidt S. Platelet factor 4 binding to lipid A of Gram-negative bacteria exposes PF4/heparin-like epitopes. Blood. 2012;14(16):3345–3352. doi: 10.1182/blood-2012-06-434985. [DOI] [PubMed] [Google Scholar]
- Shan NN, Zhu XJ, Peng J, Qin P, Zhuang XW, Wang HC, Hou M. Interleukin 18 and interleukin 18 binding protein in patients with idiopathic thrombocytopenic purpura. Br J Haematol. 2009;14(5):755–761. doi: 10.1111/j.1365-2141.2008.07520.x. [DOI] [PubMed] [Google Scholar]
- Shapiro NI, Yano K, Okada H, Fischer C, Howell M, Spokes KC, Ngo L, Angus DC, Aird WC. A prospective, observational study of soluble FLT-1 and vascular endothelial growth factor in sepsis. Shock. 2008;14(4):452–457. doi: 10.1097/SHK.0b013e31815072c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;14(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- Sanders EJ, Rigau-Perez JG, Smits HL, Deseda CC, Vorndam VA, Aye T, Spiegel RA, Weyant RS, Bragg SL. Increase of leptospirosis in dengue-negative patients after a hurricane in Puerto Rico in 1996 [correction of 1966] The American journal of tropical medicine and hygiene. 1999;14(3):399–404. doi: 10.4269/ajtmh.1999.61.399. [DOI] [PubMed] [Google Scholar]
- Tricou V, Vu HT, Quynh NV, Nguyen CV, Tran HT, Farrar J, Wills B, Simmons CP. Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis. 2010;14:142. doi: 10.1186/1471-2334-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SR, Meyer M, Semple MG, Simmons CP, Sekaran SD, Huang JX, McElnea C, Huang CY, Valks A, Young PR. et al. The diagnostic sensitivity of dengue rapid test assays is significantly enhanced by using a combined antigen and antibody testing approach. PLoS Negl Trop Dis. 2011;14(6):e1199. doi: 10.1371/journal.pntd.0001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Loyden G, Arribas J, Lopez-Casillas F. The shedding of betaglycan is regulated by pervanadate and mediated by membrane type matrix metalloprotease-1. J Biol Chem. 2004;14(9):7721–7733. doi: 10.1074/jbc.M306499200. [DOI] [PubMed] [Google Scholar]
- Levine RJ, Lam C, Qian C, Yu KF, Maynard SE, Sachs BP, Sibai BM, Epstein FH, Romero R, Thadhani R. et al. Soluble endoglin and other circulating antiangiogenic factors in preeclampsia. N Engl J Med. 2006;14(10):992–1005. doi: 10.1056/NEJMoa055352. [DOI] [PubMed] [Google Scholar]
- Venkatesha S, Toporsian M, Lam C, Hanai J, Mammoto T, Kim YM, Bdolah Y, Lim KH, Yuan HT, Libermann TA. et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med. 2006;14(6):642–649. doi: 10.1038/nm1429. [DOI] [PubMed] [Google Scholar]
- Dietmann A, Helbok R, Lackner P, Fischer M, Reindl M, Lell B, Issifou S, Kremsner PG, Schmutzhard E. Endoglin in African children with Plasmodium falciparum malaria: a novel player in severe malaria pathogenesis? The Journal of infectious diseases. 2009;14(12):1842–1848. doi: 10.1086/648476. [DOI] [PubMed] [Google Scholar]
- Silver KL, Conroy AL, Leke RG, Leke RJ, Gwanmesia P, Molyneux ME, Taylor DW, Rogerson SJ, Kain KC. Circulating soluble endoglin levels in pregnant women in Cameroon and Malawi–associations with placental malaria and fetal growth restriction. PloS one. 2011;14(9):e24985. doi: 10.1371/journal.pone.0024985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH, Busch M, Peters MG, Rosen HR, Thomas DL. et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. The Journal of infectious diseases. 2010;14(9):1371–1380. doi: 10.1086/651606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo RC, Munoz PM, de Miguel MJ, Marin CM, Blasco JM, Gortazar C, Kocan KM, de la Fuente J. Differential expression of inflammatory and immune response genes in rams experimentally infected with a rough virulent strain of Brucella ovis. Vet Immunol Immunopathol. 2009;14(3–4):295–303. doi: 10.1016/j.vetimm.2008.10.326. [DOI] [PubMed] [Google Scholar]
