Abstract
Wnt signalling is a fundamentally important signalling pathway that regulates many aspects of metazoan development and is frequently dysregulated in cancer. Although many of the core components of the Wnt signalling pathway, such as β-catenin, have been extensively studied, the broad systems level responses of the mammalian cell to Wnt signalling are less well understood. In addition, the cell- or tissue-specific protein networks that modulate Wnt signalling in the diverse tissues or developmental stages in which it functions remain to be defined. To address these questions, we undertook a broad survey of the Wnt response in different human cell lines using both interaction and expression proteomics approaches. Our data reveal both similar and divergent responses of pathways and processes in the three cell-lines analyzed as well as a marked attenuation of the response to exogenous Wnt treatment in cells harbouring a stabilizing (activating) mutation of β-catenin. We also identify cell-type specific components of the Wnt signalling network and find that by integrating expression and interaction proteomics data a more complete description of the Wnt interaction network can be achieved. Finally, our results attest to the power of LCMS/MS to reveal novel cellular responses in even relatively well studied biological pathways such as Wnt signalling.
Introduction
Wnt signalling is an evolutionarily conserved signalling pathway, regulating diverse processes in metazoan development and adult tissue homeostasis. Inappropriate activation of Wnt signalling can have profound effects on cell growth, proliferation, migration and differentiation, and is strongly linked with tumourigenesis in colorectal and other cancers (1,2).
Several related Wnt signalling pathway have been defined, including the so-called canonical, or β-catenin dependent pathway (3). Canonical or β-catenin dependent Wnt signalling is the best understood Wnt pathway, and is activated by binding of a Wnt ligand with specific cell surface receptor complexes comprised of Frizzled family members and the low density lipid receptor, LRP5/6. The resulting biochemical cascade leads to inhibition of the proteasomal degradation of cytoplasmic β-catenin, the central effecter of canonical Wnt signalling. β-catenin protein then accumulates in the cell where the formation of transcriptional complexes in the nucleus leads to activation of Wnt target genes (3). Mutations that alter the ability of the “destruction complex” to regulate the level of β-catenin, such as loss-of-function mutations of the Adenomatous Polyposis Coli (APC) tumour suppressor, or activating mutations of β-catenin itself, that stabilize the protein, are causal events in the initiation of colorectal cancer (4,5). In addition to its role as the core effecter of Wnt signalling, β-catenin also acts as a mediator of cell-cell adhesion through its interaction with cadherins at the cell surface (6). In concert with these multiple functions, a plethora of β-catenin interacting proteins have been identified, using both low-throughput and high-throughput interaction techniques (7).
Broad functional screens (8–10), transcriptomics (11,12) and proteomics approaches (13–15) have been used to define the broader Wnt signalling network, and are beginning to reveal the interconnections between Wnt signalling and other pathways and processes. However, despite our in depth understanding of many of the core Wnt signalling pathway, how the pathway controls so many varied processes during animal development and in tissue homeostasis remains poorly understood (7). Large-scale screens point to complex and context-dependent regulation of the core Wnt signalling pathways in different tissues. For example, a surprising diversity of factors have been discovered through high-throughput RNAi screens to identify Wnt modulators in different biological systems (16) and additionally using lower throughput methods (17,18).
The motivation for the study described here was two-fold. First, by comparing the proteomic response to Wnt activation in different cell-types we aimed to identify cell-specific proteins that might regulate Wnt signalling. Second, we aimed to characterize the broader systems-level effects of activation of Wnt signalling. Using label-free quantitative proteomics, we surveyed the Wnt-responsive proteome of three different human cell-lines, HEK293T, RKO and HCT116 with distinct properties. Two of the cell-lines, RKO and HCT116 cells are colorectal cancer cell lines, whilst HEK293T are derived from embryonic kidney cells. In addition, the HCT116 cell line harbours an activating mutation of β-catenin. Our results demonstrate that the global proteomic response of the cell-lines to exogenous Wnt activation differs considerably, and is markedly attenuated in the cell-line expressing stabilized β-catenin (HCT116). In addition, we that cell-type specific modulators govern the Wnt signalling response across different human cell-lines, and that integrated proteomics approaches provide a powerful means of detecting cell-specific modulators and their association protein interaction networks.
Materials and Methods
Cell cultures and WNT3A treatment
Colorectal cancer cell lines RKO and HCT116 were regularly maintained in McCoy-5A media (Life Technologies, 16600-108, Carlsbad, CA) containing 10% fetal bovine serum (Life Technologies, 10438-026, Carlsbad, CA) and 1% streptomycin-penicillin (Life Technologies, 15140-148, Carlsbad, CA) at 37°C in CO2 incubator (5% CO2, 100% H2O). Human Embryonic Kidney cell line HEK293T was regularly maintained in DMEM media (Life Technologies, 11965-092, Carlsbad, CA) containing 10% fetal bovine serum and 1% streptomycin-penicillin under the same condition. The media were removed and the cells were washed twice with serum-free McCoy5A media. Next, 1ml serum-free McCoy5A media was added in each well of 6-well cell culture plate with purified Wnt3a protein (R&D Systems, Inc. 5036-WNP-010/CF, Minneapolis, MN) at the final concentration of 0, 10, 20, 30, 40 and 50ng/ml (for dosage assays). The cells were cultured in a CO2 incubator (5% CO2, 100% H2O) at 37°C for an additional 24 hours before harvesting. Finally, the cells were harvested by scraping the cells off plates and then washed with cold PBS twice for immediate use or storage (−80°C).
Protein extraction and quantification
The harvested cells were lysed in lysis buffer (25 mM Tris-HCl, pH7.4, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 50% glycerol, Protease inhibitor cocktail) by homogenization and incubated on ice for 30 min followed by centrifugation at 13,000rpm for 30min. The supernatant (soluble fraction) was kept for further analysis. The proteins were quantified by Bio-Rad protein assay dye (500-0006, Bio-Rad, Hercules, CA) by measuring the absorbance at 595nm.
