Summary
There are no therapies that reverse the proteotoxic misfolding events that underpin fatal neurodegenerative diseases including amyotrophic lateral sclerosis (ALS) and Parkinson disease (PD). Hsp104, a conserved hexameric AAA+ protein from yeast, solubilizes disordered aggregates and amyloid, but has no metazoan homologue and only limited activity against human neurodegenerative disease proteins. Here, we reprogram Hsp104 to rescue TDP-43, FUS, and α-synuclein proteotoxicity by mutating single residues in helix 1, 2, or 3 of the middle domain or the small domain of nucleotide-binding domain 1. Potentiated Hsp104 variants enhance aggregate dissolution, restore proper protein localization, suppress proteotoxicity, and in a C. elegans PD model attenuate dopaminergic neurodegeneration. Potentiating mutations reconfigure how Hsp 104 subunits collaborate, desensitize Hsp104 to inhibition, obviate any requirement for Hsp70, and enhance ATPase, translocation, and unfoldase activity. Our work establishes that disease-associated aggregates and amyloid are tractable targets and that enhanced disaggregases can restore proteostasis and mitigate neurodegeneration.
Introduction
Protein misfolding underpins several fatal neurodegenerative disorders including amyotrophic lateral sclerosis (ALS) and Parkinson disease (PD) (Cushman et al., 2010). In PD, α-synuclein (α-syn) forms highly toxic pre-fibrillar oligomers and amyloid fibrils that accumulate in cytoplasmic Lewy bodies (Cushman et al., 2010). In ALS, TDP-43 or FUS accumulate in cytoplasmic inclusions in degenerating motor neurons (Robberecht and Philips, 2013). Unfortunately, treatments for these disorders are palliative and ineffective due to the apparent intractability of aggregated proteins. Effective therapies are urgently needed that eliminate the causative proteotoxic misfolded conformers via degradation or reactivation of the proteins to their native fold.
Inspiration can be drawn from nature where amyloidogenesis and protein misfolding have been subjugated for adaptive modalities (Newby and Lindquist, 2013). For example, beneficial yeast prions are tightly regulated by Hsp104, a hexameric AAA+ protein, which rapidly deconstructs various amyloids and pre-fibrillar oligomers (DeSantis et al., 2012; Lo Bianco et al., 2008; Newby and Lindquist, 2013). Hsp104 also reactivates proteins from disordered aggregates after environmental stress (Shorter, 2008). Hsp104 is highly conserved in eubacteria and eukaryotes except for metazoa, which bafflingly lack an Hsp104 homologue and display limited ability to disaggregate disordered and amyloid aggregates (Duennwald et al., 2012; Shorter, 2008, 2011). Thus, Hsp104 could be harnessed to augment human proteostasis and counter protein misfolding in neurodegenerative disease (Shorter, 2008). Indeed, Hsp104 synergizes with human Hsp70 and Hsp40 to resolve various misfolded species linked with human neurodegenerative disease and can partially antagonize protein misfolding and neurodegeneration in metazoa (Cushman-Nick et al., 2013; DeSantis et al., 2012; Duennwald et al., 2012; Lo Bianco et al., 2008; Shorter, 2011; Vacher et al., 2005). Hsp70 overexpression can also mitigate neurodegeneration (Cushman-Nick et al., 2013). However, these potentially therapeutic activities remain limited and vast improvements are needed to maximize therapeutic potential. Indeed, very high concentrations of Hsp104 are needed to antagonize human neurodegenerative disease proteins, which Hsp104 never ordinarily encounters, and some substrates are refractory to Hsp104 (DeSantis et al., 2012; Lo Bianco et al., 2008).
A key but elusive goal is to engineer or evolve optimized chaperones against neurodegenerative disease substrates to maximize therapeutic efficacy (Shorter, 2008). Chaperones are impractical targets for protein engineering due to their typically large size, and protein disaggregases such as Hsp104 have poorly understood structures, making rational design challenging (Saibil, 2013). Here, we broach this issue and isolate potentiated Hsp104 variants that eradicate TDP-43, FUS, and α-syn aggregates and potently suppress toxicity. We report the first artificially engineered chaperones to optimize proteostasis and thwart neurodegeneration. We suggest that neuroprotection may be possible for diverse neurodegenerative diseases via subtle structural modifications of existing chaperones.
Results
Substrate-binding tyrosines in Hsp104 pore loops are optimal for disaggregation
Hsp104 is adapted for disaggregation of the yeast proteome. We sought to engineer Hsp104 variants to disaggregate TDP-43, an RNA-binding protein with a prion-like domain (Cushman et al., 2010), which has no yeast homologue and is not a natural Hsp104 substrate. A yeast model of TDP-43 proteinopathies has been developed in which TDP-43 is overexpressed via a galactose-inducible promoter (Johnson et al., 2008). TDP-43 aggregates in the cytoplasm and is toxic to yeast, which phenocopies TDP-43 pathology in disease and has enabled identification of common ALS genetic risk factors (Elden et al., 2010). To explore Hsp104 sequence space against TDP-43 toxicity we employed Δhsp104 yeast to assess Hsp104 variants in the absence of wild-type (WT) Hsp104. TDP-43 is highly toxic in Δhsp104 yeast and Hsp104WT provides minimal rescue of toxicity (Johnson et al., 2008). Thus, Δhsp104 yeast provide a platform to isolate more active Hsp104 variants. Each Hsp104 monomer contains two nucleotide binding domains (NBD1 and NBD2), as well as an N-terminal, middle, and C-terminal domain (DeSantis and Shorter, 2012). Hsp104 forms ring-shaped hexamers with a central pore through which substrate is threaded. To alter substrate specificity we assessed Hsp104 variants bearing mutations in Hsp104’s two substrate-binding pore loops (DeSantis and Shorter, 2012). We mutated the conserved pore loop residues, Y257 and Y662, which mediate substrate binding and translocation (Tessarz et al., 2008) to all amino acids and screened this library of 400 variants for rescue of TDP-43 toxicity. After several rounds of selection, nearly all the variants possessed Y at one, or more often, both pore-loop positions. None of the pore-loop Hsp104 variants were more active than Hsp104WT in rescuing TDP-43 toxicity. Thus, Y257 and Y662 are likely optimal for disaggregation.
Select missense mutations in the middle domain potentiate Hsp104 activity
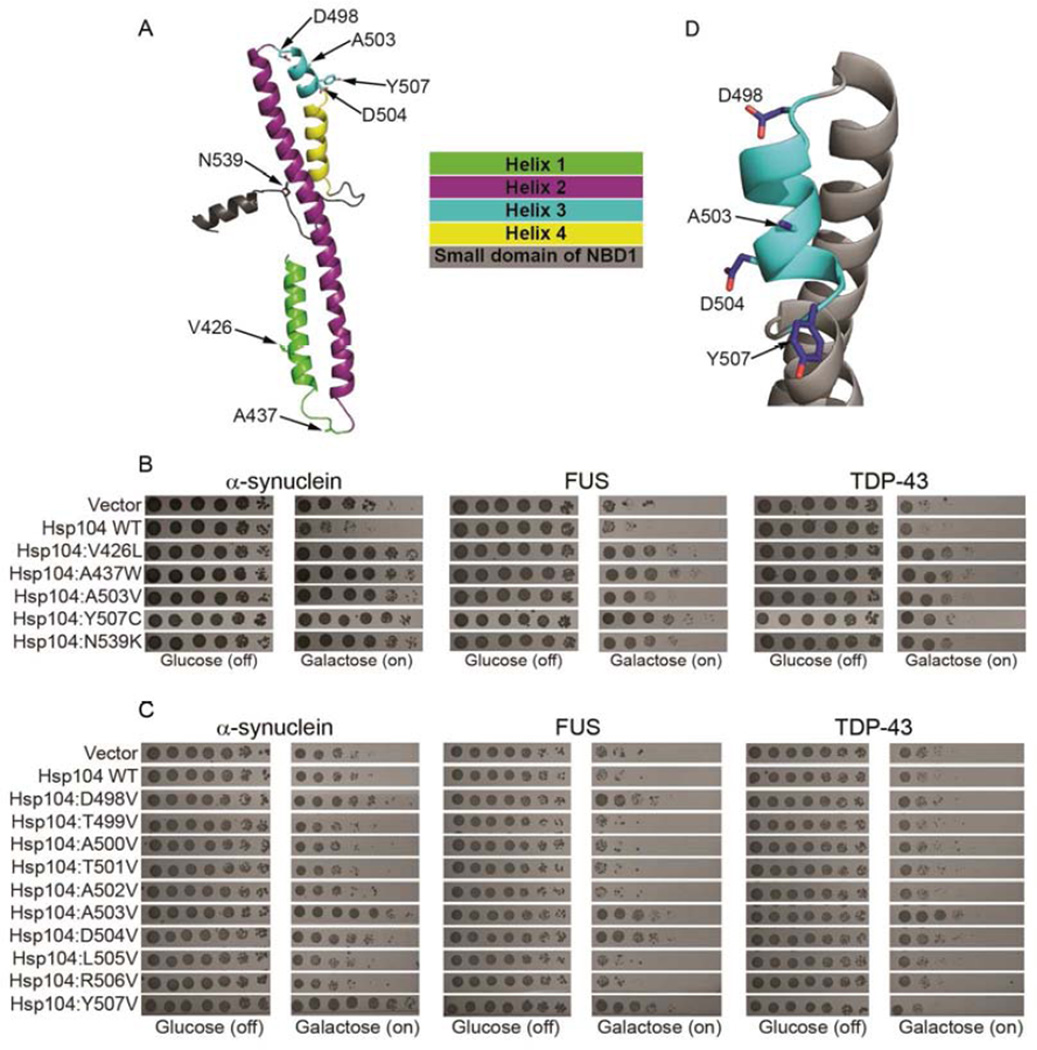
Next, we explored the coiled-coil middle domain (MD) of Hsp104, which is less conserved than the substrate-binding pore loops. MD variants can have unexpected gain of function phenotypes (Schirmer et al., 2004). The Hsp104 MD (residues 411–538; Fig. 1A) facilitates: optimal ATPase activity, communication between NBD1 and NBD2, intrinsic disaggregase activity, and interactions with Hsp70 during disordered aggregate dissolution (DeSantis and Shorter, 2012). We randomly mutagenized the MD and screened this Hsp104 library against α-syn, FUS, or TDP-43 toxicity (Johnson et al., 2008; Outeiro and Lindquist, 2003; Sun et al., 2011). We employed Δhsp104 yeast, as deletion of Hsp104 does not affect α-syn, FUS, or TDP-43 toxicity (Johnson et al., 2008; Ju et al., 2011). We identified several Hsp104 variants that potently rescued α-syn, FUS, and TDP-43 toxicity, whereas Hsp104WT was ineffective (Fig. 1B). Potentiated Hsp104 variants had a missense mutation in helix 1 (Hsp104V426L), or in the distal loop between helix 1 and 2 (Hsp104A437W), or in helix 3 (Hsp104A503V or Hsp104Y507C) (Fig. 1A, B). Unexpectedly, we uncovered an enhanced variant with a missense mutation in the NBD1 small domain (Hsp104N539K) (Fig. 1A, B). Thus, the MD or small domain of NBD1 can be mutated to potentiate Hsp104 activity against α-syn, FUS, and TDP-43.
