SUMMARY
MRE11 within the MRE11-RAD50-NBS1 (MRN) complex acts in DNA double-strand break repair (DSBR), detection and signaling; yet, how its endo- and exonuclease activities regulate DSB repair by non-homologous end-joining (NHEJ) versus homologous recombination (HR) remains enigmatic. Here we employed structure-based design with a focused chemical library to discover specific MRE11 endo- or exonuclease inhibitors. With these inhibitors we examined repair pathway choice at DSBs generated in G2 following radiation exposure. Whilst endo- or exonuclease inhibition impairs radiation-induced RPA chromatin binding, suggesting diminished resection, the inhibitors surprisingly direct different repair outcomes. Endonuclease inhibition promotes NHEJ in lieu of HR, whilst exonuclease inhibition confers a repair defect. Collectively, the results describe nuclease-specific MRE11 inhibitors, define distinct nuclease roles in DSB repair, and support a mechanism whereby MRE11 endonuclease initiates resection, thereby licensing HR followed by MRE11 exo and EXO1/BLM bidirectional resection towards and away from the DNA end, which commits to HR.
INTRODUCTION
MRE11 nuclease forms the core of the MRE11-RAD50-NBS1 (MRN) complex, which has essential roles in detecting, signaling, protecting and repairing DNA double strand breaks (DSBs) (Stracker and Petrini, 2011; Williams et al., 2007; Wyman and Kanaar, 2006). As a first responder to DSBs, MRN promotes appropriate repair by non-homologous end joining (NHEJ) or homologous recombination (HR), playing essential roles via its 3′-5′ exonuclease and single-stranded (ss) and DNA hairpin endonuclease activities (Lisby et al., 2004; Paull and Gellert, 1998; Stracker and Petrini, 2011; Trujillo et al., 2003; Williams et al., 2011). NHEJ represents the major DSB repair pathway in mammalian cells, repairing DSBs in all cell cycle phases (Rothkamm et al., 2003). HR contributes to distinct processes including meiotic recombination, replication fork stabilization and one-ended DSB repair, and overlaps with NHEJ to repair two-ended DSBs in late S/G2 phase (Jeggo et al., 2011; Schlacher et al., 2011). Current models in mammalian cells suggest that the abundant Ku70/80 heterodimer rapidly binds to all two-ended DSBs, allowing NHEJ to make the first attempt at DSB rejoining (Beucher et al., 2009; Shibata et al., 2011). Thus, even in G2 where HR functions, NHEJ rejoins most DSBs but subsequently repair switches to HR, necessitating resection (Shibata et al., 2011). Resection of two-ended DSBs is a critical step that initiates and potentially commits to repair by HR when NHEJ stalls. MRE11 nuclease activities promote resection but their roles are unclear; furthermore MRE11 exonuclease has the wrong polarity to drive resection (Llorente and Symington, 2004; Stracker and Petrini, 2011).
HR (and not NHEJ) functions during meiosis. Meiotic DSBs are introduced by Spo11, a topoisomerase II-like protein, which bridges DNA ends; DSB opening and Spo11 removal requires Mre11 nuclease activity (Garcia et al., 2011). In yeast, DSB processing creates a ssDNA nick up to 300 base pairs from the DSB end followed by bidirectional resection. Mre11 3′-5′ exonuclease activity digests towards the DSB end and Exo1 generates ssDNA moving 5′-3′. Current data suggests that Mre11 endonuclease activity makes the initial ss nick, with the combined activities promoting removal of covalently, end-bound Spo11. For HR in mitotic cells, Sae2/MRX (CtIP/MRN) initiates DSB resection, enabling 5′-3′ resection by Exo1/Sgs1 (EXO1/BLM) although further details are unclear (Mimitou and Symington, 2008; Nimonkar et al., 2011; Zhu et al., 2008).
Mre11 mutations impact either its exonuclease activity alone, both activities or disturb Mre11 interactions with interfacing Rad50 or Nbs1; mutations specifically impacting Mre11 endonuclease activity have not been described (Buis et al., 2008; Williams et al., 2011; Williams et al., 2009; Williams et al., 2008). We reasoned that unraveling the role of MRE11 nuclease activities during resection would require the ability to specifically ablate one or other activity, which in turn necessitates structural insight into regions on MRE11 required for these activities. Mirin, a characterized inhibitor of MRE11 exonuclease activity, acts by an unknown mechanism but does not disrupt the MRE11 complex (Dupre et al., 2008). Here we combined Mre11 structure determinations with focused mirin libraries to create and apply specific inhibitors to address MRE11 nuclease roles. First, we determined Mre11 structures with bound mirin, then exploited this insight and focused chemical libraries to develop inhibitors that specifically perturb MRE11 exo- or endonuclease activities. Second, we exploited these novel inhibitors to unravel MRE11’s role during resection of two-ended DSBs. Our findings support a similar mechanism to MRE11’s role during meiosis but reveal unexpected impacts on the regulation of pathway choice.
RESULTS
Structure Determination, Analysis and Derivation of Specific MRE11 Inhibitors
To develop specific Mre11 endo- and exonuclease inhibitors, we leveraged Mre11 structural data and mirin inhibitor chemistry. We created and employed a focused chemical library of mirin derivatives (PFM compounds) with different substituents in the styryl moiety and replacement of the pseudothiohydantoin ring with a substituted rodanin moiety to test structure activity relationships (SARs) (Figure 1A) in concert with structural determinations of Mre11-inhibitor complexes (Figure 1B). To define the structural basis for mirin activity, we determined Mre11 structures with bound mirin. As human MRE11 did not crystallize with mirin, we exploited the high conservation of Mre11 and produced Thermotoga maritima Mre11 (1–324) (TmMre11) (Das et al., 2010) and co-crystallized it with bound mirin (Figures 1B, S1A, S1B and Table S1). The Mre11 di-Mn active site lies in a groove at the base of the capping domain, restricting dsDNA access. Two β to α connecting loops within this groove are positioned to interact with DNA and be regulated by Rad50: one contains exonuclease-critical His61 and the other, Asn93, which we propose interacts with ssDNA (TmMre11 numbering) (Lim et al., 2011; Mockel et al., 2012; Williams et al., 2008). Our 2.3 Å resolution co-crystal structure suggested that mirin is positioned adjacent to His61 (equivalent to P. furiosus MRE11 His52, S. cerevisiae His59 and human His63) (Das et al., 2010; Hopfner et al., 2001; Lim et al., 2011; Williams et al., 2008) (Figure 1B). This His61 site controls dsDNA end access to the geometrically restricted active site by rotating the DNA phosphate backbone and opening base stacking at the DNA end (Williams et al., 2008). A similar opening of dsDNA, key for phosphate backbone access to a di-metal ion active site, occurs in FEN1 (Tsutakawa et al., 2011).
