Abstract
Background
Human heart failure (HF) increases alternative mRNA splicing of the SCN5A cardiac Na+ channel, generating variants encoding truncated, nonfunctional channels that are trapped in the endoplasmic reticulum. In this work, we tested whether truncated Na+ channels activate the unfolded protein response (UPR), contributing to SCN5A electrical remodeling in HF.
Methods and Results
UPR and SCN5A were analyzed in human ventricular systolic HF tissue samples and human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). Cells were exposed to angiotensin II (AngII) and hypoxia, known activators of abnormal SCN5A mRNA splicing, or were induced to overexpress SCN5A variants. UPR effectors, PERK, calreticulin, and CHOP, were increased in human HF tissues. Induction of SCN5A variants with AngII or hypoxia or the expression of exogenous variants induced the UPR with concomitant downregulation of Na+ current. PERK activation destabilized SCN5A and, surprisingly, Kv4.3 channel mRNAs but not TRPM7 channel mRNA. PERK inhibition prevented the loss of full-length SCN5A and Kv4.3 mRNA levels resulting from expressing Na+ channel mRNA splice variants.
Conclusions
UPR can be initiated by Na+ channel mRNA splice variants and is involved in the reduction of cardiac Na+ current during human HF. Since the effect is not entirely specific to the SCN5A transcript, the UPR may play an important role in downregulation of multiple cardiac genes in HF.
Keywords: sodium channels, heart failure, human, SCN5A, PERK, splicing variants
Human systolic heart failure (HF) is associated with decreased cardiac voltage-gated Na+ channel current,1 and these Na+ channel changes have been implicated in the increased risk of sudden death in HF.2–4 The cardiac Na+ channel is a transmembrane protein composed of four homologous domains, each containing six transmembrane segments. SCN5A, encoding the α-subunit of the Na+ channel, consists of 28 translated exons.5–7 Previously, we have shown that the Na+ channel mRNA is alternatively spliced, with two SCN5A splicing variants (E28C and E28D) increasing in HF because of an angiotensin II (AngII)- or hypoxia-mediated increase in the splicing factor complex RBM25/LUC7L3.7 These variants cause a reading frame shift resulting in premature stop codons and encode cardiac Na+ channels truncated before the pore-forming segment of domain IV of the channel. These variants cannot form functional channels and reduce Na+ current to a greater extent than their percentage of the total SCN5A mRNA (i.e. a dominant negative effect).6 The mechanism of this dominant negative effect is unclear.
The unfolded protein response (UPR) is a series of interrelated signaling pathways that occur when endoplasmic reticulum (ER) experiences excess secretory load, accumulates misfolded proteins, or is subject to other pathological conditions. UPR acts on several levels: it rapidly attenuates general protein synthesis, induces the expression of ER chaperone proteins, and enhances the degradation of misfolded proteins.8–10 These changes are presumably designed to restore protein folding and ER health. There appear to be three main sensor proteins that activate the UPR.8–10 Among these main ER stress sensors, protein kinase R-like ER kinase (PERK)-mediated UPR has been shown to be present in cardiomyocytes.11 In the heart, the UPR plays a role during development, hypertrophy, ischemia, and HF.11 HF is associated with hypoxia, elevated AngII, and increased catecholamines,12–16 all of which have been shown to activate the UPR.17–20 In this work, we investigated the role of the UPR in mediating the Na+ channel downregulation observed in HF.
Methods
Methods are described briefly and reviewed in detail in the Supplemental Material.
Differentiation and culture of human induced pluripotent stem cells-derived cardiomyocytes (hiPSC-CMs)
The human iPS cell line, DF19-9-11T,21 was differentiated into CMs using the matrix sandwich method.22 CMs differentiated for 30 days were used in this study.22
Human heart tissue samples
Human heart tissue samples were composed of end-stage cardiomyopathy hearts (n=10) and nonfailing control hearts (n=6). Normal human ventricular tissue was kindly offered by Dr J. Andrew Wasserstrom (Northwestern University, Chicago, IL). The control RNA sample was obtained from Clontech (Mountain View, CA). Samples were analyzed under University of Illinois at Chicago IRB approval (Protocol 2009-0881).