- de Fost M, Hartskeerl RA, Groenendijk MR, van der Poll T. Interleukin 12 in part regulates gamma interferon release in human whole blood stimulated with Leptospira interrogans. Clin Diagn Lab Immunol. 2003;14(2):332–335. doi: 10.1128/CDLI.10.2.332-335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SM, Mammoto T, Schultz A, Yuan HT, Christiani D, Karumanchi SA, Sukhatme VP. Excess circulating angiopoietin-2 may contribute to pulmonary vascular leak in sepsis in humans. PLoS Med. 2006;14(3):e46. doi: 10.1371/journal.pmed.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AV, Tarr PI, Watkins SL, Rajwans N, Petruzziello-Pellegrini TN, Marsden PA, Kain KC, Liles WC. Dysregulation of Angiopoietin 1 and 2 in Escherichia coli O157:H7 infection and the hemolytic-uremic syndrome. J Infect Dis. 2013;14(6):929–933. doi: 10.1093/infdis/jit268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SM, Rajwans N, Tapia KA, Jaoko W, Estambale BB, McClelland RS, Overbaugh J, Liles WC. A prospective study of endothelial activation biomarkers, including plasma angiopoietin-1 and angiopoietin-2, in Kenyan women initiating antiretroviral therapy. BMC Infect Dis. 2013;14(1):263. doi: 10.1186/1471-2334-13-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motla M, Manaktala S, Gupta V, Aggarwal M, Bhoi SK, Aggarwal P, Goel A. Sonographic evidence of ascites, pleura-pericardial effusion and gallbladder wall edema for dengue fever. Prehospital and disaster medicine. 2011;14(5):335–341. doi: 10.1017/S1049023X11006637. [DOI] [PubMed] [Google Scholar]
- De Brito T, Bohm GM, Yasuda PH. Vascular damage in acute experimental leptospirosis of the guinea-pig. J Pathol. 1979;14(4):177–182. doi: 10.1002/path.1711280403. [DOI] [PubMed] [Google Scholar]
- Fiedler U, Augustin HG. Angiopoietins: a link between angiogenesis and inflammation. Trends Immunol. 2006;14(12):552–558. doi: 10.1016/j.it.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Novotny NM, Lahm T, Markel TA, Crisostomo PR, Wang M, Wang Y, Tan J, Meldrum DR. Angiopoietin-1 in the treatment of ischemia and sepsis. Shock. 2008;14(4):1–9. doi: 10.1097/SHK.0b013e3181862c63. [DOI] [PubMed] [Google Scholar]
- Michels M, van der Ven AJ, Djamiatun K, Fijnheer R, de Groot PG, Griffioen AW, Sebastian S, Faradz SM, de Mast Q. Imbalance of angiopoietin-1 and angiopoetin-2 in severe dengue and relationship with thrombocytopenia, endothelial activation, and vascular stability. The American journal of tropical medicine and hygiene. 2012;14(5):943–946. doi: 10.4269/ajtmh.2012.12-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster A, Chao YB, Mosior M, Ford A, Gonzalez-DeWhitt PA, Hale JE, Li D, Qiu Y, Fraser CC, Yang DD. et al. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;14(11):4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Kama M, Ge G, Xie K, Tong W, Hug C, Lodish HF. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat Med. 2006;14(2):240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch G, Pisabarro MT, Sherman D, Kowalski J, Nagel M, Hass P, Xie MH, Gurney A, Bodary S, Liang XH. et al. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J Biol Chem. 2002;14(19):17281–17290. doi: 10.1074/jbc.M109768200. [DOI] [PubMed] [Google Scholar]
- Li Y, Sun L, Xu H, Fang Z, Yao W, Guo W, Rao J, Zha X. Angiopoietin-like protein 3 modulates barrier properties of human glomerular endothelial cells through a possible signaling pathway involving phosphatidylinositol-3 kinase/protein kinase B and integrin alphaVbeta3. Acta Biochim Biophys Sin (Shanghai) 2008;14(6):459–465. doi: 10.1111/j.1745-7270.2008.00421.x. [DOI] [PubMed] [Google Scholar]
- van der Heijden M, van Nieuw Amerongen GP, van Hinsbergh VW, Groeneveld AB. The interaction of soluble Tie2 with angiopoietins and pulmonary vascular permeability in septic and nonseptic critically ill patients. Shock. 2010;14(3):263–268. doi: 10.1097/SHK.0b013e3181b2f978. [DOI] [PubMed] [Google Scholar]