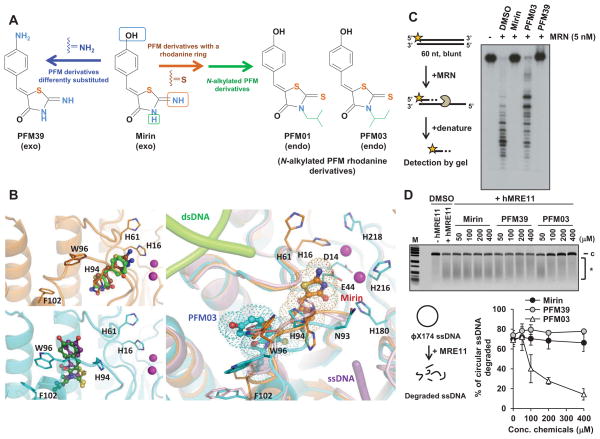
Figure 1. Development of Inhibitors against MRE11 Exo- or Endonuclease Activity.
(A) Derivation by modification of mirin. (B) TmMre11 structures in complex with exo- and endo-nuclease inhibitors. The top left panel shows an overlay of co-crystal structures of TmMre11 (cartoon, coloured brown) in complex with the exonuclease inhibitors, mirin (sticks, carbon atoms coloured brown) and PFM39 (green). The bottom left panel shows an overlay of the co-crystal structures of TmMre11 (cyan) in complex with the endonuclease inhibitors PFM01 (purple) and PFM03 (dark green). Each inhibitor type has a distinct binding site, in proximity to the active site metal ions. The right hand panel shows an overlay of the unliganded TmMre11 (cartoon, coloured pink) with mirin (brown) and PFM03 (cyan). The relative position of each inhibitor is highlighted by a dotted representation of its van der Waals surface. The endo/exo inhibitor binding sites are adjacent, but respectively positioned to distinctly disrupt dsDNA (light green tubes) end-opening by His61 or the ssDNA (violet tube) pathway adjacent to the loop containing Asn93 to Phe102, leading towards the active site metal ions. Nitrogen, oxygen and sulphur atoms are coloured blue, red and yellow respectively. Active site metal ions are shown as magenta spheres.
(C) Mirin and PFM39 but not PFM03 inhibit 5 nM MRN exonuclease activity. 100 nM labeled DNA were incubated with human MRN for 30 minutes at 37ºC. (D) PFM03, but not mirin or PFM39, inhibits human MRE11 endonuclease activity. Analysis using MRN in place of MRE11 is shown in Figure S1E. φX174 circular ssDNA was incubated with purified human MRE11 and DMSO or inhibitor at the indicated concentration. The inhibition assay was established in a time/protein concentration dependent manner to cause loss of ~70% circular DNA. The figure shows the % of circular ssDNA degraded relative to the control (DMSO-treated) sample. c: circular DNA, *: degraded DNA. PFM01 was excluded from inhibitor analysis due to inefficient solubility in vitro. Error bars represent SEM from >3 experiments. Figure S1G–I shows the impact of the inhibitors on MRN DNA binding and ATPase activity. See also Figure S1.
To test the structural implications of mirin binding, we reasoned that we should be able to replace the hydroxyl group in the styryl moiety, which does not interact with Mre11, with an amino group. Indeed, structures suggested that, like mirin, analog PFM39, which has an amino group in place of the hydroxyl group, bound in the pocket between the His61 loop and the adjacent loop (Asn93-Lys97) (Figures 1B, S1A and S1B). As for mirin, ligand binding in this site restricts phosphate rotation for dsDNA exonuclease activity. Nuclease assays using TmMre11 (Das et al., 2010) and human MRN confirmed in vitro inhibition of MRE11 exonuclease activity by mirin and PFM39 (Figures 1C, S1C and S1D). Importantly, neither compound inhibited TmMre11 or human MRE11/MRN endonuclease activity on ssDNA circles (Figures 1D, S1E and S1F).
To employ these structures to develop differential specificity, we applied the anchored plasticity approach developed for structure-based design of nitric oxide synthase isozyme-specific inhibitors (Garcin et al., 2008). The Asn93 loop, flanked by the His61 loop and a groove suitable for ssDNA binding, interacts with a flipped out base in the Mre11-DNA hairpin complex (PDB ID code 3DSD). It also binds a glycerol mimic of the phosphoribose backbone in human MRE11 (PDB ID code 3T1I) (Park et al., 2011) and sulfate ions spanning ~5 nucleotide distance, leads to bound dAMP in a Mre11 product complex (PDB ID 1II7) (Hopfner et al., 2001). Collectively, this groove provides the strongest computationally identified ssDNA-binding site. We reasoned that the flanking His61 loop provides a likely anchor point due to tight interactions at its ends in the dimer interface and the active site (by hydrophobic side chains Leu59 and Leu60, and Mn ion ligand Asp58) plus close packing along its length. We, therefore, designed N-alkyl mirin derivatives that might bind against the His61 loop anchor and shift the flexible Asn93 loop into the adjacent ssDNA-binding groove. We made complexes and determined crystal structures with two such N-alkylated derivatives: PFM01, which has a rhodanine ring plus an iso-butyl chain on the nitrogen, and PFM03, which has a sec-butyl chain. Analysis of crystal structures with Mre11 revealed that the addition of an N-alkyl group (Mre11 complex with PFM01 at 2.3Å and PFM03 at 2.4Å resolution) caused these inhibitors to bind in an adjacent site shifted ~7Å toward the dimer interface (Figures 1B, S1A and S1B). Yet, the N-alkyl mirin derivatives do anchor against the His61 loop and shift the Asn93 loop into the proposed ssDNA-binding channel. Their binding shifts residues His94-Phe102 (the loop plus start of the dimer interface helix) to geometrically interfere with the proposed ssDNA path to the active site metal ions (Figures 1B, S1A and S1B). Nuclease activity assays confirmed specific inhibition of MRE11 endonuclease activity by PFM03 with little impact on its exonuclease activity (Figures 1C, 1D and S1C–F).