Real-Time PCR quantification
Total RNA was isolated from cultured cells and human ventricular tissue using the RNeasy Mini Kit and RNeasy Lipid Tissue Mini Kit, respectively (Qiagen, Valencia, CA). The amplification conditions were a holding stage of 95°C for 20 min and 40 cycles at 95°C for 30 s and 60°C for 60 s.7
Transfection and infection assays
C-terminal GFP-tagged E28C and E28D variants constructs were transduced into hiPSC-CMs to overexpress E28C or E28D. An empty vector and a vector expressing only the fluorescent marker were used as controls.
Electrophysiology
Na+ channel currents were measured from hiPSC-CMs by using the whole-cell patch-clamp technique in the voltage-clamp configuration at room temperature.
Statistics
All data are presented as means and the 95% confidence intervals (95% CI). Means were compared using unpaired Student's t test or one-way analysis of variance (ANOVA). Time dependent variables were tested by repeated measures ANOVA. A probability value P <0.05 was considered statistically significant. Box and whisker plots show the median, second quartile, and 1.5 interquartile range. Line graphs with error bars are used to compare changes over time among multiple quantitative variables.
Results
UPR components were upregulated in human HF
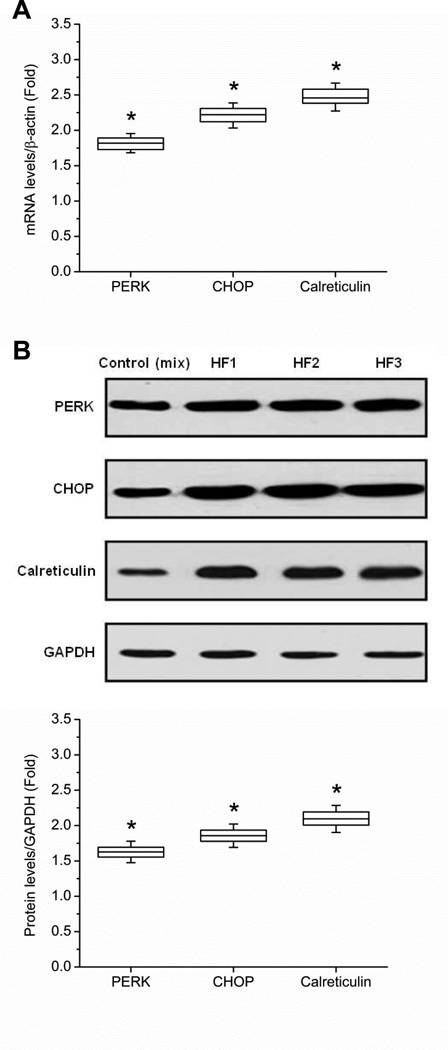
To assess UPR activation in human HF, the expressions of the major UPR pathway components were evaluated by Western blot and qPCR. Compared to normal human heart tissue, the relative mRNA abundances of PERK, CCAAT/enhancer-binding protein homologous protein (CHOP) and calreticulin were increased by 1.82- (95% CI 1.76, 1.88; P < 0.001) fold, 2.22- (95% CI 2.14, 2.29; P < 0.001) fold, and 2.47- (95% CI 2.39, 2.55; P < 0.001) fold in HF tissue, respectively (Fig. 1A). mRNA findings were correlated with protein expression by Western blots. Representative Western blots are shown in Fig. 1B. Compared to the control group, Western blot quantification showed that PERK, CHOP, and calreticulin were increased by 1.63- (95% CI 1.60, 1.65; P < 0.001) fold, 1.86- (95% CI 1.83, 1.89; P < 0.001) fold, and 2.10- (95% CI 2.06, 2.13; P < 0.001) fold in the HF tissue samples (based on three replications for each group).
Figure 1.
UPR activation in human HF tissue. (A) The fold increase in mRNA abundances of PERK, CHOP, and calreticulin in failing heart tissue (n = 3) relative to controls are shown. All mRNA abundances are normalized to β-actin. (B) Western blot quantification confirms the upregulation of PERK, CHOP, and calreticulin in human HF tissue. HF1, HF2, and HF3 represent three separate, representative HF tissue samples, respectively. Quantification is based on three replicated experiments. All protein levels are normalized by GAPDH (* P < 0.05 when compared with control, which was a mixture of 3 patient samples).
SCN5A splice variants activate UPR
We tested whether the presence of SCN5A splice variants could induce PERK. hiPSC-CMs were used because abnormal SCN5A splicing has not been shown to occur in species other than humans and primates.6 AngII and hypoxia are common pathogenic factors in HF.12 We have shown that each increases the SCN5A variants by upregulating the splicing complex RBM25/LUC7L3 7, and we used each as upstream stimuli to induce abnormal SCN5A mRNA splicing. The lack of abnormal splicing in rodents is because of a lack of upregulation of the RBM25/LUC7L3 splicing complex for unknown reasons.