Figure 1. Hsp104 MD variants rescue diverse proteotoxicity models.
(A) Homology model of the MD and a portion of the small domain of NBD1 of Hsp104. Side chains of key residues are shown as sticks. (B) Δhsp104 yeast strains integrated with galactose-inducible α-syn, FUS, or TDP-43 were transformed with the indicated Hsp104 variant or vector control. Strains were serially diluted fivefold and spotted on glucose (off) or galactose (on) media. (C) Hsp104 variants harboring missense mutations to valine ranging from residue D498 to Y507 were expressed with FUS, TDP-43, or α-syn. (D) Close-up of MD helix 3 from (A). Mutation of D498, A503, D504, or Y507 activates Hsp104.
See also Fig. S1.
Two potentiating mutations: A503V and Y507C lie in MD helix 3. Thus, we performed a valine scan of helix 3 (residues 498–507) in search of additional enhanced variants (Fig. 1C, D). Most helix-3 valine substitutions behaved like Hsp104WT (Fig. 1C). However, Hsp104D504V suppressed α-syn, FUS, and TDP-43 toxicity (Fig. 1C). Hsp104D498V and Hsp104Y507V suppressed FUS and α-syn toxicity, but not TDP-43 toxicity (Fig. 1C). Thus, select missense mutations in helix 3 engender potentiated Hsp104 variants with altered substrate specificity.
Two different Y507 mutations yielded enhanced variants. Thus, we explored other substitutions at this position. Hsp104Y507A, Hsp104Y507C, and Hsp104Y507D rescued α-syn, FUS, and TDP-43 toxicity (Fig. S1). Likewise, additional substitutions at D504 (to C), V426 (to G), or N539 (to E, D, G, or K) yielded potentiated Hsp104 variants against FUS toxicity (Fig. S1). Thus, diverse mutations at specific positions in the MD enhance Hsp104 activity.
Hsp104A503X variants suppress TDP-43 toxicity and promote its proper localization
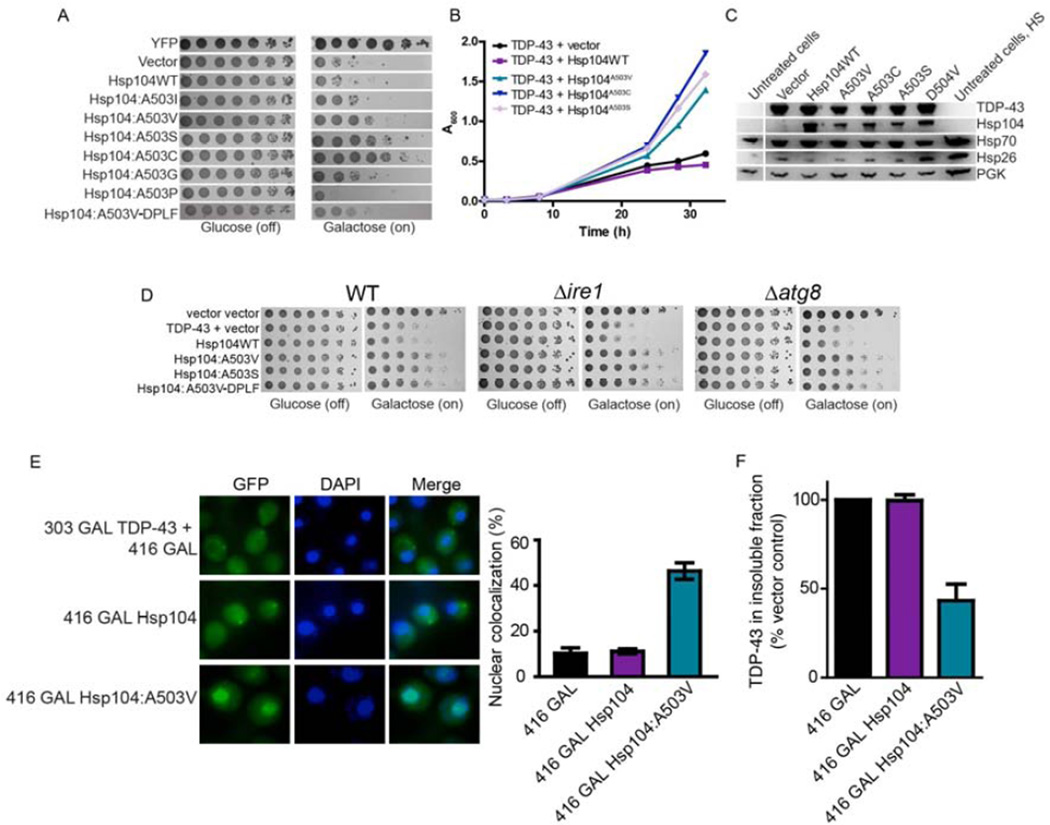
Hsp104A503V was among the strongest suppressors of α-syn, FUS, and TDP-43 toxicity, and so we explored this position further and mutated A503 to all amino acids. None of these Hsp104 variants were toxic to yeast when overexpressed at 30°C (Fig. S2). Mutation of A503 to V, S, or C suppressed TDP-43 toxicity: Hsp104A503C most strongly suppressed TDP-43 toxicity, followed by Hsp104A503S and Hsp104A503V (Fig. 2A, B, S3A). Surprisingly, mutation of A503 to nearly any residue suppressed TDP-43 toxicity, whereas Hsp104A503P enhanced toxicity (Fig. 2A, S3A). Indeed, we could now mutate the conserved pore loop Y residues (Y257 and Y662) to F (Hsp104A503V-DPLF) and retain suppression of TDP-43 toxicity (Fig. 2A). Rescue of TDP-3 toxicity was not due to lower levels of TDP-43, which was roughly equal across strains (Fig. 2C). Likewise, rescue could not be explained by higher Hsp104 expression. Hsp104 variants were expressed at slightly lower levels than Hsp104WT (Fig. 2C). Quantitative immunoblot revealed that Hsp104 hexamer:TDP-43 ratios were ~1:1.31 for Hsp104WT and ~1:2.20 for Hsp104A503V.
Figure 2. Hsp104A503X variants suppress TDP-43 toxicity, aggregation, and mislocalization.
(A) Δhsp104 yeast transformed with TDP-43 and Hsp104 variants, or YFP and vector, were serially diluted fivefold and spotted onto glucose (off) or galactose (on). (B) Selected strains from (A) were induced in liquid and growth was monitored by A600nm. (C) Strains from (B) were induced for 5h, lysed, and immunoblotted. Uninduced (untreated) and heat shocked cells (HS) serve as controls. 3-Phosphoglycerate kinase (PGK1) serves as a loading control. (D) WT, Δire1, or Δatg8 yeast were co-transformed with vector control, or TDP-43 plus vector, or the indicated Hsp104 variant and were serially diluted fivefold and spotted onto glucose (off) or galactose (on). (E) Fluorescence microscopy of cells co-expressing fluorescently tagged TDP-43 and Hsp104WT, Hsp104A503V, or vector. Cells were stained with DAPI to visualize nuclei (blue). TDP-43 localization was quantified by counting the number of cells containing colocalized nuclear staining. Values represent means±SEM (n=3). (F) Δhsp104 yeast co-transformed with TDP-43 and vector or the indicated Hsp104 variant were induced with galactose for 5h at 30°C, lysed and processed for s edimentation analysis and quantitative immunoblot. The relative amount of insoluble TDP-43 was determined as a % of the vector control. Values represent means±SEM (n=2).