Thus, crystal structures suggested inhibitors implicated to bind two distinct sites of MRE11: 1) near His61 to block DNA phosphate backbone rotation and exonuclease activity and 2) near the dimer interface to disrupt the ssDNA-binding groove and endonuclease activity. Based on nuclease activity analyses using human MRE11 and toxicity assessment in mammalian cells, we focused the ensuing cellular analyses on mirin and PFM39, which primarily inhibit MRE11 exonuclease activity, and N-alkylated PFM derivatives, PFM03 and PFM01, which primarily block endonuclease activity.
MRE11 Inhibitors Prevent DSB End Resection
HR has a major role in facilitating recovery from aberrant replication events. To focus specifically on the role of MRE11 in the interplay between HR and NHEJ at two-ended DSBs, we sought to avoid complications from its role in replication fork stabilization and/or recovery and employed the inhibitors to examine repair pathway choice at DSBs generated in G2 following exposure to ionizing radiation (IR).
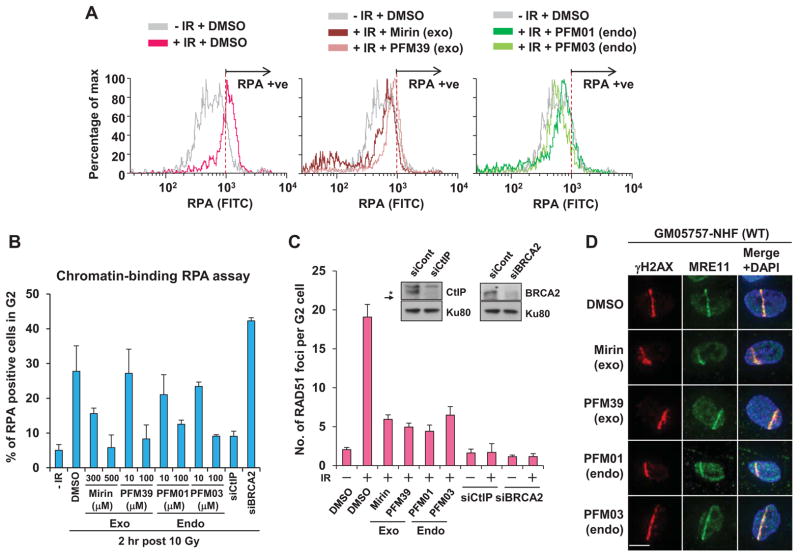
To assess IR-induced resection at two-ended DSBs, we used fluorescence activated cell-sorting (FACS) to quantify chromatin-bound RPA in G2 phase (Shibata et al., 2011). Following 10 Gy, we observed increased chromatin-bound RPA in control G2 cells but little change following treatment with MRE11 nuclease inhibitors (Figures 2A, 2B and S2A). CtIP functions with MRE11 during resection; CtIP siRNA also dramatically impairs resection (Sartori et al., 2007) (Figure 2B). Confirming an impact on resection, ssDNA formation detected using α-BrdU antibody was also significantly reduced following inhibitor treatment (Figure S2B). Inhibitor concentrations were optimized to find the lowest concentration reducing chromatin-bound RPA to levels similar to those in CtIP-depleted cells (Figure 2B). RAD51 loading onto resected DNA, which can be visualized as RAD51 foci, represents a downstream step of HR. Consistent with reduced resection, all inhibitors dramatically impaired IR-induced RAD51 foci in G2 cells (Figures 2C and S2C). For G2 immunofluorescence (IF) analysis, aphidicolin (APH), which inhibits the replicative polymerases and blocks progression of S phase cells into G2, was added. APH also induces pan-nuclear γH2AX in S phase, allowing S phase cells to be distinguished from G2 cells (Beucher et al., 2009; Shibata et al., 2011). Inhibitors did not impede MRE11 recruitment to DSBs nor significantly diminish ATM activation, assessed by IR-induced pS1981-ATM formation by Western blotting and pKAP foci formation (Figures 2D, S2D and S2E). Thus, all inhibitors impair MRE11 resection without disrupting other MRN activities, supporting the observation that ATM is activated normally in a strain expressing nuclease-dead MRE11 (Buis et al., 2008).
Figure 2. MRE11 Inhibitors Prevent DSB End Resection.
(A) MRE11 inhibitors reduce chromatin-bound RPA levels post IR. IR-induced RPA retention in A549 cells were monitored by FACS-analysis at 2 h post 10 Gy IR. G2 cells were gated using FACS Diva software (Figure S2A). (B) Quantification of RPA-FACS analysis using two different concentrations of inhibitor. (C) Either exo- or endonuclease inhibitor treatment reduces IR-induced RAD51 foci formation in G2 cells. RAD51 foci were quantified in A549 cells 2 h post 3 Gy. Knockdown efficiency of CtIP and BRCA2, included as controls, is shown. Representative images are shown in Figure S2C. (D) MRE11 recruitment to DNA damage sites is unaffected by the MRE11 inhibitors. GM05757 primary human fibroblasts were subjected to a UV-microbeam. Protein recruitment was analyzed 1h after addition of each inhibitor. γH2AX is used to identify damage sites. The scale bar is 10 μm. For G2 foci analysis, 4 μM aphidicolin (APH) was added post irradiation. APH does not affect γH2AX/RPA/RAD51 foci formation, NHEJ or HR in G2 phase cells. Full controls for using APH in the context of repair and signaling have been undertaken (Shibata et al., 2010; Shibata et al., 2011). Error bars represent SEM from >3 independent experiments. 100 μM PFM01, PFM03 or PFM39, or 500 μM mirin were added 30 min before irradiation in A, C and D. For further analysis of ATM activation after inhibitor treatment see Figures S2D and S2E. See also Figure S2.