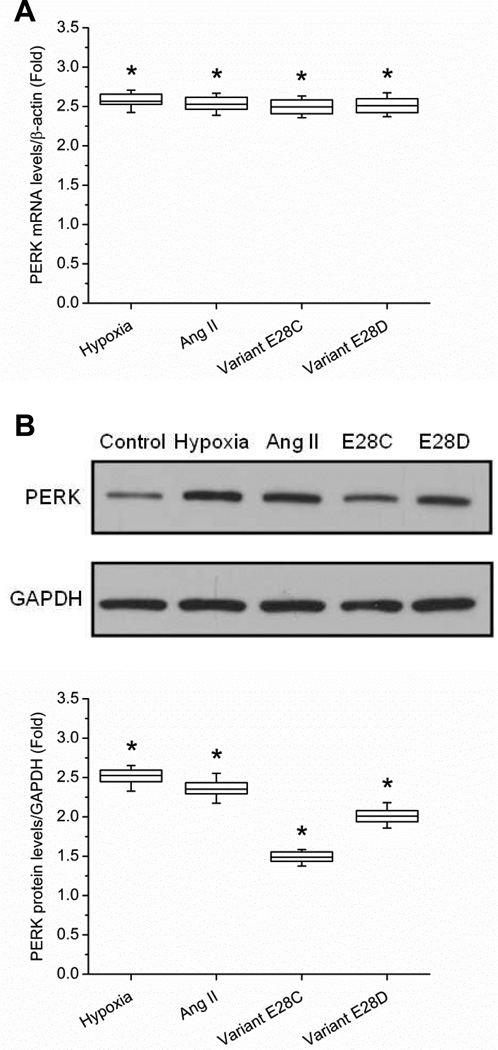
hiPSC-CMs were divided into four experimental groups: untreated control, AngII-treated (200 nmol/L), SCN5A variant E28C and variant E28D overexpression groups. The AngII dose was chosen based on previous experiments showing a maximal effect on variant abundance.7 Cells were harvested from each experimental group at 48 h, and total mRNA was extracted. The expression of PERK was examined by qPCR in each group. The results showed that PERK mRNA was increased in each experimental group compared to control, ranging from 2.5 to 2.6 fold depending on the group (P = 0.012 for each group, Fig. 2A). Western blot revealed that PERK was increased in each experimental group, ranging from 1.5 to 2.5 fold depending on the group (P = 0.033 for each group). The representative Western blots and quantification (based on three replications for each group) are shown in Fig. 2B. Overexpression of full-length SCN5A did not upregulate PERK, however (Supplemental Fig. 1).
Figure 2.
PERK is upregulated in hiPSC-CMs with treatments increasing SCN5A variants. (A) The mRNA abundance changes of PERK in AngII-treated, and cardiomyocytes overexpressing truncation variants E28C or E28D vs. normal control hiPSC-CMs are shown at 24 h. mRNA abundances are normalized to β-actin (* P < 0.05 when compared with control, n = 5 for each experimental group,). (B) Western blot quantification confirms the upregulation of PERK in the different treatment groups of hiPSC-CMs (* P < 0.05 when compared with control, a mixture of three samples; n = 5 for each experimental group).