SDS-PAGE and Immunoblotting
Equal amounts (20 μg) of proteins from different samples was loaded on precast 4–12% Bis-Tris gel (Life Technologies NP-0335, Carlsbad, CA) and subjected to electrophoresis. Afterwards, gels were either stained with Coomassie Brilliant Blue (Pierce 20278, Rockford, IL) or transferred to nitrocellulose membrane (Whatman 10402594, Dassel, Germany). Western blotting was used to detect the protein with super signal ELISA identified different pathways and processes impacted by the activation of Wnt signalling across the three cell-lines. In parallel to the global proteomic analysis of the Wnt response, we used focused interaction proteomics (affinity-purification mass-spectrometry) to identify potentially cell-specific β-catenin interacting proteins. Comparison of samples from HCT116 and HEK293T cells showed common core β-catenin interacting proteins as well as distinctly detected proteins in each cell-line. Taken together with the global proteome analysis, we conclude Pico chemiluminescent substrate. Primary antibodies anti-β-catenin (Cell Signaling Technology 9581, Danvers, MA), and anti-α-tubulin (Cell Signaling Technology, Inc., 2144, Danvers, MA) as loading control were applied at 1:1000 and secondary antibodies horseradish peroxidase (HRP)-conjugated anti-mouse (Promega W4011, Madison, WI) and HRP-conjugated anti-rabbit (Cell Signaling Technology 7074, Danvers, MA) were added at 1:20,000. Chemiluminescence detection using SuperSignal* ELISA Pico Chemiluminescent Substrate (Thermo Scientific PI-37070, Rockford, IL) was applied to all westerns.
Immunoblot quantification and statistical analysis
The bands shown in blots were then quantified by ImageJ (http://rsbweb.nih.gov/ij/) (19,20) and the mean values were plotted as well as the standard deviation for three replicates of each protein using Excel.
SDS-PAGE Gel fractionation and in-gel digestion
Protein samples separated in SDS-PAGE were fractionated into five fractions (Supplementary Figure 1) per sample/lane after Coomassie blue staining prior to tryptic digestion to increase the sensitivity for protein identification and quantification. Standard in-gel tryptic digestion was performed according to the published method (21). The combined elution fractions were lyophilized in a SpeedVac Concentrator (Thermo Electron Corporation, Milford, MA), resuspended in 100μl of 0.1% formic acid and further cleaned up by reverse phase chromatography using C18 column (Harvard, Southborough, MA). The final volume was reduced to 10 μL by vacuum centrifugation and addition of 0.1% formic acid.
Protein extraction and Affinity purification
All centrifugation and incubation are performed at 4°C and all buffers are pre-chilled on ice. For all cells, the media were removed from the 150 mm culture plates and the cells (3X107 cells per affinity purification) were collected and washed three times with 25 ml PBS. After pelleting the cells, the cells were lysed with 2 ml of cell lysis buffer (50 mM Tris-HCl, pH7.5, 1 mM EDTA, 150 mM NaCl, 0.1%NP-40, 1X protease inhibitor cocktail) with homogenization for 40 times. The lysates were incubated on ice for an additional 30 minutes before 13,000rpm centrifugation. Next, 20 μl of anti-β-catenin (Cell Signaling Technology 9581, Danvers, MA) native antibody were pre-incubated with Pierce Protein A/G Plus Agarose (Pierce 26147, Rockford, IL) for one hour followed by cross-linking by DSS (disuccinimidyl suberate) (Pierce 68528-80-3, Rockford, IL ) for extra one hour incubation at 4°C. The beads were first equilibrated with 200 μl lysis buffer before adding the cell lysate supernatant. The protein-bead mixtures were then incubated on a rotator at 4°C overnight. The beads were washed four times with cell lysis buffer and then eluted with elution buffer (Pierce 26147, Rockford, IL) or 2xSDS loading buffer with 5 min boiling.
Mass-spectrometry
Tryptic peptides were separated by online reverse phase nanoscale capillary liquid chromatography (nano-LC, DionexUltimate 3000 series HPLC system) coupled to electoral spray injection (ESI) tandem mass spectrometer (MS–MS) with octopole collision cell (Thermo-Finnegan LTQ Orbitrap).Loaded peptides were eluted on nano-LC with 90 min gradients ranging from 6 to 73% acetonitrile in 0.5% formic acid with a flow rate of 300 nL/min. Data dependent acquisition was performed on the LTQ-Orbitrap using Xcalibur software (version2.0.6, Thermo Scientific) in the positive ion mode with a resolution of 60 000 at m/z range of 325.0–1800.0, and using 35% normalized collision energy, up to five most intensive multiple charged ions were sequentially isolated, fragmented and further analyzed. Mass-spectrometry
Mass-spectrometry data processing
Raw LC-MS/MS data were processed using Mascot version 2.2.0 (Matrix Science, Boston, MA). The sequence database was searched with a fragment ion mass tolerance of 0.8Da and a parent ion tolerance of 15 PPM. The raw data were searched against the human International Protein Index database (released in 2009 and containing 74,017 protein sequences) with fixed modification carbamidomethyl (C) and variable modification oxidation (M). The five fractions for each sample were combined as a single search in Mascot. Peptides were filtered at a significance threshold of P < 0.05 (Mascot). Raw mass spectrometry chromatograms were processed and analyzed using Xcalibur Qual Browser software (Thermo Fisher Scientific Inc. Version 2.0.7) and then manually annotated and verified. Scaffold (Proteome Software Inc., Portland, OR, USA; version 3.00.04) was used to analyze LC-MS/MS-based peptide and protein identifications(22). Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (23). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides (23). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. To compare ratios across cell-lines, the spectral counts were normalized according to the Normalized Spectral Abundance Factors (NSAF) (24). In addition to the spectral counting approach, relative quantification was performed using ion peak intensity measures. Raw LCMS/MS data were analyzed with the Rosetta Elucidator software version 3.3.0.1 (Rosetta Inpharmatics LLC, Seattle, WA) employing the following shotgun workflow: (i) DTA files creation on monoisotopic peak; (ii) embedded Mascot search with IPI-Human database (89,652 protein entries, released in June 2010) (iii) feature alignment across all 6x5 MS runs for each cell line and filtering for features with p < 0.05; (iv) feature annotation; (v) expression data were built as combined data for each dataset. For both spectral count and ion current quantifications, the log2 ratio of stimulated/control samples and p-values computed (Wilcoxon signed rank test). AP-MS data were analyzed using the SAINTexpress software (Hyungwon Choi et al, in prep) using spectral counts and computing ratios and significance of differences between prey proteins observed in bait experiments and control experiments. Non-specific prey proteins were further filtered by excluding proteins found as contaminants in a previous large-scale AP-MS study (25) and by comparison to the CRAPome compendium of AP-MS experiments (26). Significant proteins were then used in subsequent interaction network analysis and visualization using the Ingenuity Pathways Analysis (Ingenuity) and Pathway Studio (Ariadne Genomics) tools. Hierarchical clustering of protein expression data was performed using UPGMA method with Euclidean distance as the distance metric.