MRE11 Inhibitors Confer Distinct DSB Repair Phenotypes
X-ray induced DSB repair monitored by enumerating γH2AX foci loss occurs with two-component kinetics in G2-phase (Beucher et al., 2009; Shibata et al., 2011). About 80% of X-ray induced DSBs are repaired with fast kinetics by NHEJ, hence NHEJ mutants show a repair defect 2 h post IR, while ~20% of DSBs are repaired with slow kinetics by HR. To investigate HR, γH2AX foci were enumerated at 8 h post IR, when loss of HR proteins in mutants or following siRNA confers a DSB repair defect (longer times were avoided due to mitotic progression despite incomplete repair (Deckbar et al., 2007)). Addition of mirin or PFM39 caused a G2 repair defect at 8 h resembling that shown by HR-defective ATMi-treated or HSC62 (BRCA2-defective) cells (Howlett et al., 2002) (Figures 3A, 3B and S3A). In contrast, PFM01 or PFM03 treatment allowed normal DSB repair. None of the MRE11 inhibitors affected γH2AX foci numbers at 2 h post IR suggesting that they do not affect canonical NHEJ, the fast DSB repair process (Beucher et al., 2009) (Figure S3B). The impact on DSB repair was confirmed by enumerating IR-induced chromosomal breaks in G2 cells (Figure 3C). ATM is required for the slow DSB repair component in G1 and G2, partly to promote heterochromatin (HC) relaxation by phosphorylating KAP-1, an HC-component (Beucher et al., 2009; Goodarzi et al., 2008). To substantiate that defective repair is not due to a failure to promote HC relaxation, we examined repair following KAP-1 siRNA, which alleviates the need for ATM to effect HC relaxation (Goodarzi et al., 2008). KAP-1 depletion did not relieve the repair defect caused by MRE11 exonuclease inhibition but overcame the defect from ATM inhibition (Figure 3D). Collectively, these findings show that MRE11 exo- or endonuclease inhibitors confer distinct DSB repair phenotypes.
Figure 3. Distinct Phenotypes of MRE11 Inhibitors in DSB repair.
(A–B) Exo but not endo-inhibitor treatment causes a DSB repair defect in G2. DSB repair in G2 (CENPF+) cells was investigated by γH2AX foci analysis. 1BR3 (WT) hTERT were fixed and stained at 8 h post 3 Gy. Inhibitors were added 30 min before IR. The full time course for DSB repair is shown in Figure S3A. (C) DSB repair defects following inhibitor addition are consolidated by chromosomal break analysis. Irradiated G2 cells were collected at 12–16 h post 6 Gy. (D) Depletion of the HC building factor, KAP-1, does not alleviate the DSB repair defect in exo-inhibitor treated cells. 1BR3 hTERT cells were subjected to KAP-1 siRNA, and irradiated with 3 Gy. KAP-1 knockdown efficiency is shown. To prevent progression of S-phase cells into G2 during analysis, we added replicative polymerase inhibitor APH in all foci assays (A, B and D). The dots in the box plot were created using SigmaPlot 12.0 and represent outliers, >95% or <5% of samples. A line in each box represents the median. Error bars represent SEM from >3 experiments. See also Figure S3.
MRE11 Endonuclease Activity Initiates Resection to Direct Repair towards HR
CtIP siRNA prevents HR in G2 cells but repair can proceed by NHEJ (Shibata et al., 2011). Hence CtIP siRNA does not cause a DSB repair defect in G2 cells. In contrast, loss of downstream HR components (BRCA2 or RAD51) confers a DSB repair defect, suggesting that CtIP licenses HR and that CtIP itself or an immediate downstream step (prior to RAD51 loading) commits to HR and precludes usage of NHEJ (Shibata et al., 2011). Since inhibition of MRE11 endo-activity does not confer a DSB repair defect but impedes resection, we hypothesized that MRE11 endonuclease functions similarly to CtIP, i.e. initiates HR. We reasoned that if MRE11 endonuclease inhibition allows NHEJ to proceed, it would overcome the requirement for BRCA2, an essential HR protein that functions downstream of resection, for G2-phase DSB repair. BRCA2-deficient HSC62 fibroblasts show impaired repair in G2 at 8 h post IR (Beucher et al., 2009; Howlett et al., 2002). Addition of PFM01 or PFM03 but not mirin relieved this defect, which was confirmed by chromosomal break analysis (Figures 4A, 4B and S4A). These results argue that inhibition of MRE11 endonuclease activity, like depletion of CtIP, allows the usage of NHEJ. In contrast, the exonuclease activity functions downstream of this step, when there has been a commitment to HR. Thus, MRE11 endonuclease inhibition allows repair by NHEJ but this possibility is precluded following MRE11 exonuclease inhibition. Consistent with this model, RAD51 foci did not form following exo- or endo-inhibitor treatment (mirin or PFM01) (Figure 4C). To substantiate that repair occurs by NHEJ, we added PFM01 to cells lacking XLF, an NHEJ protein, and observed an additive repair defect, similar to the impact of siRNA CtIP (Figure 4D). Examination of DSB repair pathway usage in cells with chromosomally integrated I-SceI inducible DSBs substantiated that adding PFM01 or PFM03 enhances NHEJ and reduces HR whereas PFM39 inhibits HR without significantly increasing NHEJ (Figure 4E). Addition of the inhibitors did not alter the cell cycle distribution during analysis (Figure S4B). These data demonstrate that both MRE endo- and exonuclease activities are required for HR, and that MRE11 endonuclease activity initiates resection, licensing repair for HR. MRE11 exonuclease activity functions downstream when there has been a commitment to HR.
Figure 4. MRE11 Endonuclease Activity Commits to Homologous Recombination.