See also Fig. S2, S3, and S4.
Hsp70 and Hsp26 levels were also similar for all strains, indicating that Hsp104 variants do not induce a heat shock response (HSR; Fig. 2C). Hsp104A503V expression from the native Hsp104 promoter (which is weaker than the galactose promoter) suppressed TDP-43 toxicity (Fig. S4A, B). Here, quantitative immunoblot revealed that Hsp104 hexamer:TDP-43 ratios were ~1:1.70 for Hsp104WT and ~1:4.55 for Hsp104A503V. Thus, even low Hsp104A503V levels rescued TDP-43 toxicity. Finally, Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF rescued TDP-43 toxicity in Δire1 (to disrupt the unfolded protein response [UPR]) and Δatg8 (to disrupt autophagy) strains (Fig. 2D). Thus, neither the UPR nor autophagy, are needed for enhanced Hsp104 variants to rescue TDP-43 toxicity.
TDP-43 normally shuttles between the nucleus and cytoplasm. However, in ALS, TDP-43 is usually depleted from the nucleus and aggregated in the cytoplasm (Robberecht and Philips, 2013). Indeed, cytoplasmic TDP-43 aggregates persist upon Hsp104WT overexpression (Fig. 2E). By contrast, Hsp104A503V eliminated cytoplasmic TDP-43 aggregates and ~46% of cells had nuclear TDP-43 localization (Fig. 2E). Accordingly, Hsp104A503V reduced the amount of insoluble TDP-43 by ~57%, whereas Hsp104WT was ineffective (Fig. 2F). Thus, Hsp104A503V eliminates TDP-43 aggregation and toxicity and restores TDP-43 to the nucleus. These phenotypes are a therapeutic goal for ALS and other TDP-43 proteinopathies. Several suppressors of TDP-43 toxicity have been isolated in yeast, but none clear cytoplasmic TDP-43 aggregates (Sun et al., 2011). Thus, our enhanced Hsp104 variants are the first genetic suppressors that eradicate TDP-43 aggregates and restore TDP-43 to the nucleus.
Hsp104A503X variants suppress FUS toxicity and aggregation
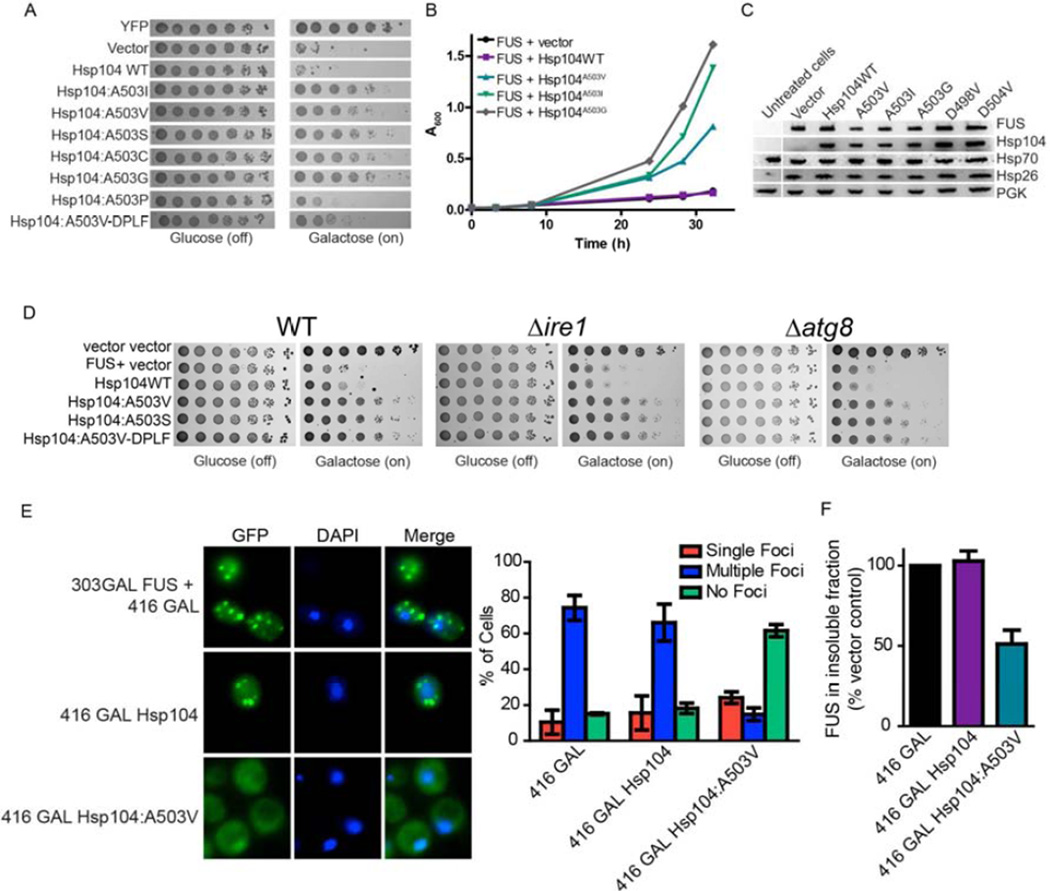
Next, we tested Hsp104A503X variants for rescue of FUS toxicity in yeast. FUS, like TDP-43 is a nuclear RNA-binding protein with a prion-like domain that forms cytoplasmic aggregates in degenerating neurons of FUS proteinopathy patients and in yeast (Ju et al., 2011; Robberecht and Philips, 2013; Sun et al., 2011). As for TDP-43, mutation of A503 to any amino acid except P strongly suppressed FUS toxicity, as did Hsp104A503V-DPLF (Fig. 3A, B, S3B). Hsp104A503G most strongly suppressed FUS toxicity (Fig. 3A, B, S3B). Rescue of FUS toxicity by Hsp104A503X variants (or Hsp104D498V or Hsp104D504V) could not be explained by: lower FUS levels, induction of Hsp70 or Hsp26 in a HSR, or higher Hsp104 levels (Fig. 3C). Indeed, quantitative immunoblot revealed that Hsp104 hexamer:FUS ratios were ~1:5.13 for Hsp104WT and ~1:3.25 for Hsp104A503V. Even low Hsp104A503V levels expressed from the natural Hsp104 promoter suppressed FUS toxicity (Fig. S4C, D). Here, quantitative immunoblot revealed that Hsp104 hexamer:FUS ratios were ~1:5.21 for Hsp104WT and ~1:9.58 for Hsp104A503V. Rescue of FUS toxicity by Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF occurred in Δire1 strains and Δatg8 strains (Fig. 3D). Thus, the UPR and autophagy are not required for potentiated Hsp104 variants to suppress FUS toxicity.
Figure 3. Hsp104A503X variants suppress FUS toxicity and aggregation.
(A) Δhsp104 yeast transformed with FUS and the Hsp104 variants, or YFP and vector, were serially diluted fivefold and spotted onto glucose (off) or galactose (on). (B) Selected strains from (A) were induced in liquid and growth was monitored by A600nm. (C) Strains from (B) were induced for 5h, lysed, and immunoblotted. (D) WT, Δire1, or Δatg8 yeast were co-transformed with vector control, or FUS plus vector, or the indicated Hsp104 variant and were serially diluted fivefold and spotted onto glucose (off) or galactose (on). (E) Fluorescence microscopy of cells co-expressing FUS-GFP and Hsp104WT, Hsp104A503V, or vector. Cells were stained with Hoechst dye to visualize nuclei (blue). FUS aggregation was quantified by counting the number of cells containing 0, 1, or more than 1 foci. Values represent means±SEM (n=3). (F) Δhsp104 yeast co-transformed with FUS and vector or the indicated Hsp104 variant were induced with galactose for 5h at 30°C, lysed and processed for sedimentation analysis and quantitative immunoblot. The relative amount of insoluble FUS was determined as a % of the vector control. Values represent means±SEM (n=2).
See also Fig. S3 and S4.
Hsp104A503V eliminated FUS aggregates, whereas Hsp104WT had no effect (Fig. 3E). In contrast to TDP-43, FUS was now diffuse in the cytoplasm (Fig. 3E) because the yeast nuclear transport machinery fails to decode the FUS PY-NLS (Ju et al., 2011). Hsp104A503V reduced the amount of insoluble FUS by ~49%, whereas Hsp104WT was ineffective (Fig. 3F). Genome-wide overexpression screens have yielded several suppressors of FUS toxicity in yeast, but none that solubilize FUS inclusions (Ju et al., 2011; Sun et al., 2011). Thus, potentiated Hsp104 variants are the first genetic suppressors that eradicate FUS aggregates.