Results and Discussion
Experimental design
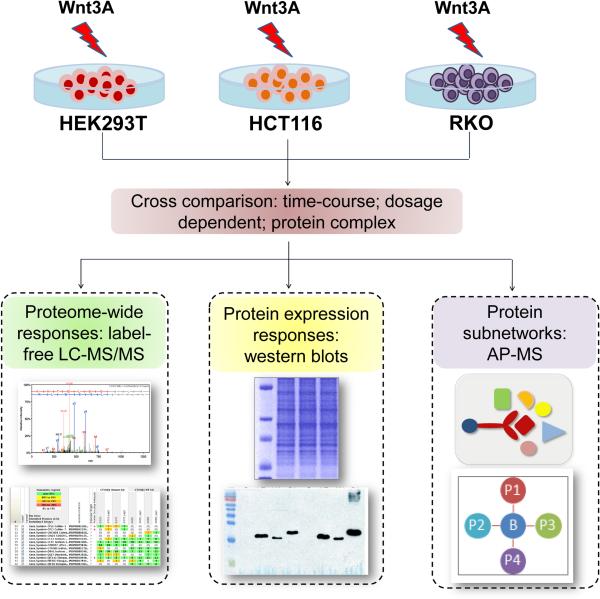
The overall scheme of the study is illustrated in Figure 1. We used both proteomic profiling and interaction proteomics approaches to study the Wnt response in different human cell-lines. To map the proteomic responses to activation of Wnt signalling, we selected 3 human cell lines for study. First, HEK293T (human embryonic kidney cell line) cells exhibit a robust response to exogenous activation of Wnt signalling and have previously served as a model for understanding the dynamics of the Wnt response (11,13). Second, to analyze the Wnt-responsive proteome in the context of colorectal cancer cells, we selected two colorectal cancer cell-lines, RKO and HCT116. Importantly, these two cell lines differ according to their β-catenin status; HCT116 cells are heterozygous for a stabilizing mutation of β-catenin (27,28), that results in loss of a serine residue at position 45, whereas RKO cells are wild-type β-catenin and for the APC tumour suppressor. The response to exogenous activation of Wnt signalling is therefore quite distinct; RKO cells exhibit a robust response to exogenous activation of Wnt signalling (14) whereas HCT116 cells have an endogenously higher level of Wnt activation due to the accumulation of stabilized β-catenin in the cell.
Figure 1.
Schematic overview. Wnt signalling was activated in three different human cell-lines (HEK293T, HCT116 and RKO) by treatment with Wnt3A ligand. Protein samples extracted from the treated cells were prepared for global protein expression analysis using either label free LC-MS/MS or immune blots with native antibodies as shown. Affinity-purification mass-spectrometry (AP-MS) was used to characterize β-catenin associated proteins in the same cell lines. Protein sub-networks were then generated according to individual cell line.
Cell cultures were treated with purified Wnt3A protein, an activator of the canonical Wnt signalling pathway. Previous studies have used different dosages of Wnt proteins to activate Wnt signalling. For example, a previous study of the rapid responses (< 30mins) of the phosphoproteome in HEK293 cells treated the cells with 100ng/ml Wnt3a (29). In another study, the dynamics of the Wnt activated gene-expression response were studied in HEK293 cells following treatment with 200ng/ml Wnt3A (Gujral & MacBeath, 2010a) and reprogramming of glioblastoma cancer cells occurs following treatment of cells with 30ng/ml or less of Wnt ligands (30). In addition, cells were incubated for 24 hours following Wnt3A treatment since a previous analysis of the dynamics of transcriptional activation of Wnt target genes identified early and late phases of activation between 0-24 hours following treatment (11). Similarly, other studies using cell-models treated with Wnt ligands have shown transcriptional response in a similar time frame (31). We reasoned that analysis of the proteomic response after 24 hours would allow us to potentially capture proteomic responses that encompass the earlier phases of transcriptional activation whilst not extending the time course too far such that the cells return to their pre-treatment states.
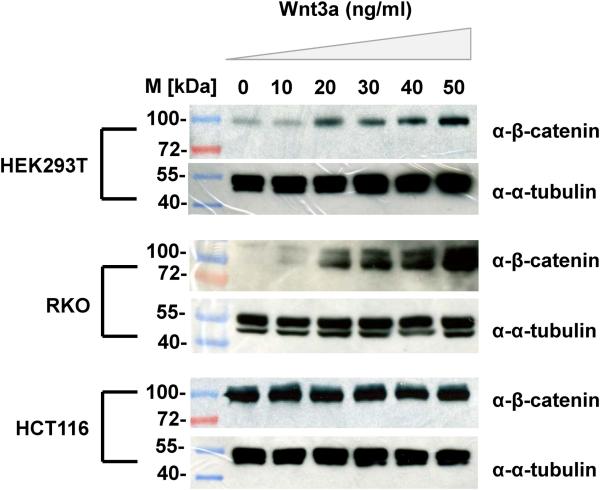
In order to establish appropriate experimental conditions for activating Wnt signalling across the 3 cell-lines, we first analyzed the levels of β-catenin to different dosages of Wnt3A. Initial experiments indicated that a robust response of β-catenin occurred with ~30ng/ml Wnt3A, and so we focused on a range of dosages between 0-50ng/ml. Immunoblots using anti-β-catenin antibodies were used to quantitate β-catenin levels in response to this Wnt3a dosage range as shown in Figure 2. Notably, there is a marked dose-dependent response of HEK293T and RKO cells within the range of Wnt3A treatments used. However, in the HCT116 cells the levels of β-catenin are high even in untreated cells, and the levels of β-catenin do not noticeably respond to Wnt3A treatment, within the dosage regime used.
Figure 2.