- Conroy AL, Glover SJ, Hawkes M, Erdman LK, Seydel KB, Taylor TE, Molyneux ME, Kain KC. Angiopoietin-2 levels are associated with retinopathy and predict mortality in Malawian children with cerebral malaria: a retrospective case–control study. Crit Care Med. 2012;14(3):952–959. doi: 10.1097/CCM.0b013e3182373157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL, Reusch P, Barleon B, Hang C, Dobbs N, Marme D. Soluble Tie2 and Flt1 extracellular domains in serum of patients with renal cancer and response to antiangiogenic therapy. Clin Cancer Res. 2001;14(7):1992–1997. [PubMed] [Google Scholar]
- Engin H, Ustundag Y, Tekin IO, Gokmen A, Ertop S, Ilikhan SU. Plasma concentrations of angiopoietin-1, angiopoietin-2 and Tie-2 in colon cancer. European cytokine network. 2012;14(2):68–71. doi: 10.1684/ecn.2012.0308. [DOI] [PubMed] [Google Scholar]
- Pousa ID, Mate J, Salcedo-Mora X, Abreu MT, Moreno-Otero R, Gisbert JP. Role of vascular endothelial growth factor and angiopoietin systems in serum of Crohn’s disease patients. Inflamm Bowel Dis. 2008;14(1):61–67. doi: 10.1002/ibd.20269. [DOI] [PubMed] [Google Scholar]
- Gomez RM, Vieira ML, Schattner M, Malaver E, Watanabe MM, Barbosa AS, Abreu PA, de Morais ZM, Cifuente JO, Atzingen MV. et al. Putative outer membrane proteins of Leptospira interrogans stimulate human umbilical vein endothelial cells (HUVECS) and express during infection. Microb Pathog. 2008;14(5–6):315–322. doi: 10.1016/j.micpath.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Vieira ML, D’Atri LP, Schattner M, Habarta AM, Barbosa AS, de Morais ZM, Vasconcellos SA, Abreu PA, Gomez RM, Nascimento AL. A novel leptospiral protein increases ICAM-1 and E-selectin expression in human umbilical vein endothelial cells. FEMS Microbiol Lett. 2007;14(2):172–180. doi: 10.1111/j.1574-6968.2007.00924.x. [DOI] [PubMed] [Google Scholar]
- Turner GD, Ly VC, Nguyen TH, Tran TH, Nguyen HP, Bethell D, Wyllie S, Louwrier K, Fox SB, Gatter KC. et al. Systemic endothelial activation occurs in both mild and severe malaria. Correlating dermal microvascular endothelial cell phenotype and soluble cell adhesion molecules with disease severity. Am J Pathol. 1998;14(6):1477–1487. [PMC free article] [PubMed] [Google Scholar]
- Paris DH, Jenjaroen K, Blacksell SD, Phetsouvanh R, Wuthiekanun V, Newton PN, Day NP, Turner GD. Differential patterns of endothelial and leucocyte activation in ‘typhus-like’ illnesses in Laos and Thailand. Clin Exp Immunol. 2008;14(1):63–67. doi: 10.1111/j.1365-2249.2008.03673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathirat P, Isarangkura P, Srichaikul T, Suvatte V, Mitrakul C. Abnormal hemostasis in dengue hemorrhagic fever. Southeast Asian J Trop Med Public Health. 1993;14(Suppl 1):80–85. [PubMed] [Google Scholar]
- Lin YP, McDonough SP, Sharma Y, Chang YF. Leptospira immunoglobulin-like protein B (LigB) binding to the C-terminal fibrinogen alphaC domain inhibits fibrin clot formation, platelet adhesion and aggregation. Molecular microbiology. 2011;14(4):1063–1076. doi: 10.1111/j.1365-2958.2010.07510.x. [DOI] [PubMed] [Google Scholar]
- Nascimento EJ, Silva AM, Cordeiro MT, Brito CA, Gil LH, Braga-Neto U, Marques ET. Alternative complement pathway deregulation is correlated with dengue severity. PloS one. 2009;14(8):e6782. doi: 10.1371/journal.pone.0006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avirutnan P, Hauhart RE, Marovich MA, Garred P, Atkinson JP, Diamond MS. Complement-mediated neutralization of dengue virus requires mannose-binding lectin. mBio. 2011;14(6):e00276-11. doi: 10.1128/mBio.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AS, Abreu PA, Vasconcellos SA, Morais ZM, Goncales AP, Silva AS, Daha MR, Isaac L. Immune evasion of leptospira species by acquisition of human complement regulator C4BP. Infect Immun. 2009;14(3):1137–1143. doi: 10.1128/IAI.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri T, Murgia R, Stefanel P, Meri S, Cinco M. Regulation of complement activation at the C3-level by serum resistant leptospires. Microb Pathog. 2005;14(4):139–147. doi: 10.1016/j.micpath.2005.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Table S2 accompany the manuscript. They provide statistical validation for the two logistic regression models presented in Figure 1.