PERK causes SCN5A variant-mediated downregulation of the full-length SCN5A mRNA
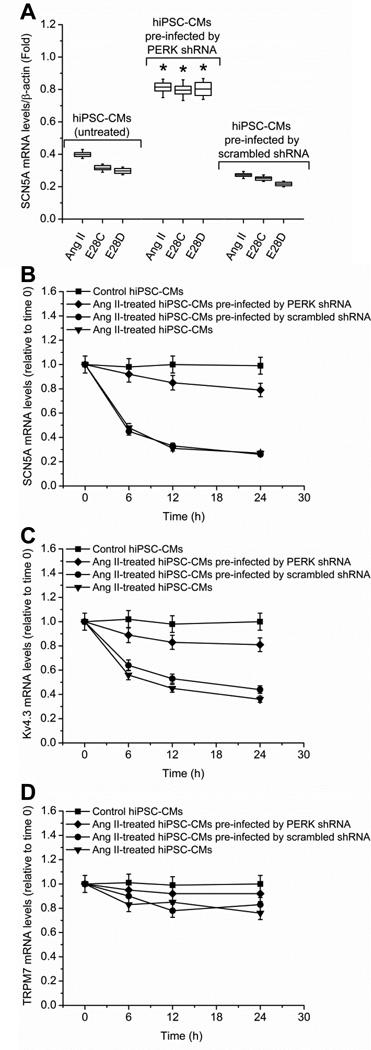
Activated PERK phosphorylates eukaryotic Initiation Factor 2α (eIF2α), increasing mRNA instability by inhibiting ribosomal association.23, 24 To test if PERK activation affected SCN5A stability, hiPSC-CMs cells were divided into three experiment groups: AngII-treated cells, AngII-treated cells pre-infected by pGIPZ lentiviral anti-PERK shRNAmir, and AngII-treated cells pre-infected by scrambled shRNA. AngII (200 nmol/L) treatment was given to all experiment groups for 48 h until cell harvesting. The infection rate was above 90%, evaluated by the ratio of GFP positive cells (pGIPZ lentiviral infected cells) to total cells. PERK knockdown efficiency was 60.0% (95% CI 55.3%, 64.7%; P = 0.017) at 48 h, evaluated by Western blot (Supplemental Fig. 2). Compared to the AngII-treated group, the expression of the full-length SCN5A mRNA was increased by 1.42- (95% CI 1.40, 1.44; P = 0.031) fold when cells were pre-infected by anti-PERK shRNA. Scrambled shRNA did not change the AngII effect. The results indicated that AngII-mediated SCN5A mRNA downregulation was dependent on PERK (Fig. 3A). Similar results were observed when hiPSC-CMs were tranfected with variant E28C or E28D constructs rather than treated with AngII. Full-length SCN5A was increased by 1.54- (95% CI 1.49, 1.59; P = 0.044) fold and 1.47- (95% CI 1.38, 1.56; P = 0.042) fold respectively after pre-infected with anti-PERK shRNA at 48 h. Again, scrambled shRNA did not prevent the effect of the variants.
Figure 3.
PERK is involved in the SCN5A variant-mediated downregulation of full-length SCN5A. (A) Hypoxia, AngII (200 nmol/L), or overexpression of variant E28C or E28D constructs reduced full-length SCN5A mRNA. In each case, this reduction was inhibited by anti-PERK shRNAmir. Pre-infection by scrambled shRNA had no effect on the SCN5A mRNA reduction by any treatment. qPCR measurements by three duplicates are shown at 24 h in each treatment group and normalized by β-actin (* P < 0.05 compared with control group, n = 5 for each experimental group). PERK-mediated mRNA decay assays for SCN5A, Kv4.3 and TRPM7 are shown in panels B-D, respectively. Control (closed squares), AngII-treated (200 nmol/L, inverted triangles), AngII-treated with pre-infection by anti-PERK shRNAmir (closed diamonds), and AngII-treated with pre-infection by scrambled shRNA (closed circles) groups are shown. mRNA was harvested for each group at 0, 6 h, 12 h, and 24 h. The target genes were measured by qPCR and normalized to β-actin. The error bars in panels B-D represented standard error (SE). AngII treatment reduced SCN5A (P = 0.019, n = 5 for each experimental group) (B) and Kv4.3 (P = 0.042, n = 5 for each experimental group) (C) mRNA stability when compared to control while having no effect on TRPM7 (P = 0.078, n = 5 for each experimental group) (D). Pre-infection by pGIPZ lentiviral anti-PERK shRNAmir but not scrambled shRNA could prevent mRNA instability (P = 0.027, n = 5 for each experimental group).