Calibrating the Wnt-response in different cell lines (HEK293T, RKO, HCT116). Immunoblots show β-catenin protein expression. Cells were treated with varying doses of Wnt3A (0-50ng/ml), incubated for 24 hours and twenty micrograms of total proteins extracted from each cell lysate were subjected to SDS-PAGE followed by immunoblotting with β-catenin antisera or α-tubulin (as loading control). M refers to standard protein marker.
Proteome-wide responses to Wnt3a
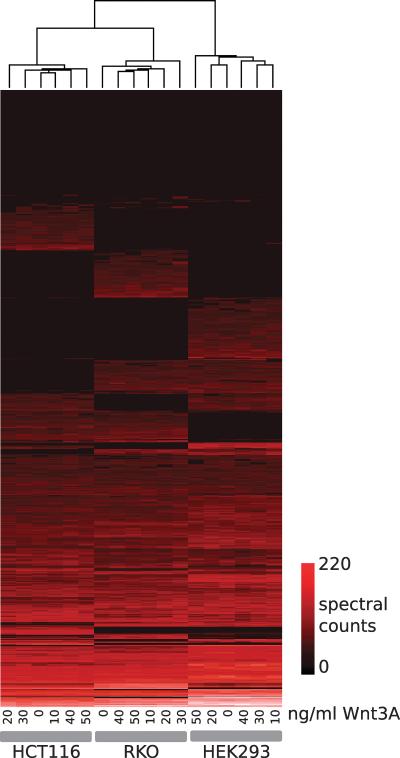
To broadly catalogue the proteomic response to Wnt3A in HEK293T, RKO and HCT116 cells, we used LC-MS/MS to analyze the Wnt response. To capture as much of the proteomic response to Wnt as possible, cell cultures were treated with the range of Wnt3A doses established in Figure 2 (cultures of each cell-line were treated with 0, 10, 20, 30, 40 or 50ng/ml of Wnt3a). Whole cell lysates from each of the 18 cell-line/treatment samples were then analyzed using 1D SDS-PAGE LC-MS/MS. Fractions from each cell-line/treatment were combined and peptides and proteins quantified using two methods: spectral counts, with subsequent NSAF normalization (24), and elution peak/ion current analysis. (supplementary data) summarize all of the data from the three cell-lines. We first analyzed the complete matrix (1039 proteins, 18 samples) of spectral counts by hierarchically clustering the data (by proteins and by sample) as shown in Figure 3. Notably, all samples from each cell-line cluster together (6 samples per cell-line), indicating that the distinct cell-line proteomes and not the Wnt3A treatments is the primary driver of the protein profiles observed. In addition, as expected given their respective origins, the colorectal cancer cell lines (HCT116, RKO) cluster together, indicating that their proteomic profiles show greater similarity than either does to HEK293T.
Figure 3.
Clustered heat map for the Wnt protein expression study. For each sample, spectral counts (Normalized Spectral Abundance Factors) were computed for all proteins. The complete dataset was hierarchically clustered (Euclidean distance, UPGMA) and represented as a heat map as shown. Sample dendrogram is shown at the top of the figure; the protein dendrogram has been omitted for clarity. Samples cluster according to cell-line, and distinct cell-line specific profiles are visible. As colorectal cancer derivatives, samples from the HCT116 and RKO cell lines are more similar to one another than to HEK293 samples.
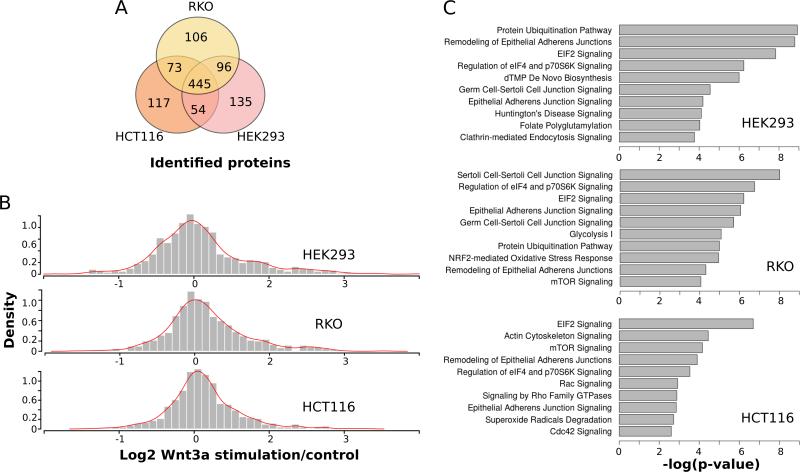
The overlap between each of the cell-line proteomes is shown in Figure 4A. To analyze the proteomic response of each cell-line to Wnt3a, the (log2) ratio and p-value (Wilcoxon signed rank test) of stimulated samples compared to control samples was computed for all proteins. For each cell-line, the distribution of ratios is plotted as a histogram as shown in Figure 4B. Interestingly, the distribution of HCT116 Wnt stimulation ratios shows considerably less dispersion than the equivalent distributions from the HEK293T and RKO datasets. The HCT116 and HEK293T distributions are most statistically different (p= 0.000113; D=0.1324; two-sample Kolmogorov-Smirnov test), suggesting that the global response of the HCT116 proteome is attenuated with respect to HEK293T and RKO cells. The attenuation of the proteomic response of HCT116 cells to Wnt treatment is in line with the attenuated response of β-catenin levels to Wnt stimulation as observed by immunoblot analysis (Figure 2). HCT116 cells harbour a heterozygous activating mutation of β-catenin, with endogenously high basal levels of Wnt signalling. Thus, the HCT116 cells may be less capable of the dynamic responses observed for HEK293T and RKO cells to Wnt activation, at least at the concentrations of Wnt used in the study. Finally, the pathways represented by proteins identified in each of the three cell-lines were compared. All proteins significantly perturbed in response to Wnt treatment (up or down, p<0.1), were mapped to known pathways (Figure 4C). Despite the different cell lines used, several common pathways were observed including epithelial adherens signalling and remodelling, EIF2 signalling and protein ubiquitination. Notably, many of the pathways observed in the analysis are signalling pathways. Of particular interest, signalling by Rho family GTPases and Cdc42 signalling are important mechanisms for regulation of processes such as cell morphology, migration and cell-cycle progression (32). These are important cancer-related processes, and thus significant that these pathways are associated in particular with HCT116 cells (Figure 4C).
Figure 4.