(A) Addition of MRE11 endo inhibitors rescues the repair defect in HR-defective cells. γH2AX foci were enumerated in 48BR (WT) and HSC62 (BRCA2-defective) primary cells at 8 h post 3 Gy. The full time course for DSB repair and cell cycle profiles are shown in Figure S4. (B) The chromosome break defect observed in BRCA2 siRNA treated 1BR3 (WT) hTERT was alleviated following treatment with endo inhibitor. (C) RAD51 foci formation is diminished in HSC62 (BRCA2-defective) cells with or without inhibitor treatment. RAD51 foci formation was analyzed 2 h post 3 Gy. (D) Treatment with an endo-inhibitor causes an additive repair defect in XLF but not control cells, demonstrating NHEJ usage. γH2AX analysis was performed in XLF-defective hTERT cells post 2 Gy. 2 Gy was used (rather than 3 Gy) to enhance accuracy in scoring the greater number of DSBs remaining in XLF-defective cells. (E) Inhibitor treatment impairs HR but endo-inhibitor addition only enhances NHEJ usage. 50 μM of PFM39, PFM01 or PFM03 was added 8 h post I-SceI transfection until cell fixation. The frequency of DSB repair usage was measured using the reporter cell lines, U2OS DR-GFP (HR) or H1299 dA3 (NHEJ), respectively. The percentage of repair was normalized to the DMSO treated control. Error bars represent SEM from >3 experiments. See also Figure S4.
MRE11 Endonuclease Functions Upstream of Exonuclease Activity in HR
Our data suggests that MRE11 endonuclease initiates resection, and functions upstream of MRE11 exonuclease activity. To test this, we examined whether inhibition of MRE11 endo-activity could relieve the need for MRE11 exo-activity for DSB repair, i.e. allow NHEJ to ensue. As anticipated, PFM01 or PFM03 addition substantially (although not completely) relieved the DSB repair defect conferred by mirin or PFM39 (Figures 5A and 5B). In contrast, a repair defect remained following combined addition of mirin and PFM39 but not PFM01 + PFM03 suggesting that the impact of PFM01/03 is not explained simply by enhancing the level of nuclease inhibition (Figure 5B). Further, RAD51 foci did not form, and the defect was additive in XLF cells supporting the notion that NHEJ is utilized for DSB repair (Figures 5C and 5D). Co-addition of PFM01 and PFM39 enhanced NHEJ usage in the I-SceI assay resembling that observed following endonuclease inhibition alone (Figure 5E). Inhibitor titrations verified that specific inhibition of MRE11 endonuclease activity determines pathway choice rather than the magnitude of resection (Figure S5). Collectively, these findings argue that MRE11 endonuclease functions upstream of its exonuclease activity, licensing repair by HR
Figure 5. MRE11 Endonuclease Functions Upstream of Exonuclease Activity in Homologous Recombination.
(A–B) Endo-inhibitors alleviate the repair defect conferred by the exo-inhibitors. DSB repair was analyzed by γH2AX foci in irradiated G2 cells at 2 and 8 h post 3 Gy. The repair defect is only observed at 8 h, consistent with the fact that at 2 h DSB repair occurs by MRE11-independent NHEJ. (C) A defect in IR-induced RAD51 foci formation was confirmed in the presence of double inhibitor treated 1BR3 (WT) hTERT cells. (D) Cells treated with MRE11 exo- and endo-inhibitors repair DSBs by NHEJ, similar to the result in cells treated with MRE11 endonuclease inhibitor alone. (E) HR and NHEJ repair frequency following single or combined endo- and exo-inhibitor treatment was measured by the U2OS DR-GFP (HR) or H1299 dA3 (NHEJ) assay as described in Figure 4E. Error bars represent SEM from >3 experiments. For further evidence showing that inhibition of MRE11 endo and not exonuclease activity allows NHEJ usage. See also Figure S5.
ATLD Patient Cells can Switch to NHEJ Usage Verifying That MRE11 Regulates Pathway Choice
Ataxia telangiectasia like disorder (ATLD) is a human syndrome with hypomorphic mutations in MRE11 (Stewart et al., 1999). We exploited a primary fibroblast from an ATLD patient (ATLD2) to analyze DSB repair pathway choice. Consistent with previous reports, ATLD2 cells show significantly reduced MRN levels, impairing IR-induced ATM activation and signaling (Uziel et al., 2003) (Figure 6A). Since ATLD2 cells show reduced ATM signaling and pKAP-1 foci formation, they are, like ATM, defective in the repair of HC DSBs (Noon et al., 2010). This role of MRE11 is not affected by the inhibitors (Figure S6). Thus, we relieved this role of ATLD by carrying out KAP-1 siRNA, which constitutively relaxes HC and bypasses this function of ATM/MRE11 (Noon et al., 2010) (Figures 6B and S6). Although KAP-1 siRNA alleviates the DSB repair defect in G2 phase ATLD2 cells, it does not rescue the defect in HSC62 (BRCA2- defective) cells demonstrating the specificity of the impact (Figure 6B). However, significantly, the impaired RPA/RAD51 foci formation in ATLD2 cells is not rescued by KAP-1 siRNA although these end-points are not affected in KAP1 siRNA treated control cells (Figure 6C), demonstrating: 1) that MRE11 is essential for resection and 2) that NHEJ can proceed in ATLD cells. HSC62 cells show, as expected, normal resection but no RAD51 loading irrespective of KAP1 siRNA (Figure 6C). These results are consistent with previous findings that inhibition of ATM causes a G2 phase DSB repair defect that can be relieved by KAP1 siRNA (Shibata et al., 2011). Further, to consolidate the impact of MRE11 inhibitors, we treated ATLD cells with the inhibitors +/− KAP-1 siRNA (Figure 6D). As expected, no additional impact on DSB repair was observed following addition of either exo- or endonuclease inhibitor, demonstrating that the repair defect caused by the exonuclease inhibitor is mediated via MRE11 inhibition. Based on our findings above, we suggest that ATLD2 cells subjected to KAP-1 siRNA cells can utilize NHEJ instead of HR. To consolidate this, we added a DNA-PK inhibitor (DNA-PKi) at 3.5 h when most NHEJ in G2 is completed. DNA-PKi addition prevented the rescue of DSB repair (Figure 6E).
Figure 6. Analysis of DSB repair pathway choice in MRE11-defective ATLD cells.