Hsp104A503X variants suppress α-syn toxicity and promote its proper localization
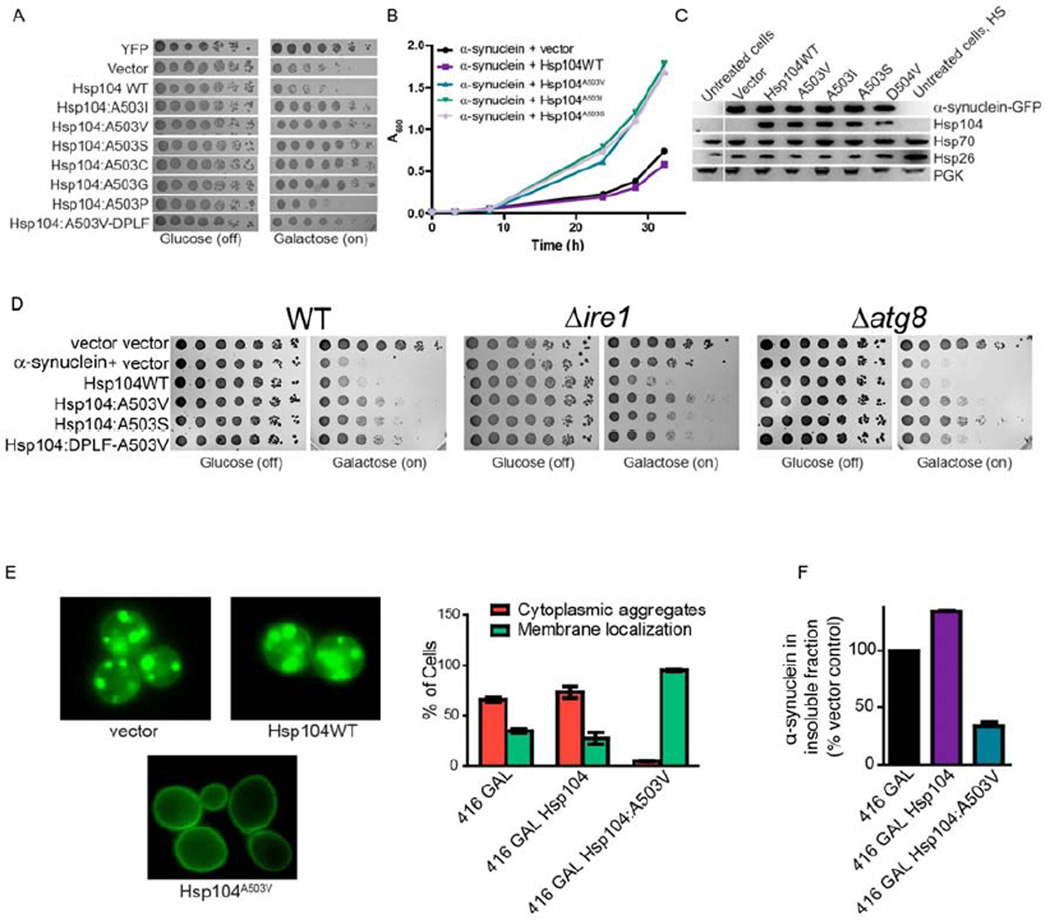
Next, we tested Hsp104A503X variants against α-syn toxicity in yeast. α-Syn is a lipid-binding protein that localizes to the plasma membrane, but forms cytoplasmic inclusions in degenerating dopaminergic neurons in PD and in yeast (Cushman et al., 2010; Outeiro and Lindquist, 2003). Nearly all Hsp104A503X variants suppressed α-syn toxicity except Hsp104A503P, which had no effect (Fig. 4A, B, S3C). By contrast, Hsp104WT slightly enhanced α-syn toxicity (Figure 4A, B). Hsp104A503V-DPLF suppressed α-syn toxicity, though not as strongly as Hsp104A503V (Fig. 4A). Rescue of α-syn toxicity by Hsp104A503X variants (or Hsp104D504V) could not be explained by: lower α-syn levels, induction of Hsp70 or Hsp26 in a HSR, or higher Hsp104 levels (Fig. 4C). Quantitative immunoblot indicated that the Hsp104 hexamer:α-syn ratios were ~1:2.43 for Hsp104WT and ~1:2.84 for Hsp104A503V. Expression of Hsp104A503V from the Hsp104 promoter suppressed α-syn toxicity, whereas Hsp104WT had no effect (Fig. S4E, F). Here, quantitative immunoblot indicated that the Hsp104 hexamer:α-syn ratios were ~1:3.03 for Hsp104WT and ~1:5.79 for Hsp104A503V. Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF rescued α-syn toxicity in Δire1 and Δatg8 strains (Fig. 4D). Thus, the UPR and autophagy are not required for rescue.
Figure 4. Hsp104A503X variants suppress α-syn toxicity, aggregation, and mislocalization.
(A) Δhsp104 yeast co-transformed with two copies of α-syn-YFP and the Hsp104 variants, or YFP and vector, were serially diluted fivefold and spotted onto glucose (off) or galactose (on). (B) Selected strains from (A) were induced in liquid and growth was monitored by A600nm. (C) Strains from (B) were induced for 8h in galactose, lysed, and immunoblotted. (D) WT, Δire1, or Δatg8 yeast were co-transformed with vector controls, or α-syn plus vector or the indicated Hsp104 variant and were serially diluted fivefold and spotted onto glucose (off) or galactose (on). (E) Fluorescence microscopy of cells co-expressing α-syn-YFP and Hsp104WT, Hsp104A503V, or vector. α-Syn localization was quantified by counting the number of cells with plasma membrane fluorescence or cytoplasmic aggregates. Values represent means±SEM (n=3). (F) Δhsp104 yeast co-transformed with α-syn and vector or the indicated Hsp104 variant were induced with galactose for 8h at 30°C, lysed a nd processed for sedimentation analysis and quantitative immunoblot. The relative amount of insoluble α-syn was determined as a % of the vector control. Values represent means±SEM (n=2).
See also Fig. S3 and S4.
Hsp104A503V eliminated cytoplasmic α-syn inclusions and restored plasma membrane α-syn localization, whereas Hsp104WT had no effect (Fig. 4E). Indeed, Hsp104A503V reduced the amount of insoluble α-syn by ~66%, whereas Hsp104WT increased it by ~33.9% (Fig. 4F). Thus, potentiated Hsp104 variants eradicate α-syn inclusions and restore α-syn localization.
Potentiated Hsp104 variants prevent neurodegeneration in a C. elegans PD model
To test potentiated Hsp104 variants in a metazoan nervous system, we used a transgenic C. elegans PD model, which has illuminated mechanisms and modifiers of α-syn-induced neurodegeneration (Cao et al., 2005; Cooper et al., 2006; Tardiff et al., 2013). We selected Hsp104A503S and Hsp104A503V-DPLF to study in this context, which displayed strong (Hsp104A503S) and moderate (Hsp104A503V-DPLF) rescue of α-syn toxicity (Fig. 4A). We focused on these variants because unlike Hsp104A503V they conferred greater than WT levels of thermotolerance and were less toxic to yeast at 37°C when expressed from the galactose promoter (Fig. S5A, B).
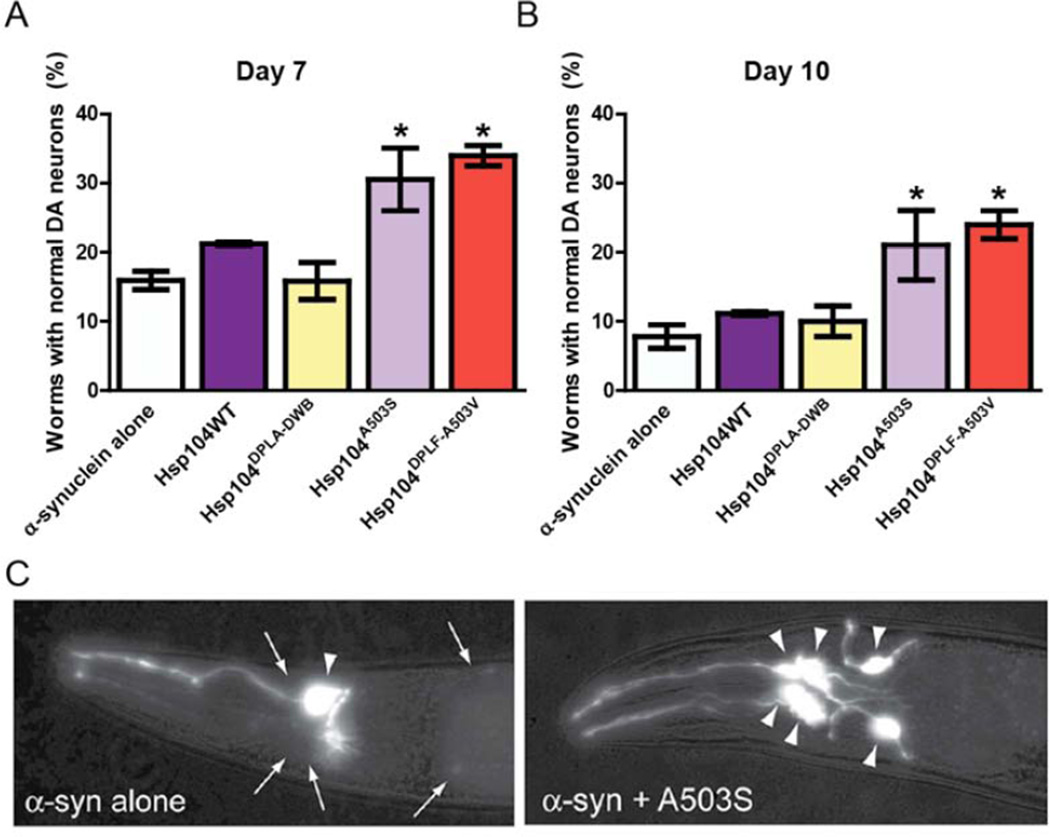
The dopamine transporter (dat-1) gene promoter was used to direct expression of Hsp104 variants and α-syn to dopaminergic (DA) neurons. Expression of α-syn alone resulted in ~16% of animals with normal numbers of DA neurons after 7 days and ~8% of animals after 10 days compared to controls (Fig. 5A–C). Coexpression of Hsp104WT or an ATPase-dead, substrate binding-deficient Hsp104DPLA-DWB (which bears the “double pore loop” and “double Walker B” mutations: Y257A:E285Q:Y662A:E687Q) did not rescue neurodegeneration (Fig. 5A, B). C. elegans expressing Hsp104A503S and Hsp104A503V-DPLF displayed significant protection (30.5% and 34% normal worms, respectively) compared to the null Hsp104 variant or α-syn alone at day 7 (Fig. 5A). This trend continued at day 10 (Fig. 5B), when Hsp104A503S (21%) and Hsp104A503V-DPLF (24%) expressing worms had significantly more normal DA neurons compared to α-syn alone (7.8%), Hsp104DPLA-DWB (10%), or Hsp104WT (11%). Hsp104 variants did not alter α-syn mRNA levels (Fig. S5C). Thus, Hsp104A503S and Hsp104A503V-DPLF remain significantly neuroprotective against α-syn toxicity even as animals age.