Global analyses of proteome expression datasets. (A) Venn diagram of sets of proteins identified in each cell-line proteome (B) Histograms of proteome response to Wnt treatment in each cell-line. Each histogram represents the complete distribution of all protein ratios (Log2 stimulated /control) from spectral count analysis. The density of each distribution is plotted. Of note, the HCT116 distribution is markedly attenuated (Kolmogorov-Smirnov test – see text), with fewer proteins exhibiting positive stimulated/control ratios in response to Wnt treatment (C) significantly enriched pathways and processes within each cell-line proteome. Wnt responsive proteins (p<0.1) for each cell line were analyzed using Ingenuity Pathways Analysis, and the top ten most enriched pathways plotted as shown. Significantly enriched pathways include pathways in common to each cell-line as well as those that are distinct.
Comparative analysis of Wnt networks
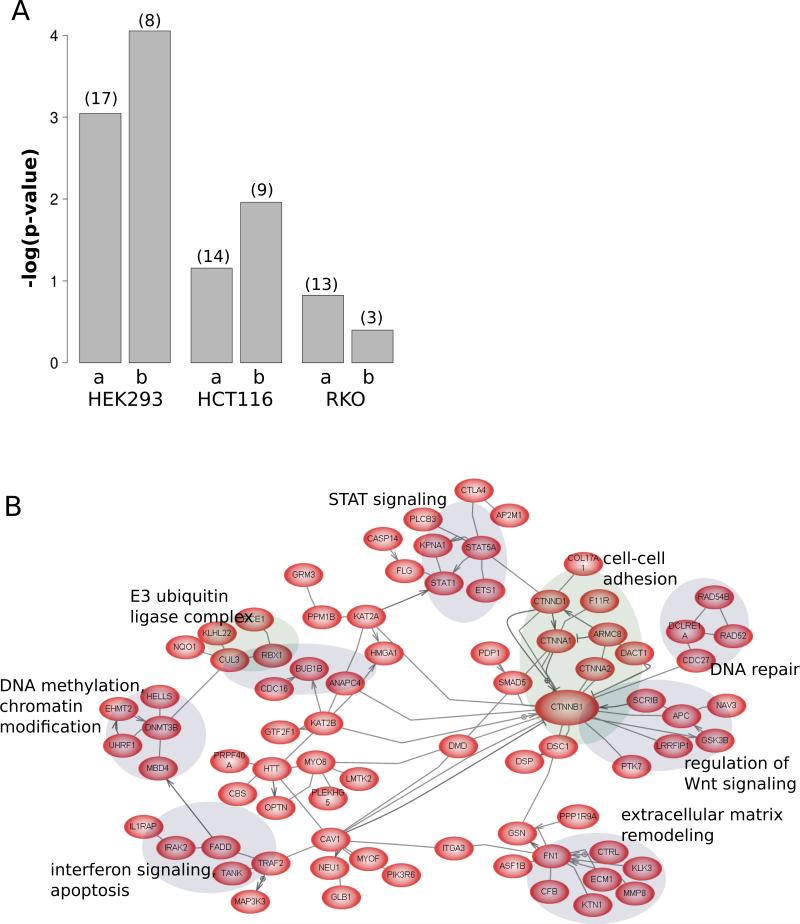
To identify how the Wnt pathway interaction networks might differ between the cell lines, the sets of proteins from each cell line were analyzed as follows. Each cell-line proteome (from total ion current analysis of the data) was classified according to pathway and process (Pathway Studio). Figure 5A shows the relative enrichment for proteins annotated as Wnt pathway components. Both the total sets of proteins and those proteins specific to each cell-line were analyzed. As shown in the bar plot, the trends are similar for the complete or specific sets, with the HEK293T proteome being the most well represented in terms of Wnt pathway components followed by HCT116 and RKO proteomes. Whilst conducting this analysis, we noted that the HCT116 proteome contained several core Wnt components, including CTNNB1/β-catenin that were not identified in either HEK293T or RKO cells. This is not unexpected, since the HCT116 cells express a mutant, stabilized form of β-catenin. Taking the set of 630 proteins identified only in HCT116 and subjecting them to sub-network analysis (Pathway Studio), we observed a large connected component as shown in Figure 5B. Interestingly, this unbiased analysis identified a connected subnetwork focused on CTNNB1/β-catenin, and included several known Wnt regulatory components as well as clusters of proteins functioning in signalling and cancer-related pathways, such as DNA repair and Stat signalling. Thus, the β-catenin associated sub-networks shown in Figure 5B are likely to be strongly enriched in HCT116 cells as compared to the other two cell-lines.
Figure 5.
(A) Wnt pathway coverage in cell-line proteomes. For each cell-line, the enrichment (Fisher's exact test p-value) is plotted (expressed as –log10 *p-value), for either all of the proteins detected (a) or those proteins detected specifically in each cell-line (b). The number in parentheses above each bar indicates the actual number of Wnt pathway proteins detected. (B) Known interactions amongst the set of proteins specific to HCT116. The network shown is the largest connected component of known interactions within the set of 630 proteins identified (ion current analysis) only in HCT116 cells, showing many CTNNB1/β-catenin interacting proteins as well as selected protein modules involved in important tumourigenesis-related processes. For clarity, only edges representing binding, protein modification or direct regulation between proteins are shown.