The specificity of the PERK effect on the downregulation of full-length SCN5A was evaluated by analyzing mRNA stability of two other channels regulated by AngII, Kv4.3, 25, 26 and TRPM7.22 These two channels were chosen as representatives because both are present in cardiomyocytes25, 26 and because Kv4.3 but not TRPM7 is known to be regulated in HF.27 hiPSC-CMs were divided into four experimental groups: control (untreated), AngII-treated (200 nmol/L) cells, AngII-treated (200 nmol/L) cells pre-infected by pGIPZ lentiviral anti-PERK shRNAmir, and AngII-treated (200 nmol/L) cells pre-infected by a vector containing scrambled shRNA. Except for the control groups, AngII treatments were given to the all experiment groups for 24 h. mRNA was extracted at 0, 6 h, 12 h, and 24 h after AngII treatment. Compared to the untreated cells, the expression of both SCN5A (Fig. 3B) and Kv4.3 (Fig. 3C) were reduced about 60%, 70%, and 70% at 6 h, 12 h, and 24 h, respectively when cells were treated with AngII or treated with AngII and pre-infected by scrambled shRNA, while the expression of both SCN5A and Kv4.3 mRNAs were only reduced about 20%, 30%, 30% at 6 h, 12 h, and 24 h, respectively when cells were treated with the anti-PERK shRNAmir, implying that the downregulation of PERK prevented mRNA decay of both SCN5A and Kv4.3. There was no statistical difference between the SCN5A and Kv4.3 mRNA decay curves before or after PERK inhibition (P = 0.108), suggesting that both ion channels mRNA abundances were affected equivalently by PERK activation. There is no statistical difference in TRPM7 mRNA (Fig. 3D) between the different treatment groups and at the various time points (P = 0.075), suggesting that the PERK-mediated UPR showed some degree of specificity.
Truncated Na+ channels were localized to the endoplasmic reticulum (ER)
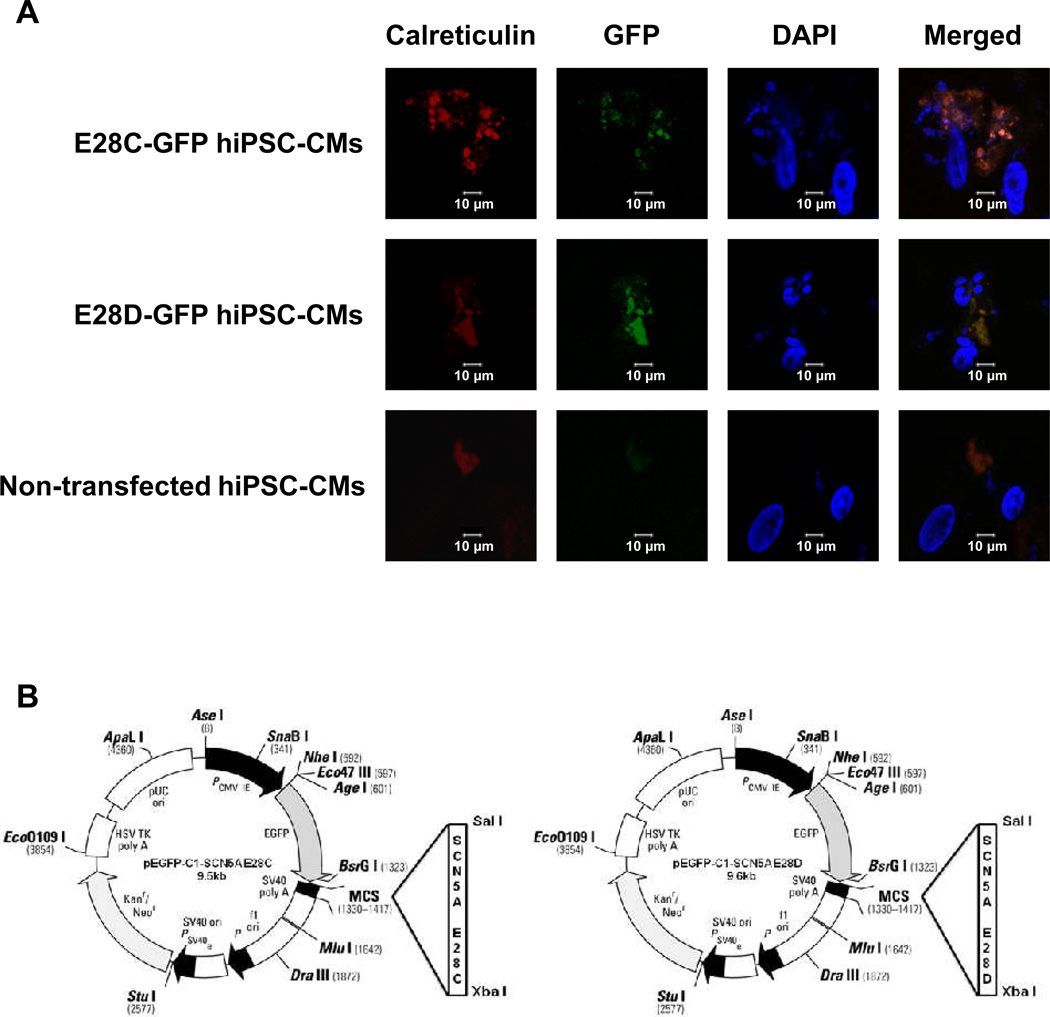
To examine the subcellular localization of truncated Na+ channels, we overexpressed pEGFP-C1-SCN5A E28C and pEGFP-C1-SCN5A E28D in hiPSC-CMs (Fig. 4A). The detail information of both constructs is shown in Fig. 4B. In each case, the truncation variant was linked to GFP at the 5’ end. Therefore, green immunofluorescence indicated the location of truncated Na+ channel encoded by E28C or E28D. The transfected cells were studied at 48 h and stained with the ER marker calreticulin (in red, Fig. 4A). The merged images showed the co-localization of truncated Na+ channel variants with calreticulin. This pattern was distinct from the membrane localization of full-length SCN5A (Supplemental Fig. 3).