(A) ATLD2 cells show significantly reduced MRN levels, causing impaired IR-induced ATM activation and downstream signaling. Cells were harvested 30 min post 3 Gy (right panel). (B) Depletion of KAP-1 alleviates the DSB repair defect in ATLD2 cells, consolidating that the repair defect is caused by impaired chromatin modification (see also Figure S6). DSB repair in G2 cells are examined by γH2AX foci analysis post 3 Gy. Box plot with statistical analysis is shown in right panel. (C) Impaired RPA/RAD51 foci formation in ATLD2 G2 cells is not rescued by KAP-1 siRNA, demonstrating that MRE11 is essential for resection. (D) Exo or endonuclease inhibitor treatment shows no additive impact on DSB repair in ATLD + KAP-1 siRNA cells. (E) The fraction rescued by KAP-1 siRNA is prevented by a DNA-PKi. DNA-PKi is added at 3.5 h post 3 Gy when most NHEJ has been completed in G2. Error bars represent SEM from >3 experiments.
Thus, consistent with the notion that MRE11 endo-activity functions upstream of resection, ATLD cells can utilize NHEJ instead of HR provided the ATM-dependent chromatin relaxation defect is bypassed by KAP-1 siRNA. Under these conditions, DSB repair can ensue by NHEJ. These results therefore substantiate our findings with MRE11 inhibitors providing evidence that their impact is due to specific MRE11 nuclease inhibition. With the inhibitors, however, it is unnecessary to utilize KAP1 siRNA since they do not inhibit ATM activation (Figure S2).
Combined Exonuclease Inhibition Allows DSB Repair by NHEJ
Next, we tested whether residual resection activity in exonuclease inhibitor-treated cells could be attributed to EXO1/BLM-dependent nuclease activity, since bidirectional exonuclease digestion 3′-5′ by MRE11 and 5′-3′ by EXO1/BLM was proposed to promote resection during yeast meiotic recombination (Garcia et al., 2011). Either MRE11 exo-activity inhibition or depletion of EXO1/BLM reduced RPA foci formation at DSBs (Figure 7A). Importantly, combined inhibition/depletion caused a greater reduction, as substantiated by pRPA immunoblotting (Figures 7A and 7B). Examination of DSB repair by enumerating γH2AX foci in EXO1/BLM depleted cells with or without PFM39 or PFM01 revealed, unexpectedly, that PFM39 relieves the repair defect caused by EXO1/BLM depletion suggesting that combined loss of exonuclease activities allows DSB repair by NHEJ (Figure 7C). To substantiate this, we added DNA-PKi at 3.5 h post IR. Although DNA-PKi does not affect DSB repair in control cells (since repair after 3.5 h occurs by HR), repair was significantly impaired when PFM39 was added to EXO1/BLM siRNA-treated cells (Figures S7A and S7B). Further, the I-SceI based reporter assay revealed increased NHEJ usage following EXO1/BLM siRNA + PFM39 compared to control cells or following EXO1/BLM siRNA alone (Figure 7D). These results suggest that neither NHEJ nor HR can proceed following loss of either MRE11 3′-5′ exo or 5′-3′ EXO1/BLM since ssDNA gaps arise from one or other exonuclease activity; however loss of both exonuclease activities allows repair by NHEJ. We suggest that the ss nick generated by MRE11 endonuclease activity is not a barrier to NHEJ most likely because it can undergo repair. Thus, whilst MRE11 endonuclease activity is critical in initiating resection and directing the switch from NHEJ to HR (i.e. it licenses HR), failure to enlarge the nick due to combined loss of MRE11 and EXO1/BLM exonuclease activities also allow the progression of NHEJ (Figures 7E and S7C). Thus, the generation of a ssDNA region rather than the endonucleolytic nick is the step that prevents the usage of NHEJ and, therefore represents the commitment step.
Figure 7. Combined MRE11 Exo 3′-5′ and EXO1/BLM 5′-3′ Loss Allows Repair Switch to NHEJ.
(A) MRE11 3′-5′ exonuclease and EXO1/BLM 5′-3′, which digest in opposite directions, both contribute to IR-induced RPA foci formation. 1BR3 (WT) hTERT cells were fixed and stained with RPA (a resection marker) and 53BP1 (a DSB marker) at 2 h post 3 Gy. RPA signal intensity was measured by ImageJ. The quantified signal intensity of RPA foci at 53BP1 foci is shown in the right panel. 200–300 foci were analyzed for each sample. (B) MRE11 exo and EXO1/BLM activities redundantly contribute to RPA Ser4/8 phosphorylation. A549 cells are harvested at 2 h post 30 Gy. The pRPA signal was quantified by ImageJ. Since phosphorylated RPA migrates distinctly to unmodified RPA migration, the pRPA signal was normalized to Ku80 instead of total RPA. (C) Inhibition of MRE11 exonuclease activity in EXO1/BLM depleted cells allows normal DSB repair kinetics. γH2AX foci analysis was performed in A549 cells following EXO1/BLM siRNA +/− inhibitors post 3 Gy. (D) HR and NHEJ repair frequency following EXO1/BLM siRNA alone or EXO1/BLM siRNA + PFM39 was measured by the U2OS DR-GFP (HR) or H1299 dA3 (NHEJ) assay. Error bars represent SEM from >3 experiments.
For evidence that repair after endonuclease inhibition occurs by NHEJ. (E) Model for repair pathway choice at two-ended DSBs in G2. 1) in WT cells, ssDNA regions are expanded by MRE11 3′-5′ exo and EXO1/BLM ssDNA. 2) Loss of either MRE11 3′-5′ exo or EXO1/BLM results in a DSB repair defect since neither HR nor NHEJ can ensue. 3) The DSB repair pathway can be switched from HR to NHEJ in the absence of CtIP, MRE11 endo or combined loss of MRE11 exo + EXO1/BLM activity. See also Figure S7.
DISCUSSION
Understanding the regulation of pathway choice and the influence of nucleolytic steps and initiation of committed nuclease steps for DSB repair is of substantial biological and medical importance (Ciccia and Elledge, 2010). Combined structural and biochemical analyses allowed us to employ a focused chemical library to develop MRE11 exo- and novel endonuclease inhibitors. These findings show that rather than binding to MRE11’s nuclease active site, mirin and its analogues instead impede the path of DNA towards the active site. Although positioning of exo inhibitors mirin and PFM39 is based upon weak electron density, which does not unambiguously distinguish orientation (Figure S1), both are implicated to bind the same site, consistent with blocking MRE11’s ability to open dsDNA for access to the nuclease active site and causing the observed exonuclease inhibition (Figure S1B). These data suggest that MRE11, like the damage response proteins, Atl1 and FEN1, must sculpt and open dsDNA (Tsutakawa et al., 2011; Tubbs et al., 2009). Mirin has revealed unexpected roles for MRE11 nuclease activity, such as processing replication forks (Schlacher et al., 2011). Our findings using specific MRE11 endo- and exonuclease inhibitors provide novel and unexpected insights into the regulation of pathway choice.