Figure 5. Hsp104A503S and Hsp104A503V-DPLF protect against α-syn toxicity and dopaminergic neurodegeneration in C. elegans.
(A) Hsp104 variants and α-syn were coexpressed in the dopaminergic (DA) neurons of C. elegans. Hermaphrodite nematodes have six anterior DA neurons, which were scored at day 7 post-hatching. Hsp104A503S and Hsp104A503-VDPLF have significantly greater protective activity than both α-syn alone and the null variant. Normal worms have a full complement of DA neurons at this time. (B) At day 10, there is a decline in worms with normal DA neurons. Hsp104A503S and Hsp104A503V-DPLF exhibit greater protective activity when compared to Hsp104WT and the null variant. Values represent means±SEM (of 3 independent experiments, n=30 per replicate with 3–4 replicates per independent experiment. * p<.05 One-Way ANOVA group). Normal worms have a full complement of DA neurons at this time. (C) Photomicrographs of the anterior region of C. elegans co-expressing GFP with α-syn. Worms expressing α-syn alone (left) exhibit an age dependent loss of DA neurons. Worms expressing α-syn plus Hsp104A503S (right) exhibit greater neuronal integrity. Arrows indicate degenerating or missing neurons. Triangles indicate normal neurons.
See also Fig. S5.
Potentiated Hsp104 variants typically have elevated ATPase activity
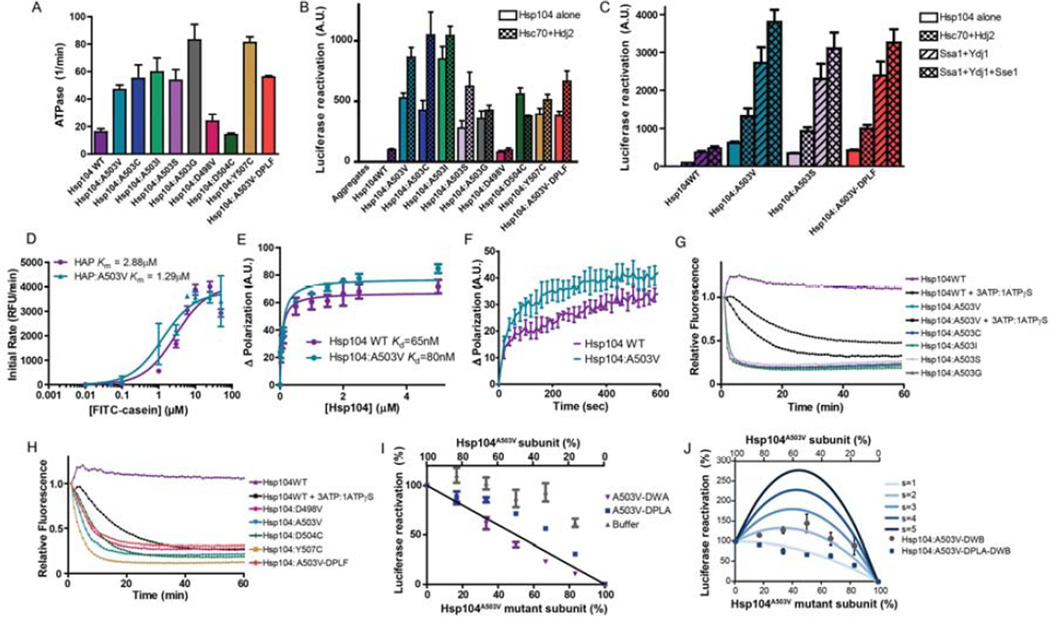
Nearly all of the Hsp104A503X variants suppressed α-syn, FUS, and TDP-43 toxicity in yeast. This unexpected degeneracy is intriguing as there are few, if any, examples of missense mutations to nearly any class of residue that lead to a therapeutic gain of function. To explore the mechanism behind this gain of function, we assessed the biochemical properties of several Hsp104 variants that suppressed toxicity. Each of Hsp104A503X variant and Hsp104Y507C exhibited ~2-4-fold higher ATPase activity than Hsp104WT (Fig. 6A). Hsp104D498V has higher ATPase activity than Hsp104WT, though not as high as the Hsp104A503X variants (Fig. 6A). Hsp104D504C had similar ATPase activity to Hsp104WT (Fig. 6A). Thus, enhanced Hsp104 variants typically have higher ATPase activity than Hsp104WT. However, Hsp104D504C illustrates that elevated ATPase activity is not absolutely required for potentiation.
Figure 6. Potentiated Hsp104 variants are tuned differently than Hsp104WT.
(A) ATPase activity of Hsp104 variants. Values represent means±SEM (n=3). (B) Luciferase aggregates were incubated with Hsp104 variant plus (checkered bars) or minus (clear bars) Hsc70 (0.167µM) and Hdj2 (0.167µM). Values represent means±SEM (n=3) (C) Luciferase aggregates were incubated with Hsp104 variant plus or minus: Hsc70 (0.167µM) and Hdj2 (0.073µM); Ssa1 and Ydj1; or Ssa1, Ydj1, and Sse1. Values represent means±SEM (n=3). (D) Increasing concentrations of FITC-casein were incubated with ClpP plus HAPWT or HAPA503V. Initial degradation rates were plotted against FITC-casein concentration to determine Km. Values represent means±SEM (n=3). (E) FITC-casein was incubated with increasing concentrations of Hsp104WT or Hsp104A503V. Change in fluorescence polarization was plotted against Hsp104 concentration to determine Kd. Values represent means±SEM (n=3). (F) Kinetics of Hsp104WT (1µM) or Hsp104A503V (1µM) binding to FITC-casein (0.1µM) assessed by fluorescence polarization. Values represent means±SEM (n=3). (G, H) RepA1–70-GFP was incubated with Hsp104 variant and GroELtrap plus ATP or ATP:ATPγS (3:1). GFP unfolding was measured by fluorescence. Representative data are shown. (I) Buffer, Hsp104A503V-DWA, or Hsp104A503V-DPLA was mixed in varying ratios with Hsp104A503V to create heterohexamer ensembles and luciferase disaggregase activity was assessed. Values represent means±SEM (n=3). Black line denotes the theoretical curve of a probabilistic mechanism where only a single WT subunit is required for disaggregation. (J) Experiments were performed as in (I) for Hsp104A503V-DWB and Hsp104A503VDPLA-DWB. Theoretical curves are shown wherein adjacent pairs of A503V:A503V or A503V:mutant subunits confer hexamer activity, while adjacent mutant subunits have no activity. Each adjacent A503V:A503V pair has an activity of 1/6. Adjacent A503V:mutant pairs have a stimulated activity (s), and the effect of various s values are depicted. Values represent means±SEM (n=3).
Potentiated Hsp104 variants do not require Hsp70 and Hsp40 for disaggregation
Rescue of toxicity by enhanced Hsp104 variants might reflect an altered mechanism of disaggregation. Thus, we assessed activity against disordered luciferase aggregates (DeSantis et al., 2012). Hsp104WT was inactive alone and required Hsp70 and Hsp40, which could be from human (Hsc70 and Hdj2) or yeast (Ssa1 and Ydj1; Fig. 6B, C). By contrast, potentiated Hsp104 variants were extremely active without Hsp70 and Hsp40, and with the exception of Hsp104D504C, Hsc70 and Hdj2 further increased activity (Fig. 6B, C). Typically, in the absence of Hsc70 and Hdj2, potentiated Hsp104 variants were ~3-9-fold more active than Hsp104WT plus Hsc70 and Hdj2 (Fig. 6B). The only exception was Hsp104D498V, which in the absence of Hsc70 and Hdj2 was still as active as Hsp104WT plus Hsc70 and Hdj2 (Fig. 6B). Hsp104WT was most active in the presence of Ssa1, Ydj1, and the Hsp110, Sse1 (Fig. 6C) (Shorter, 2011). However, even here, Hsp104WT luciferase reactivation activity only reached Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF activity in the absence of Ssa1, Ydj1, and Sse1 (Fig. 6C). In the presence of Ssa1, Ydj1, and Sse1, the luciferase reactivation activity of Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF was ~7-8-fold higher than Hsp104WT (Fig. 6C). Potentiated Hsp104 variants are highly active without Hsp70 and Hsp40 (Fig. 6B, C). Thus, absolute dependence on Hsp70 and Hsp40 hinders Hsp104 from rescuing α-syn, FUS, and TDP-43 toxicity. Independence from Hsp70 and Hsp40 is promising for applying Hsp104 variants to reverse protein misfolding in diverse systems, such as purification of aggregation-prone recombinant proteins from E. coli where DnaK incompatibility is an issue (DeSantis and Shorter, 2012).