Figure 4.
Truncated Na+ channels encoded by mRNA splicing variants E28C or D accumulate in ER. hiPSC-CMs were transfected with GFP-labeled E28C or E28D constructs. (A) The truncated Na+ channel protein is shown in green, the location of ER marker, calreticulin, is shown in red, and nuclei are shown in blue. The co-localization of truncated Na+ channels and calreticulin is shown in the merged images. Calreticulin staining in nontransfected cells was used as a negative control. (B) The E28C and D constructs are shown in panel B. The gene sequences of E28C and D have been previously reported 7.
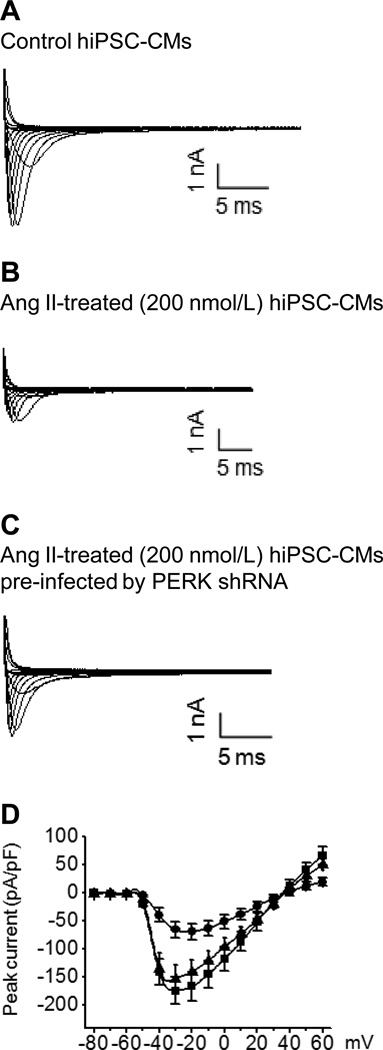
PERK activation reduces Na+ current
The functional consequence of PERK–mediated destabilization of SCN5A mRNA was tested by measuring Na+ current in hiPSC-CMs. The cells were divided into three experimental groups: control, AngII-treated (200 nmol/L), and AngII-treated (200 nmol/L) cells pre-infected by anti-PERK shRNA. Cells were used for measuring Na+ current at 48 h. The representative current traces from each group are shown in Fig. 5. The peak Na+ current density at -30 mV in AngII-treated human cardiomyocytes was reduced by 62% (95% CI 58%, 66%; P = 0.027) when compared to control cells. The effect of AngII on peak current was abrogated by the pre-treatment of anti-PERK shRNA. Scrambled shRNA lentiviral particles had no effect on Na+ channel current regulation by AngII (P = 0.091, data not shown). As found previously, gating changes could not explain the magnitude of peak current changes seen (Supplemental Table 1 and Supplemental Fig. 4).6
Figure 5.
PERK mediates downregulation of Na+ currents after AngII treatment. The representative current traces without normalization for membrane surface area and normalized current-voltage curves are shown from control (■), AngII-treated (●), and AngII-treated cells with anti-PERK shRNA (▲). Current-voltage curves using current density are based on five cells in each group. shRNA lentiviral particles had no effect on AngII-mediated Na+ channel regulation (P = 0.094, data not shown).