Differential β-catenin associated proteins
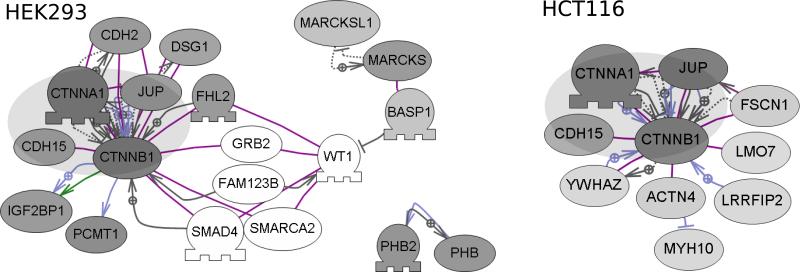
We next used an interaction proteomics to compare β-catenin associated proteins between cell-lines. HCT116 and HEK293T cells were used in AP-MS experiments since, based on the experiments already described, these cell-lines are most distinct in their proteomic response to Wnt3A. Cells were treated with Wnt3A and anti- β-catenin AP-MS experiments performed, as previously described (33). Proteins were quantified (by spectral counts), and comparisons between bait and control experiments used to exclude non-specific binding proteins. The sets of proteins are provided in the supplementary data respectively for the significant HEK293T and HCT116 proteins. Sub-network analysis of each of these sets of proteins identified the connected components that are shown in Figure 6. A core set of β-catenin associated proteins was identified in both cell-lines, and these correspond to several well-known β-catenin-interacting proteins. Several of these proteins (CTNNA1, CDH15, JUP) are cadherin-associated proteins, functioning to regulate cell-cell adhesion. The direct role of these proteins in regulation of Wnt signalling is less clear, although plakoglobin (JUP) may mediate Wnt signalling, particularly in the absence of β-catenin (34). Beyond the common core of β-catenin associated proteins in HEK293T and HCT116 cells, several other proteins were identified as shown in Figure 6. These sets of proteins are quite distinct, and indicate that β-catenin-associated protein complexes might differ considerably between these cells. In HCT116 cells, the AP-MS experiments identified 14-3-3ζ (YWHAZ) as a β-catenin interacting protein. It was recently shown that the association between YWHAZ/14-3-3ζ and β-catenin stabilizes β-catenin and promotes malignancy in lung tumours (35). The association between β-catenin and YWHAZ/14-3-3ζ, is not likely specific to cancer cells, however, since YWHAZ/14-3-3ζ proteins were previously identified by LC-MS/MS in anti-β-catenin immunoprecipitates from HEK293T cells and from DLD1 cells (another colorectal adenocarcinoma cell line) (36). Leucine-rich repeat flightless interacting protein 2 (LRRFIP2) was also identified in the HCT116 AP-MS experiments. Intriguingly the LRRFIP2 protein was identified in a genome-wide screen for novel Wnt modulators, and shown to interact with Dishevelled (Dvl), an upstream regulator of the Wnt signalling pathway (37). In addition, as the name implies, LRRFIP2 protein is a Flightless (FLII) interacting protein. Our previous work, using epitope-tagged Adenomatous Polyposis Coli (APC) as bait in AP-MS experiments, identified FLII protein as an APC interactor (33). APC is a well characterized β-catenin interacting protein, and was identified in both the HEK293T and HCT116 AP-MS experiments. Finally, several connected proteins were detected only in HEK293T experiments as shown in Figure 6. Several of these (BASP1/MARCKS/MARCKSL1) interact, and although not known as direct interactors of β-catenin, may function (or interact) in concert with β-catenin as shown in Figure 6. BASP1 is a well-characterized interactor of WT1/Wilms Tumor 1, functioning as a transcriptional cosuppressor (38,39). For example, FAM123B/WTX (Wilms tumor on the X) is a WT1 associated protein, that also directly interacts with APC and β-catenin (40,41). Although the analysis presented here is not comprehensive, these results point to significant differences between β-catenin or Wnt-associated protein complexes in different cell types. In addition, these differences in protein associations may indicate different functional or tissue-specific properties.
Figure 6.
Comparative analysis of β-catenin associated proteins in HEK293 and HCT116 cells. Sub-network analysis (Pathway Studio) was performed on the sets of specific interactors from anti- β-catenin AP-MS experiments. The most significant connected components are shown for each cell-line. A common core of proteins (represented by the large gray oval) was detected as present in each cell line, whereas other proteins, either known or not known as β-catenin interactors were unique. The shade of each protein node represents the specificity of detection (the ratio of anti- β-catenin affinity-purifications / control purifications). Darker shaded nodes represent proteins with higher ratios. Non-shaded/White nodes represent proteins not detected in the AP-MS experiments, but included in the figure as potential network components.
Conclusions
Here we describe a multi-pronged proteomic analysis of the Wnt response in human cell lines. The analysis revealed both common and divergent changes in protein abundance across the cell-lines in response to Wnt treatment. By analyzing the Wnt-responsive proteome of 3 cell-lines, we showed that there are Wnt-responsive pathways and processes common to all 3 cell lines analysed, as well as responses unique to each cell-line. In particular, the colorectal cancer cell line, HCT116, which harbours a heterozygous stabilizing β-catenin mutation, exhibits a markedly attenuated response to Wnt treatment. Levels of β-catenin respond only marginally in HCT116 cells to the Wnt treatments in our study. Interestingly, comparison of HCT116 proteomic activation with RKO and HEK293T cells showed that the whole proteome responds less in HCT116 cells consistent with the notion that β-catenin is the central effecter for Wnt-driven proteomic responses in HCT116 cells. We recognize that the interaction and expression proteomics presented here are not comprehensive and future more detailed biochemical and proteomic analysis will be required to fully reconstruct cell or tissue-specific Wnt signalling networks. In addition, extended proteomic analyses that allow greater depths of proteomic sampling, will allow for more comprehensive profiles of cell-line proteomes to be acquired. Extensive fractionation for example, can greatly increase the numbers of proteins identified and quantified (42). Extending the Wnt signalling network and identifying cell or tissue-specific modulators will also be enhanced by integration of different types of large-scale screen. For example, the integration of RNAi and proteomics approaches provided for the identification of several new modulators of Wnt/β-catenin in colorectal cancer cell lines (43).
In summary, the results presented here show that both the overall proteomic response to Wnt activation differs between cell lines, and by extension, is likely to significantly differ between different tissues. In addition, detecting differences between Wnt pathway components such as β-catenin associated protein complexes, strongly suggests that cell-specific modulators interact with the core Wnt signalling components to mediate the response in different cells. Thus future work must focus on defining these cell-specific modulators and networks.
Supplementary Material
Acknowledgements
R.M.E and Z.W. acknowledge NCI award 1R21CA16006 that supported in part the work described here. Z.W. acknowledges support from NIH awards R21CA160060, R01CA127590, P50CA150964 and P30 CA043703 and J.S. acknowledges support from the Cancer Pharmacology Training Program (R25CA148052) at Case Western Reserve University.