Potentiated Hsp104 variants translocate substrate faster than Hsp104WT
We next determined that potentiated Hsp104 variants displayed accelerated substrate translocation. Thus, we used an Hsp104 variant, termed HAP, where G739-K741 is mutated to IGF, which enables association with the chambered peptidase ClpP (Tessarz et al., 2008). In the presence of ClpP, translocated substrates are degraded rather than released. Thus, HAP translocates FITC-casein for degradation by ClpP thereby releasing FITC and increasing fluorescence. In the presence of ClpP, HAPA503V (Km~1.29µM) is a more effective FITC-casein translocase than HAPWT (Km ~2.88µM) (Fig. 6D). The lower Km for HAPA503V might reflect differences in substrate recognition rather than translocation speed. However, the Kd of Hsp104WT (Kd~65nM) and Hsp104A503V (Kd ~80nM) for FITC-casein were similar (Fig. 6E) as were binding kinetics (Fig. 6F). Thus, substrate recognition by Hsp104WT and Hsp104A503V is very similar. Hence, we suggest that Hsp104A503V translocates substrate more rapidly than Hsp104WT. Accelerated translocation likely enables potentiated variants to avoid kinetic traps and exert additional force to unfold stable substrates.
Potentiated Hsp104 variants are enhanced unfoldases
Next, we established that enhanced Hsp104 variants had enhanced unfoldase activity using a RepA1–70-GFP substrate (Doyle et al., 2007). To assess RepA1–70-GFP unfolding in the absence of spontaneous refolding we added GroELtrap, which captures unfolded proteins and prevents refolding (Weber-Ban et al., 1999). Hsp104WT unfolds RepA1–70-GFP but only in the presence of a permissive ratio of ATP and ATPγS (Doyle et al., 2007) (Fig. 6G, H). Thus, with ATP alone, Hsp104WT did not unfold RepA1–70-GFP (Fig. 6G). By contrast, Hsp104A503X variants rapidly unfolded RepA1–70-GFP in the presence of ATP (Fig. 6G). Hsp104WT unfolded RepA1–70-GFP in the presence of an ATP:ATPγS (3:1) mixture. By contrast, ATP:ATPγS slightly inhibited Hsp104A503V unfoldase activity, but even here, Hsp104A503V unfolded RepA1–70-GFP more rapidly than Hsp104WT (Fig. 6G). Hsp104A503X variants had very similar unfoldase kinetics (Fig. 6G). By contrast, Hsp104D498V, Hsp104D504C, and Hsp104A503V-DPLF were slightly slower unfoldases than Hsp104A503V, whereas Hsp104Y507C was slightly faster (Fig. 6H). These differences could reflect changes in substrate recognition or turnover or both. Regardless, potentiated Hsp104 variants are enhanced unfoldases that are intrinsically primed to unfold substrates and do not have to wait for regulatory events (mimicked here by ATPγS addition).
Hsp104A503V hexamers are tuned differently than Hsp104WT hexamers
Do potentiated Hsp104 variants employ the same mechanism of intersubunit collaboration as Hsp104WT to disaggregate proteins? How Hsp104 subunits within the hexamer collaborate to promote disaggregation can be interrogated via mutant subunit doping. Here, mutant subunits defective in ATP hydrolysis, substrate binding, or both, are mixed with WT subunits to generate heterohexamer ensembles according to the binomial distribution (DeSantis et al., 2012). Hsp104 forms dynamic hexamers that exchange subunits on the minute timescale, which ensures statistical incorporation of mutant subunits (DeSantis et al., 2012). The disaggregase activity of various heterohexamer ensembles enables determination of the number of mutant subunits that inactivate the WT hexamer. Thus, we can determine if subunit collaboration within Hsp104 hexamers is probabilistic (six mutant subunits are required to abolish activity), subglobally cooperative (two to five mutant subunits abolish activity), or globally cooperative (one mutant subunit abolishes activity) (DeSantis et al., 2012). Incorporation of Hsp104A503V-DWA subunits (which bear the “double Walker A” [DWA] K218T:K620T mutations and cannot bind ATP) or Hsp104A503V-DPLA subunits (which bear the “double pore loop” [DPL] Y257A:Y662A mutations and cannot bind substrate) into Hsp104A503V hexamers caused a roughly linear decline in luciferase disaggregase activity (Fig. 6I). This linear decline indicates that like Hsp104WT, Hsp104A503V hexamers resolve disordered aggregates via a probabilistic mechanism (DeSantis et al., 2012). Thus, a single Hsp104A503V subunit per hexamer able to hydrolyze ATP and engage substrate can drive disaggregation.
However, Hsp104A503V hexamers operate differently than Hsp104WT hexamers. A single Hsp104DWB subunit (which bear the “double Walker B” [DWB] E285Q:E687Q mutations and can bind but not hydrolyze ATP) inactivates the Hsp104WT hexamer (DeSantis et al., 2012). By contrast, the luciferase disaggregase activity of Hsp104A503V was stimulated by Hsp104A503V-DWB subunits (Fig. 6J). FRET studies confirmed that Hsp104A503V-DWB subunits incorporated into Hsp104A503V hexamers. The FRET efficiency was 0.36 (compared to 0.38 for mixing Hsp104WT with Hsp104DWB (DeSantis et al., 2012)) using the conditions employed for luciferase reactivation. In high salt buffer (1M NaCl), hexamerization is inhibited and FRET efficiency decreased to 0.24. At higher Hsp104 concentration (1µM), which favors hexamerization, FRET efficiency increased to 0.43. We could model the stimulatory effect of Hsp104A503V-DWB subunits if we imposed rules whereby an Hsp104A503V-DWB subunit stimulates activity of an adjacent Hsp104A503V subunit ~2-fold (Fig. 6J). This stimulation depended on substrate-binding by Hsp104A503V-DWB as Hsp104A503V-DPLA-DWB subunits (which bear the “double pore loop” and “double Walker B” Y257A:E285Q:Y662A:E687Q mutations and can bind but not hydrolyze ATP and cannot bind substrate) failed to stimulate adjacent Hsp104A503V subunits (Fig. 6J). Thus, Hsp104A503V hexamers operate via principles distinct from those of Hsp104WT hexamers. The Hsp104A503V hexamer displays greater plasticity and tolerates a wider variety of subunit-inactivating events to maintain a robust disaggregase activity. Thus, an Hsp104A503V subunit that: (1) binds but cannot hydrolyze ATP and (2) engages substrate, stimulates the disaggregase activity of an adjacent Hsp104A503V subunit. In Hsp104WT, a single subunit with these properties inactivates the hexamer. The increased resilience of Hsp104A503V hexamers to subunit-inactivating events likely empowers facile resolution of recalcitrant substrates.
Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF disaggregate preformed α-syn fibrils more efficaciously than Hsp104WT
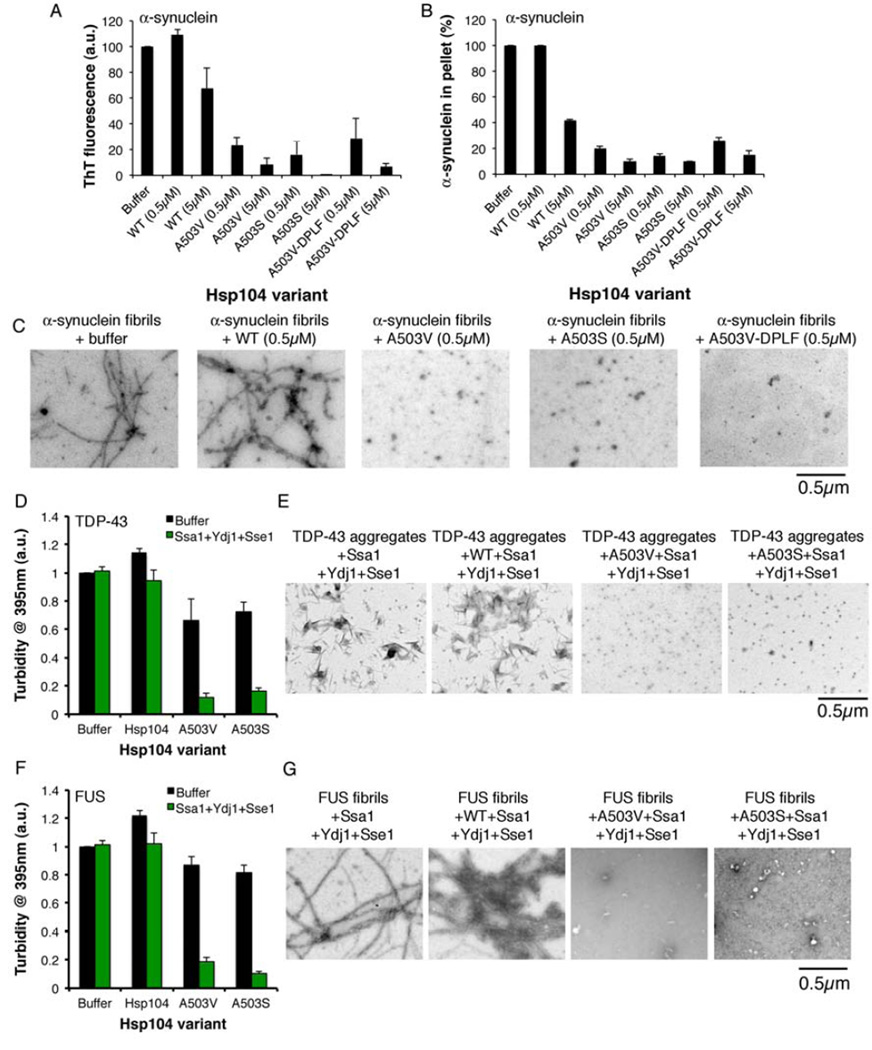
To test Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF in comparison to Hsp104WT against a recalcitrant PD-associated substrate we employed α-syn fibrils, allowing us to distinguish if Hsp104 prevented amyloid formation or eliminated preformed amyloid. Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF disaggregated preformed α-syn fibrils at concentrations where Hsp104WT was inactive (Fig. 7A–C). Indeed, electron microscopy (EM) revealed that α-syn fibrils were converted to small structures by low concentrations of Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF, whereas Hsp104WT left fibrils intact (Fig. 7C). Thus, Hsp104A503V, Hsp104A503S, and Hsp104A503V-DPLF are more powerful amyloid disaggregases than Hsp104WT.
Figure 7. Potentiated Hsp104 variants disaggregate preformed α-syn, TDP-43, and FUS fibrils more efficaciously than Hsp104WT.
(A–C) α-syn fibrils were incubated without or with Hsp104WT, Hsp104A503V, Hsp104A503S, or Hsp104A503V-DPLF for 1h at 30°C. Fiber disassembly was assessed by ThT fluorescence (A), sedimentation analysis (B), or (C) EM (Bar, 0.5µm). (A, B) Values represent means±SEM (n=2). (D, E) TDP-43 aggregates were incubated with buffer, Hsp104WT, Hsp104A503V, or Hsp104A503S plus or minus Ssa1, Ydj1, and Sse1 for 1h at 30°C. (D) Aggregate dissolution assessed by turbidity. Values represent means±SEM (n=3). (E) Aggregate dissolution assessed by EM. Bar, 0.5µm. (F, G) FUS aggregates were incubated with buffer, Hsp104WT, Hsp104A503V, or Hsp104A503S plus or minus Ssa1, Ydj1, and Sse1 for 1h at 30°C. (F) Aggregate dissolution assessed by turbidity (absorbance at 395nm). Values represent means±SEM (n=3). (G) Aggregate dissolution assessed by EM. Bar, 0.5µm.
Hsp104A503V and Hsp104A503S disaggregate preformed TDP-43 and FUS aggregates more efficaciously than Hsp104WT
Next, we tested whether Hsp104A503V and Hsp104A503S were more potent disaggregases of TDP-43 and FUS (Johnson et al., 2009; Sun et al., 2011). Hsp104WT was unable to resolve TDP-43 aggregates and slightly enhanced TDP-43 aggregation in the absence of Ssa1, Ydj1, and Sse1 (Fig. 7D). By contrast, Hsp104A503V and Hsp104A503S partially resolved TDP-43 aggregates in the absence of Ssa1, Ydj1, and Sse1 (Fig. 7D). Hsp104A503V and Hsp104A503S in the presence of Ssa1, Ydj1, and Sse1, but not by Hsp104WT, effectively dissolved short TDP-43 filaments and amorphous structures (Fig. 7D, E). Very similar results were obtained with preformed FUS fibrils (Fig. 7F, G). Hsp104WT slightly increased FUS aggregation in the absence of Ssa1, Ydj1, and Sse1, whereas Hsp104A503V and Hsp104A503S modestly reduced aggregation (Fig. 7F). Hsp104A503V and Hsp104A503S effectively disaggregated FUS in the presence of Ssa1, Ydj1, and Sse1, whereas Hsp104WT was ineffective (Fig. 7F). Indeed, Hsp104A503V and Hsp104A503S eradicated FUS fibrils (Fig. 7G). Thus, Hsp104A503V and Hsp104A503S disaggregate preformed TDP-43 and FUS aggregates more efficaciously than Hsp104WT.
Discussion
Here, we demonstrate that Hsp104, a protein disaggregase from yeast, can be modified to powerfully eradicate diverse substrates implicated in ALS and PD. We have developed the first, to our knowledge, disaggregases (or even chaperones) engineered to optimize proteostasis. Indeed, enhanced Hsp104 variants are the first agents defined to reverse TDP-43 and FUS aggregation. They not only suppress toxicity and eliminate protein aggregates, but also restore proper protein localization. Importantly, these Hsp104 variants are not overtly toxic like other MD mutants (Lipinska et al., 2013). Thus, potentiated Hsp104 variants can be uncovered that are not invariably toxic and which rescue various toxic neurodegenerative disease proteins in vitro and in vivo under conditions where Hsp104WT is impotent. Potentiated Hsp104 variants suppress neurodegeneration in a C. elegans PD model. Thus, we provide the first example of engineered disaggregases rescuing neurodegeneration in a metazoan nervous system under conditions where the WT disaggregase is ineffective. Our findings suggest that general neuroprotection via activated protein disaggregases may be possible for diverse neurodegenerative diseases.
We have identified the MD as a key region governing Hsp104 function. It is perplexing and unprecedented that missense mutations to nearly any residue at specific and disparate positions (e.g. A503, Y507) confers a therapeutic gain of function. Potentiation stems from loss of amino acid identity rather than specific mutation. Thus, Hsp104 activity is likely tightly constrained but can be unleashed by subtle changes to side chains at specific positions. These constraints are too tight for Hsp104WT to counter TDP-43, FUS, and α-syn aggregation and toxicity under the conditions employed in our experiments. Thus, we reveal a surprising inimical deficit in existing disaggregase functionality. We suggest that the MD functions as a capacitor braced to unleash Hsp104 activity. Missense mutations at specific positions in MD helix 1, 2, or 3 or the small domain of NBD1 (immediately C-terminal to the MD) likely destabilize autoinhibitory interactions that dampen Hsp104 activity or induce conformational changes that mimic or aid in an allosteric activation step. Potentiating mutations obviate any absolute requirement for Hsp70 and enhance Hsp104 ATPase activity, substrate translocation speed, unfoldase activity, and amyloid disaggregase activity. Additionally, Hsp104A503V hexamers display enhanced plasticity and are more resistant to defective subunits than Hsp104WT. Thus, enhanced variants possess a more robust disaggregase activity that is desensitized to inhibition. Irrespective of the mechanism of activation, we have established that seemingly minor structural modulation of a disaggregase can suppress a constellation of otherwise intractable proteotoxicities in vivo. We are unaware of any precedent for attaining such a wide-reaching set of gain of therapeutic functions via such minor changes in primary sequence, e.g. by removing a single methyl group (A503G) or by adding a single methylene bridge (V426L).
Further engineering to develop enhanced variants that specifically target single proteins (e.g. disaggregate FUS but not TDP-43) will prove valuable to minimize any off-target effects. Hsp104 could be potentiated against any protein, which might find key applications in purification of troublesome recombinant proteins. Irrespective of the feasibility of introducing Hsp104 as a therapeutic, our work suggests that protein aggregates are not intractable and that general neuroprotection via altered proteostasis is achievable. Ultimately, we envision introducing potentiated Hsp104 variants in short transient bursts to restore natural proteostasis. In this way, long-term expression of an exogenous protein is avoided. Reactivation of disease-associated proteins to their non-pathogenic states suggests that Hsp104 variants and other agents that achieve this goal may be highly promising for halting and reversing neurodegenerative disease. Nonetheless, caution is needed and many barriers must be breached to translate Hsp104 variants into disruptive technologies and potential therapeutics.
Experimental Procedures
Yeast strains and media
Yeast were WT W303a or the isogenic W303aΔhsp104 strain. Δire1 and Δatg8 were in BY4741. Standard methods were used for transformation and spotting. See Extended Experimental Procedures.
Library Construction and Screening
The pore loop variant library was constructed via QuikChange mutagenesis (Agilent) and DNA shuffling to obtain randomly combined residues at positions Y257 and Y662. The MD variant library was constructed using GeneMorph II EZClone Domain Mutagenesis kit (Agilent) with modifications. Libraries were transformed into yeast harboring pAG303GAL-TDP-43, pAG303GAL-FUS, or pAG303GAL-α-syn. Yeast were grown overnight in raffinose-containing media and plated on galactose-containing media for selection. Select colonies were sequenced by colony PCR. Isolated Hsp104 variants were cloned independently and transformed into yeast to ensure they suppressed toxicity. See Extended Experimental Procedures.