Discussion
Recently, we reported that two SCN5A mRNA splicing variants are upregulated in end stage, systolic human HF tissue. These variants reach greater that >50% of the total SCN5A mRNA and do not produce functional channels.6, 7 The expression of these variants in cells stably expressing the full-length channel causes a dose-dependent reduction in the transcripts of full-length SCN5A, as well as the reduction in functional Na+ current expression.6, 7 Moreover, a mutation in a single allele of SCN5A mimicking a variant causes an 86% reduction in Na+ current, greater than the 50% predicted reduction.6 These data suggest that the variants have a dominant negative effect on the full-length channel, but the underlying mechanism is unclear. We hypothesized that UPR might be responsible for the variant-initiated, dominant negative reduction in Na+ current.
Our data show that UPR was activated in end stage, systolic human heart failure. While the human heart experiments do not establish which cell type is experiencing UPR activation, the cardiomyocyte experiments establish that UPR regulates Na+ current in these cells. We show that truncated Na+ channels, encoded by SCN5A variants, become trapped in the ER and activate the UPR. UPR activation resulted in decreased full-length Na+ channel mRNA, protein, and current. Previously, we have shown that full-length, C-terminal GFP-tagged SCN5A targets to the sarcolemma and functions normally and untagged variants cause a dose dependent reduction in SCN5A transcript,6 making unlikely the possibility that UPR activation was related to GFP tagging of the variants. Moreover, elevated AngII and hypoxia, conditions that exist in HF12 and are known to increase the SCN5A truncation variants,6, 7 had similar effects on PERK activation, suggesting that variant induction may contribute to the UPR with these stimuli. Inhibition of PERK prevented full-length SCN5A mRNA degradation and Na+ current reduction, suggesting that UPR activation is downstream of variant translation and required for the variant-mediated reduction in Na+ current.
UPR activation is thought to cause translational inhibition affecting a significant number of, but not all, proteins. The spectrum of proteins affected is unknown as is the mechanism by which some proteins avoid UPR-mediated inhibition. In our case, we showed that AngII-induced PERK activation was responsible for degradation of mRNA encoding the K+ channel α-subunit, Kv4.3, which encodes the cardiac transient outward current. Since this K+ channel is downregulated in HF or by AngII,27–32 it seems likely that a mechanism of this downregulation is UPR activation. It is possible that UPR activation explains, at least partially, the co-regulation of Kv4.3 and Na+ channels observed in HF.33 Another AngII-regulated ion channel, TRPM7, was not affected by PERK regulation, confirming that UPR has certain selectivity. While we did not measure currents of Kv4.3 or TRPM7, the changes in mRNA are consistent with those seen in HF.25, 32, 33 Therefore, the results make it clear that UPR does not affect all protein expression equally, but the basis for selectivity between proteins is unknown.