We propose a two-step mechanism for MRE11’s role in resection during HR; an upstream endonucleolytic incision step followed by MRE11’s exonuclease activity digesting 3′-5′ towards the DNA end coupled with EXO1/BLM activity carrying out 5′-3′ resection away from the end. These findings reinforce and extend studies on HR during meiosis in yeast, similarly supporting a key role for MRE11 endonuclease activity upstream of its exonuclease activity. Yet, our study reveals a unique role of MRE11 nuclease activities in regulating DSB repair pathway choice with marked mechanistic differences from its role in yeast meiosis. In meiosis, Spo11 is covalently bound to DSB ends necessitating its physical removal prior to repair. Further, repair must progress by HR since Spo11 removal initiates resection. By contrast in mitotic cells, Ku is likely bound at DSB ends, suggesting that NHEJ acts first to try DSB repair (Dobbs et al., 2010). If NHEJ fails, then, we propose, MRE11 endonuclease activity nicks adjacent to the DSB. Subsequently, resection proceeds bidirectionally via MRE11 3′-5′ and EXO1/BLM 5′-3′ exonuclease activities. Our model implies that endonucleolytic incision is restricted to the 5′ strand and cleaves dsDNA. RAD50, CtIP or additional factors may facilitate dsDNA opening to allow this directional cleavage. Ku may be subsequently ejected from DSB ends by MRE11 nuclease activities or by proteolytic degradation (Feng and Chen, 2012; Postow et al., 2008). Additionally, the high abundance of Ku and presence of DNA-PKcs in mammalian cells could influence the regulation of pathway choice between mammalian and yeast cells.
Evaluation of MRN functions is complex due to its multiple roles, including functions at the replication fork (activating ATR or one-ended DSB repair), which may be distinct to its roles in two-ended DSB repair. Here, we dissected MRE11’s function in two-ended DSB repair, excluding complications from replication. This type of specific analysis is essential to unearth the details of MRE11’s complex roles. Our description of novel MRE11 inhibitors and their cellular impact in this specific context opens the door for translational applications. Resection is an important step promoting translocations that drive carcinogenesis. There is emerging evidence that resection occurs more avidly in tumor cells due, at least in part, to CDK upregulation. Thus, it is likely that these inhibitors and the ability to manipulate DSB repair pathway choice will have biological and clinical value.
EXPERIMENTAL PROCEDURES
Exonuclease Assay
MRE11-RAD50-NBS1 was purified as described previously (Yu et al., 2012). Exonuclease assays were performed in Exo-buffer (25 mM MOPS (morpholine-propanesulfonic acid) pH 7.0, 60 mM KCl, 0.2% Tween-20, 2 mM DTT, 2 mM ATP, 5 mM MnCl2). Double-stranded (ds) DNA substrates were generated by annealing purified oligonucleotides JYM925 (5′-gggtgaacctgcaggtgggcaaagatgtcctagcaatgtaatcgtcaagctttatgccgt-3′) and JYM945 (5′-acggcataaagcttgacgattacattgctaggacatctttgcccacctgcaggttcaccc-3′). 5 nM MRE11-RAD50-NBS1 (MRN), 0.8 mM of the indicated inhibitors, and labelled DNA (100 nM) were incubated in Exo buffer for 30 minutes at 37ºC, followed by deproteinization in one-fifth volume of stop buffer (20 mM Tris-Cl pH 7.5 and 2 mg/mL proteinase K) for 15 minutes at 37ºC. Reactions were loaded on a 8 % acrylamide/urea gel, run at 75W for 60 minutes, dried onto DE81 filter paper and autoradiographed.
Endonuclease Assays
Endonuclease assays were performed with φX174 circular ssDNA Virion DNA (New England Biolabs), 100 ng substrate was incubated with 300 ng purified WT hMRE11 in a 20 μl reaction (30 mM Tris-HCl pH 7.5, 1 mM dithiothreitol, 25 mM KCl, 200 ng acetylated bovine serum albumin (BSA), 0.4% DMSO and 5 mM MnCl2) at 37ºC for 30 minutes with inhibitors as described. For MRN assays, 100 ng substrate was incubated with 400 ng protein for 45 minutes in a 20 μl reaction (30 mM Tris-HCl pH 7.5, 1 mM dithiothreitol, 25 mM KCl, 200 ng acetylated BSA, 0.5%DMSO and 5 mM MnCl2) Reactions were terminated by addition of 1/10 volume of stop solution (3% SDS, 50 mM EDTA) and proteinase K to a final concentration of 0.1 mg/ml and incubation for 10 minutes at 37ºC. The reaction products were run in a 0.8% agarose gel (1xTAE) for 90 minutes at 100 mA. DNA was stained with Ethidiumbromide and visualized using a Typhoon9200 scanner. Quantification was performed with ImageQuant 5.2 software.
Protein Crystallization and Crystal Structure Determinations
Mre11 was expressed, purified, and crystallized. Diffraction data were collected, processed, and refined as described in the Supp. Materials. X-ray data for crystallography was collected at the Advanced Light Source (Lawrence Berkeley National Laboratory) on SIBYLS beamline 12.3.1 or 8.3.1. Refined structures have good statistics and geometry (Table S1).
Cell Culture and Irradiation
48BR (human primary fibroblasts), ATLD2 (primary fibroblasts), 1BR3 hTERT, HSC62 hTERT (BRCA2-defective) and 2BN hTERT (XLF-defective), were cultured in Dulbecco’s modification of Eagles medium with 15% fetal calf serum (FCS). A549 cells were cultured in minimal essential medium (MEM) with 10% FCS. Cells were irradiated in medium using a 137Cs source (Gammacell 1000; dose rate, 7.5 Gy/min). MRE11 inhibitor concentrations were 500 μM Mirin, 100 μM PFM39, 100 μM PFM01 and 100 μM PFM03, unless otherwise stated. 10 μM ATM inhibitor (KU55933) or 10 μM DNA-PKcs inhibitor (NU7441) (both Merck Chemicals, Darmstadt, Germany) were added 30 min prior to irradiation unless indicated.