Hsp104 Variant Toxicity and Thermotolerance
W303Δhsp104 yeast were transformed with the indicated 416GAL-Hsp104 plasmid. Cultures were grown in synthetic raffinose medium to A600nm=2.0, spotted onto SD-Ura or SGal-Ura, and incubated at 30°C or 37°C for 48–72h. For thermotol erance, yeast were grown to saturation in synthetic raffinose media and diluted to A600nm=0.3 in galactose-containing media. After 4h at 30°C, cells were heat shocked at 50°C for 0–30min, cooled for 2min on ice, serially diluted, and spotted on synthetic dropout media containing galactose. Plates were incubated at 30°C for 48–72h.
Sedimentation Analysis
Yeast were induced in galactose-containing medium for 5h (TDP-43 and FUS) or 8h (α-syn). Cells were lysed, separated into soluble and insoluble fractions by sedimentation, and processed for quantitative immunoblot. See Extended Experimental Procedures.
Fluorescence Microscopy
After 5h induction at 30°C (8h for α-syn strains), yeast cultures were processed for fluorescence microscopy. For TDP-43, cells were fixed and stained with 4’,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. For FUS, live cells were used and nuclei were visualized with Hoechst dye. See Extended Experimental Procedures.
Analysis for Dopaminergic Neuron Death in C. elegans
Three distinct C. elegans stable lines were created for each Hsp104 variant. Age synchronized worms were generated by allowing 50 transgenic adults on a NGM plate to lay eggs for 3h. Adults were then removed (day 0). At day 7 and 10 of analysis, 40 randomly selected transgenic worms were placed in 3mM Levamisol for paralysis and transferred to a 2% agarose pad on a glass microscope slide. Worms have 8 dopaminergic neurons visible through Pdat-1::gfp, which fade in an age-dependent manner due to α-syn accumulation. Only the 6 anterior neurons of the worm were analyzed. Each worm was scored “Wild type” when there was a full complement of visible, anterior dendritic processes. Worms missing one or more dendritic processes was scored “Not Wild Type”. In total three separate stable lines were analyzed. See Extended Experimental Procedures.
Protein Purification
Proteins were purified as recombinant proteins in E. coli using standard techniques. See Extended Experimental Procedures.
ATPase activity
Hsp104 (0.042µM hexamer) was incubated with ATP (1mM) for 5min at 25°C. ATPase activity was assessed by inorganic phosphate release using a malachite green detection kit (Innova).
Luciferase Reactivation
Aggregated luciferase (50nM) was incubated with Hsp104 (0.167 µM hexamer) with ATP (5.1mM) and an ATP regeneration system (ARS; 1mM creatine phosphate, 0.25µM creatine kinase) plus or minus Hsc70 (0.167µM) and Hdj2 (0.167µM) for 90min at 25°C (DeSantis et al., 2012). In some reactions (Fig. 6C), Hsc70 concentration was 0.167µM and Hdj2 concentration was 0.073µM. In other reactions, Hsc70 and Hdj2 were replaced with Ssa1 (0.167µM) and Ydj1 (0.073µM) or Ssa1 (0.167µM), Ydj1 (0.073µM), and Sse1 (0.043µM). Luciferase activity was assessed by luminescence. Mutant doping experiments were as described (DeSantis et al., 2012). Hsp104A503V variants: Hsp104A503V-DWA (K218T:A503V:K620T), Hsp104A503V-DPLA (Y257A:A503V:Y662A), Hsp104A503V-DWB (E285Q:A503V:E687Q), Hsp104A503V-DPLA-DWB (Y257A:E285Q:A503V:Y662A:E687Q) were mixed with Hsp104A503V in varying ratios to give a total concentration of 0.5µM Hsp104 hexamer. Hsc70 and Hdj2 were omitted for these experiments. Random subunit mixing was confirmed by FRET. See Extended Experimental Procedures.
RepA1–70-GFP Unfolding
RepA1–70-GFP unfolding was as described (Doyle et al., 2007).
FITC-Casein Degradation and Binding
FITC-casein (0.1–50µM) was incubated at 25°C with HAP or HAPA503V (1µM hexamer) and ClpP (21µM monomer) plus ATP (5mM) and ARS. Degradation of FITC-casein was monitored by fluorescence (excitation 490nm, emission 520nm). To calculate initial rate, a linear fit of the first 2.5min of the reaction was constructed and the slope was calculated. To assess binding, FITC-casein (6nM) was incubated with increasing concentrations (0–5µM) of Hsp104WT or Hsp104A503V with 2mM ATPγS for 10min at 25°C. Fluorescence polarization was measured (excitation 470nm, emission 520nm).
α-syn fibril disaggregation
α-syn (80µM) was assembled into fibrils via incubation in 40mM HEPES-KOH, pH 7.4, 150mM KCl, 20mM MgCl2, 1mM DTT for 48h at 37°C with agitation. α-syn fibrils (0.5µM monomer) were incubated without or with Hsp104WT, Hsp104A503V, Hsp104A503S, orHsp104A503V-DPLF (0.5 or 5µM) plus ATP (10mM) and ARS (20mM creatine phosphate and 0.5µM creatine kinase) for 1h at 30°C. Disaggregation was assessed by Thioflavin-T (ThT) fluorescence, sedimentation analysis, and EM (Lo Bianco et al., 2008).
TDP-43 and FUS disaggregation
To generate TDP-43 and FUS aggregates, GST-TEV-TDP-43 (6µM) or GST-TEV-FUS (6µM) were incubated with TEV protease in 50mM TrisHCl pH 7.4, 50mM KCl, 5mM MgCl2, 0.2M trehalose, and 20mM glutathione. FUS was aggregated for 90min at 25°C without agitation, by which time all the FUS had aggregated (Sun et al., 2011). TDP-43 was aggregated for 4h at 25°C with agitation, by which time all the TDP-43 h ad aggregated (Johnson et al., 2009). TDP-43 or FUS aggregates (3µM monomer) were incubated for 1h at 30°C with Hsp104WT, Hsp104A503V, or Hsp104A503S (1µM) plus or minus Ssa1 (1µM), Ydj1 (0.44µM), and Sse1 (0.26µM) plus ATP (10mM) and ARS (20mM creatine phosphate and 0.5µM creatine kinase). Disaggregation was assessed via turbidity (absorbance at 395nm) and EM (Johnson et al., 2009; Sun et al., 2011).
Supplementary Material
We reprogram Hsp104 to rescue TDP-43, FUS, and α-synuclein proteotoxicity
Potentiated Hsp104 variants clear inclusions and restore proper protein localization
Potentiating mutations reconfigure how Hsp104 hexamers operate
Enhanced disaggregases can restore proteostasis and mitigate neurodegeneration
Acknowledgements
We thank Sue Lindquist, Johannes Buchner, Walid Houry, Aaron Gitler, Martin Duennwald, and Brad Johnson for kindly sharing reagents; Beatrice Razzo and Nabeel Akhtar for help in library construction and screening; Mark Lemmon, Mariana Torrente, and Liz Sweeny for critiques. Our studies were supported by: an American Heart Association Post-Doctoral Fellowship (M.E.J); NIH training grant (T32GM071339) and NRSA predoctoral fellowship F31NS079009 (M.E.D.); NSF Graduate Research Fellowship DGE-0822 (L.M.C.); NSF CAREER Award 0845020 (K.A.C.); NIH grant R15NS075684 (G.A.C); NIH Director's New Innovator Award DP2OD002177, NIH grants R21NS067354, R21HD074510, and R01GM099836, a Muscular Dystrophy Association Research Award (MDA277268), Packard Center for ALS Research at Johns Hopkins University, Target ALS, and an Ellison Medical Foundation New Scholar in Aging Award (J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Cao S, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J Neurosci. 2005;25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman-Nick M, Bonini NM, Shorter J. Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model. PLoS genetics. 2013;9:e1003781. doi: 10.1371/journal.pgen.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis ME, Shorter J. The elusive middle domain of Hsp104 and ClpB: location and function. Biochim Biophys Acta. 2012;1823:29–39. doi: 10.1016/j.bbamcr.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, Bosco DA, Hayward LJ, Brown RH, Jr, Lindquist S, et al. A Yeast Model of FUS/TLS-Dependent Cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska N, Zietkiewicz S, Sobczak A, Jurczyk A, Potocki W, Morawiec E, Wawrzycka A, Gumowski K, Slusarz M, Rodziewicz-Motowidlo S, et al. Disruption of ionic interactions between the nucleotide binding domain 1 (NBD1) and middle (M) domain in Hsp100 disaggregase unleashes toxic hyperactivity and partial independence from Hsp70. J Biol Chem. 2013;288:2857–2869. doi: 10.1074/jbc.M112.387589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, Aebischer P. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby GA, Lindquist S. Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol. 2013;23:251–259. doi: 10.1016/j.tcb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Outeiro TF, Lindquist S. Yeast Cells Provide Insight into Alpha-Synuclein Biology and Pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;13:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Homann OR, Kowal AS, Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol Biol Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PloS one. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular Determinants and Genetic Modifiers of Aggregation and Toxicity for the ALS Disease Protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, et al. Yeast Reveal a "Druggable" Rsp5/Nedd4 Network that Ameliorates alpha-Synuclein Toxicity in Neurons. Science. 2013 doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Molecular Microbiology. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Human molecular genetics. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.