References
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology [Internet] 2004 Jan;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [cited 2012 Jul 13] Available from: http://www.ncbi.nlm.nih.gov/pubmed/15473860. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell [Internet] 2006 Nov;127(3):469–80. doi: 10.1016/j.cell.2006.10.018. Available from: http://dx.doi.org/10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol [Internet] 2009 Jul;10(7):468–77. doi: 10.1038/nrm2717. Available from: http://dx.doi.org/10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 4.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997 Mar;275(5307):1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 5.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996 Oct;87(2):159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 6.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science [Internet] 2004 Mar;303(5663):1483–7. doi: 10.1126/science.1094291. Available from: http://dx.doi.org/10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. The EMBO journal [Internet]. European Molecular Biology Organization. 2012 Jun 13;31(12):2714–36. doi: 10.1038/emboj.2012.150. [cited 2013 Feb 27] Available from: http://dx.doi.org/10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasgupta R. Functional genomic approaches targeting the wnt signaling network. Curr Drug Targets. 2009 Jul;10(7):620–31. doi: 10.2174/138945009788680455. [DOI] [PubMed] [Google Scholar]
- 9.Berndt JD, Biechele TL, Moon RT, Major MB. Integrative analysis of genome-wide RNA interference screens. Sci Signal [Internet] 2009;2(70):pt4. doi: 10.1126/scisignal.270pt4. Available from: http://dx.doi.org/10.1126/scisignal.270pt4. [DOI] [PubMed] [Google Scholar]
- 10.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A [Internet] 2008 Jul;105(28):9697–702. doi: 10.1073/pnas.0804709105. Available from: http://www.pnas.org/content/105/28/9697.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gujral TS, MacBeath G. A system-wide investigation of the dynamics of Wnt signaling reveals novel phases of transcriptional regulation. In: Isalan M, editor. PLoS One [Internet] 4. Vol. 5. Public Library of Science; Jan, 2010. p. e10024. [cited 2013 Feb 27] Available from: http://dx.doi.org/10.1371/journal.pone.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijayakumar S, Liu G, Rus IA, Yao S, Chen Y, Akiri G, et al. High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/β-catenin target gene, CDC25A. Cancer cell [Internet] 2011 May 17;19(5):601–12. doi: 10.1016/j.ccr.2011.03.010. [cited 2013 Mar 1] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3116447&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang L- Y, Deng N, Wang L- S, Dai J, Wang ZL, Jiang X- S, et al. Quantitative phosphoproteome profiling of Wnt3a-mediated signaling network: indicating the involvement of ribonucleoside-diphosphate reductase M2 subunit phosphorylation at residue serine 20 in canonical Wnt signal transduction. Molecular & cellular proteomics : MCP [Internet] 2007 Nov 1;6(11):1952–67. doi: 10.1074/mcp.M700120-MCP200. [cited 2013 Feb 27] Available from: http://www.mcponline.org/content/6/11/1952.full. [DOI] [PubMed] [Google Scholar]
- 14.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science [Internet] 2007 May;316(5827):1043–6. doi: 10.1126/science/1141515. Available from: http://dx.doi.org/10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 15.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, et al. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol [Internet] 2006 Apr;8(4):348–57. doi: 10.1038/ncb1381. Available from: http://dx.doi.org/10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 16.Pancratov R, Dasgupta R. Postgenomic technologies targeting the Wnt signaling network. Wiley Interdiscip Rev Syst Biol Med [Internet] 2011 Mar; doi: 10.1002/wsbm.140. Available from: http://dx.doi.org/10.1002/wsbm.140. [DOI] [PubMed]
- 17.Hatini V, Bokor P, Goto-Mandeville R, DiNardo S. Tissue- and stage-specific modulation of Wingless signaling by the segment polarity gene lines. Genes & Dev. [Internet] 2000 Jun 1;14(11):1364–76. [cited 2013 Aug 15] Available from: http://genesdev.cshlp.org/content/14/11/1364.full. [PMC free article] [PubMed] [Google Scholar]
- 18.Roël G, Hamilton FS, Gent Y, Bain AA, Destrée O, Hoppler S. Lef-1 and Tcf-3 transcription factors mediate tissue-specific Wnt signaling during Xenopus development. Current biology : CB [Internet] 2002 Nov 19;12(22):1941–5. doi: 10.1016/s0960-9822(02)01280-0. [cited 2013 Aug 15] Available from: http://www.ncbi.nlm.nih.gov/pubmed/12445388. [DOI] [PubMed] [Google Scholar]
- 19.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis [Internet] 2009 Jun;30(11):1845–55. doi: 10.1002/elps.200800720. [cited 2013 Mar 1] Available from: http://www.ncbi.nlm.nih.gov/pubmed/19517440. [DOI] [PubMed] [Google Scholar]
- 20.Tan HY, Ng TW. Accurate step wedge calibration for densitometry of electrophoresis gels. Optics Communications [Internet] 2008 May;281(10):3013–7. [cited 2013 Feb 28] Available from: http://dx.doi.org/10.1016/j.optcom.2008.01.012. [Google Scholar]
- 21.Jiménez CR, Huang L, Qiu Y, Burlingame AL. In-gel digestion of proteins for MALDI-MS fingerprint mapping. Current protocols in protein science / editorial board, John E. Coligan ... [et al.] [Internet] 2001 May; doi: 10.1002/0471140864.ps1604s14. [cited 2013 Mar 30];Chapter 16:Unit 16.4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18429131. [DOI] [PubMed]
- 22.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics [Internet] 2010 Mar;10(6):1265–9. doi: 10.1002/pmic.200900437. [cited 2013 Mar 5] Available from: http://www.ncbi.nlm.nih.gov/pubmed/20077414. [DOI] [PubMed] [Google Scholar]