Immunofluorescence and Immunoblotting
Immunofluorescence staining and immunoblotting were as described previously (Shibata et al., 2011). Antibodies are listed in Supplemental Information.
SiRNA Knockdown
siRNA transfection of 48BR, ATLD2, 1BR3 hTERT, XLF hTERT and A549 cells was undertaken using Hiperfect (Qiagen, Hilden, Germany). siRNA oligonucleotides for scrambled control, Ku80, BRCA2, EXO1 and BLM were from Dharmacon SMARTpool siRNA, and KAP-1 siRNA is used as described (Shibata et al., 2011). siRNA was carried out as described (Shibata et al., 2011).
Chromosomal Break Analysis
24 h following BRCA2 siRNA transfection, cells were irradiated (6 Gy) with/without inhibitors. 4 μM APH was added 10 min before IR. Medium was refreshed 12 h post IR, and cells were incubated with 4 μM APH, 0.1 mg/mL Colcemid and 600 μM UCN-01, which abolishes G2/M checkpoint arrest, 12–16 h post-IR to collect irradiated G2 cells in mitosis. Metaphase spread and giemsa staining were as described (Yamauchi et al., 2011).
IR-induced RPA Retention Assay
The assay was as previously described (Shibata et al., 2011).
γH2AX/RPA/RAD51 Foci Analysis
Foci (>800 foci/sample) were scored blindly. Unless stated otherwise, all foci analysis represents the mean and SEM of 3 experiments. Usually, results were scored by >2 individuals. To examine IR-induced foci in G2 cells (Figures 2C, 3A–B, 3D, 4A, 4C–D, 5A–D, 6B–E, 7C), 4 μM APH was added post irradiation. Cells were stained with γH2AX and CENP-F (G2 marker).
Homologous Recombination and Non-homologous End Joining Assay
1 x 105 DR-GFP U2OS or 1.25 x 105 H1299 dA3-1 cells were plated into 6 well dishes, 24 h before I-SceI transfection. 1.0 or 1.25 μg I-SceI vector (pSceI) was transfected by GeneJuice or NanoJuice™ transfection kit (Novagen). After 8 h, medium was refreshed and DMSO or MRE11 inhibitors added. After 40 h, cells are trypsinized and GFP positive cells measured by FACS (FACSCanto, BD Biosciences) with FACS Diva software..
UV Laser Track
UV-microbeams were performed as described (Harding and Bristow, 2012). Briefly, cells were treated with a 355 nm laser set at 4 mW following 10 mM BrdU treatment for 24 h.
Statistical Analysis
All data were derived from 3–4 independent experiments unless stated. Box plot was created by SigmaPlot 12.0. Statistical significance was determined using Student’s two-tailed t test or Mann-Whitney U test by SigmaPlot 12.0. Asterisks represent: *P<0.05, **P<0.01, ***P<0.001.
Supplementary Material
HIGHLIGHTS.
Designed inhibitors specifically block MRE11 endo- or exonuclease activity.
MRE11 endonuclease initiates resection, licensing repair by HR at two-ended DSBs.
MRE11 exo and EXO1/BLM resect bidirectionally towards and away from the DNA end.
SsDNA formed by either 5′-3′ or 3′-5′ exonuclease activities commits to HR.
Acknowledgments
We thank Drs. A. Carr, A. Oliver and T. Paull for discussions, H. Ogiwara and M. Jasin for cell lines and A. Rodrigue, Y. Coulombe and C. Charbonnel for technical help, the National Institutes of Health [CA117638 to J.A.T.], National Cancer Institute [P01 CA092584 to C.W. and J.A.T)], and the Netherlands Organization for Scientific Research (VICI 700.56.441 to C.W.) for support. X-ray diffraction technologies at SIBYLS beamline 12.3.1 at the Advanced Light Source are partly supported by the U.S. Department of Energy program Integrated Diffraction Analysis Technologies (IDAT). JYM is a FRSQ Senior investigator and is supported by the CIHR. MMG is a CIHR Vanier scholar. PAJ is supported by the Medical Research Council, Association for International Cancer Research, Department of Health and the Wellcome Research Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Moiani D, Axelrod HL, Miller MD, McMullan D, Jin KK, Abdubek P, Astakhova T, Burra P, Carlton D, et al. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J Mol Biol. 2010;397:647–663. doi: 10.1016/j.jmb.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, Stiff T, Jeggo P, Lobrich M. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin ED, Arvai AS, Rosenfeld RJ, Kroeger MD, Crane BR, Andersson G, Andrews G, Hamley PJ, Mallinder PR, Nicholls DJ, et al. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat Chem Biol. 2008;4:700–707. doi: 10.1038/nchembio.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Harding SM, Bristow RG. Discordance between phosphorylation and recruitment of 53BP1 in response to DNA double-strand breaks. Cell Cycle. 2012;11:1432–1444. doi: 10.4161/cc.19824. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Jeggo PA, Geuting V, Lobrich M. The role of homologous recombination in radiation-induced double-strand break repair. Radiotherapy and oncology : J Europ Soc for Therapeu Radiol Oncol. 2011;101:7–12. doi: 10.1016/j.radonc.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockel C, Lammens K, Schele A, Hopfner KP. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, Goodarzi AA, Lobrich M, Jeggo PA. Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol Cell Biol. 2010;30:3371–3383. doi: 10.1128/MCB.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol = Biochimie et biologie cellulaire. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annual Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Yamauchi M, Suzuki K, Oka Y, Suzuki M, Kondo H, Yamashita S. Mode of ATM-dependent suppression of chromosome translocation. Biochem Biophys Res Commun. 2011;416:111–118. doi: 10.1016/j.bbrc.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Yu Z, Vogel G, Coulombe Y, Dubeau D, Spehalski E, Hebert J, Ferguson DO, Masson JY, Richard S. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 2012;22:305–320. doi: 10.1038/cr.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.