- 23.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003 Sep;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 24.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst [Internet] 2007 May;3(5):354–60. doi: 10.1039/b701483j. Available from: http://dx.doi.org/10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 25.Ewing RM, Chu P, Li H, Taylor P, Climie S, McBroom L, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Molecular Systems Biology [Internet] 2007;3:89. doi: 10.1038/msb4100134. Available from: http://www.nature.com/msb/journal/v3/n1/full/msb4100134.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellacheruvu D, Wright Z, Couzens AL, Lambert J- P, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nature Methods [Internet] 2013 Jul 7;10(8):730–6. doi: 10.1038/nmeth.2557. [cited 2013 Aug 7] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23921808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan TA, Wang Z, Dang LH, Vogelstein B, Kinzler KW. Targeted inactivation of CTNNB1 reveals unexpected effects of beta-catenin mutation. Proc Natl Acad Sci U S A [Internet] 2002 Jun;99(12):8265–70. doi: 10.1073/pnas.082240999. Available from: http://dx.doi.org/10.1073/pnas.082240999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilyas M. beta -Catenin mutations in cell lines established from human colorectal cancers. Proceedings of the National Academy of Sciences [Internet] 1997 Sep 16;94(19):10330–4. doi: 10.1073/pnas.94.19.10330. [cited 2013 Apr 16] Available from: http://europepmc.org/abstract/MED/9294210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang L- Y, Deng N, Wang L- S, Dai J, Wang ZL, Jiang X- S, et al. Quantitative phosphoproteome profiling of Wnt3a-mediated signaling network: indicating the involvement of ribonucleoside-diphosphate reductase M2 subunit phosphorylation at residue serine 20 in canonical Wnt signal transduction. Molecular & cellular proteomics : MCP [Internet] 2007 Nov;6(11):1952–67. doi: 10.1074/mcp.M700120-MCP200. [cited 2012 Apr 17] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17693683. [DOI] [PubMed] [Google Scholar]
- 30.Rampazzo E, Persano L, Pistollato F, Moro E, Frasson C, Porazzi P, et al. Wnt activation promotes neuronal differentiation of Glioblastoma. Cell death & disease [Internet]. Macmillan Publishers Limited. 2013 Jan 21;4:e500. doi: 10.1038/cddis.2013.32. [cited 2013 Feb 27] Available from: http://dx.doi.org/10.1038/cddis.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klapholz-Brown Z, Walmsley GG, Nusse YM, Nusse R, Brown PO. Transcriptional program induced by Wnt protein in human fibroblasts suggests mechanisms for cell cooperativity in defining tissue microenvironments. In: Zwaka T, editor. PloS one [Internet] 9. Vol. 2. Public Library of Science; Jan, 2007. p. e945. [cited 2013 Oct 23] Available from: http://dx.plos.org/10.1371/journal.pone.0000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leve F, Morgado-Díaz JA. Rho GTPase signaling in the development of colorectal cancer. Journal of cellular biochemistry [Internet] 2012 Aug;113(8):2549–59. doi: 10.1002/jcb.24153. [cited 2013 Oct 25] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22467564. [DOI] [PubMed] [Google Scholar]
- 33.Song J, Hao Y, Du Z, Wang Z, Ewing RM. Journal of proteome research [Internet] American Chemical Society; Nov 7, 2012. Identifying Novel Protein Complexes in Cancer Cells Using Epitope-Tagging of Endogenous Human Genes and Affinity-Purification Mass Spectrometry. [cited 2012 Nov 14]; Available from: http://pubs.acs.org/doi/abs/10.1021/pr300598t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda O, Usami N, Kondo M, Takahashi M, Goto H, Shimokata K, et al. Plakoglobin (gamma-catenin) has TCF/LEF family-dependent transcriptional activity in beta-catenin-deficient cell line. Oncogene [Internet] 2004 Jan 29;23(4):964–72. doi: 10.1038/sj.onc.1207254. [cited 2013 Apr 29] Available from: http://www.ncbi.nlm.nih.gov/pubmed/14661054. [DOI] [PubMed] [Google Scholar]
- 35.Chen C- H, Chuang S- M, Yang M- F, Liao J- W, Yu S- L, Chen JJW. A novel function of YWHAZ/β-catenin axis in promoting epithelialmesenchymal transition and lung cancer metastasis. Molecular cancer research : MCR [Internet] 2012 Oct;10(10):1319–31. doi: 10.1158/1541-7786.MCR-12-0189. [cited 2013 Aug 20] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22912335. [DOI] [PubMed] [Google Scholar]
- 36.Tian Q, Feetham MC, Tao WA, He XC, Li L, Aebersold R, et al. Proteomic analysis identifies that 14-3-3zeta interacts with beta-catenin and facilitates its activation by Akt. Proc Natl Acad Sci U S A [Internet] 2004 Oct;101(43):15370–5. doi: 10.1073/pnas.0406499101. Available from: http://dx.doi.org/10.1073/pnas.0406499101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Bang AG, Kintner C, Orth AP, Chanda SK, Ding S, et al. Identification of the Wnt signaling activator leucine-rich repeat in Flightless interaction protein 2 by a genome-wide functional analysis. Proceedings of the National Academy of Sciences of the United States of America [Internet] 2005 Mar 8;102(6):1927–32. doi: 10.1073/pnas.0409472102. [cited 2013 Aug 30] Available from: http://www.pnas.org/content/102/6/1927.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter B, Hill KJ, Charalambous M, Wagner KJ, Lahiri D, James DI, et al. BASP1 is a transcriptional cosuppressor for the Wilms’ tumor suppressor protein WT1. Molecular and cellular biology [Internet] 2004 Jan;24(2):537–49. doi: 10.1128/MCB.24.2.537-549.2004. [cited 2013 Sep 2] Available from: http://www.pubmedcentral.nih.gov/articlerende r.fcgi?artid=343806&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodfellow SJ, Rebello MR, Toska E, Zeef LAH, Rudd SG, Medler KF, et al. WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. The Biochemical journal [Internet] 2011 Apr 1;435(1):113–25. doi: 10.1042/BJ20101734. [cited 2013 Sep 2] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3062854&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science (New York, N.Y.) [Internet] 2007 May 18;316(5827):1043–6. doi: 10.1126/science/1141515. [cited 2013 Mar 28] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17510365. [DOI] [PubMed] [Google Scholar]
- 41.Rivera MN, Kim WJ, Wells J, Stone A, Burger A, Coffman EJ, et al. he tumor suppressor WTX shuttles to the nucleus and modulates WT1 activity. Proc Natl Acad Sci U S A [Internet] 2009 May;106(20):8338–43. doi: 10.1073/pnas.0811349106. Available from: http://dx.doi.org/10.1073/pnas.0811349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, et al. Molecular systems biology [Internet] 1. Vol. 7. Nature Publishing Group; Jan 8, 2011. The quantitative proteome of a human cell line. p. 549. [cited 2013 Sep 6] Available from: http://www.nature.com/msb/journal/v7/n1/full/msb201182.html#Materials-and-methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, et al. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal [Internet] 2008 Jan;1(45):ra12. doi: 10.1126/scisignal.2000037. [cited 2012 Apr 27] Available from: http://www.ncbi.nlm.nih.gov/pubmed/19001663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.