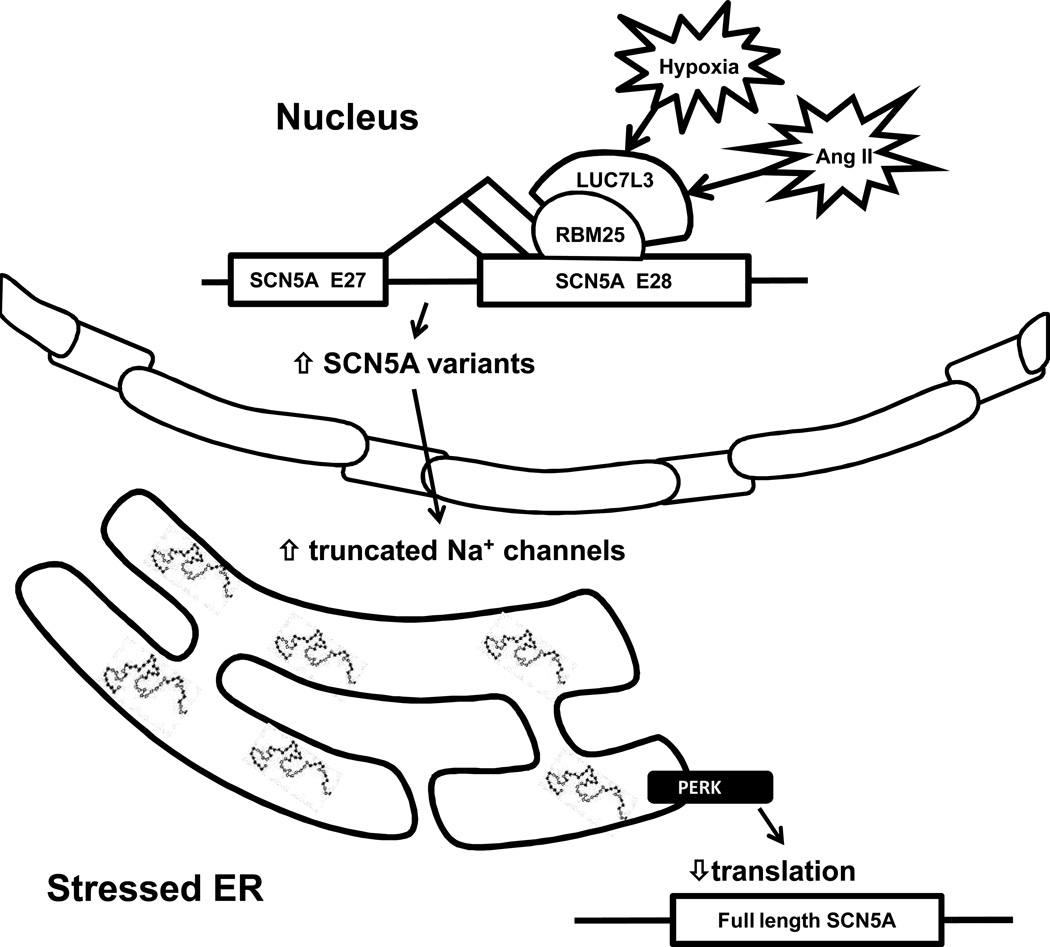
In summary, this and our previous results6, 7 suggest that the reduction in Na+ current during systolic human HF results in part from abnormal SCN5A mRNA splicing to produce nonfunctional channels that become trapped in the ER and activate the UPR. UPR activation leads to destabilization of the remaining full-length SCN5A mRNA, exacerbating of the reduction in Na+ currents. Fig. 6 shows a current hypothesis for the effect of HF on SCN5A mRNA handling based on this and previous work. While we were not able to test the extent to which the Na+ channel reductions mediated in this way contribute to arrhythmia because the splice variants occur only in humans,6 the changes in current were in the range known to contribute to arrhythmic risk. The broad nature of the UPR effect may contribute to the downregulation of other critical proteins in end stage, systolic HF.
Figure 6.
A summary of the effects of abnormal Na+ channel splicing in end stage, systolic HF. AngII and hypoxia are two regulators for RBM25/LUC7L3-mediated SCN5A splicing regulation 7. HF is associated with an increase in Na+ channel mRNA variants resulting from splicing at cryptic splice sequences in the terminal exon of SCN5A (i.e. exon 28). These variants encode non-functional cardiac Na+ channels. Variant levels reach greater that >50% of the total SCN5A mRNA 6, 7. Abnormal mRNA splicing and variant-mediated UPR activation contribute to a decreased in full-length Na+ channel mRNA, protein, and current in HF.
Supplementary Material
Acknowledgments
Funding Sources: National Institutes of Health grants P01 HL058000, R01 HL1024025, R01 HL106592, Veterans Administration Merit Award, and R41 HL112355 to SCD.
Footnotes
Conflict of Interest Disclosures: SCD is the inventor on patent applications: 1) SCN5A Splice Variants for Use in Methods Relating to Sudden Cardiac Death and Need for Implanted Cardiac Defibrillators, PCT/US2012/20564 and 2) SCN5A Splicing Factors and Splice Variants For Use in Diagnostic and Prognostic Methods, 13/291,826.
References
- 1.Valdivia CR, Chu WW, Pu J, Foell JD, Haworth RA, Wolff MR, Kamp TJ, Makielski JC. Increased late sodium current in myocytes from a canine heart failure model and from failing human heart. J Mol Cell Cardiol. 2005;38:475–483. doi: 10.1016/j.yjmcc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Akai J, Makita N, Sakurada H, Shirai N, Ueda K, Kitabatake A, Nakazawa K, Kimura A, Hiraoka M. A novel SCN5A mutation associated with idiopathic ventricular fibrillation without typical ECG findings of Brugada syndrome. FEBS Lett. 2000;479:29–34. doi: 10.1016/s0014-5793(00)01875-5. [DOI] [PubMed] [Google Scholar]
- 3.Brugada P, Brugada R, Brugada J. The Brugada syndrome. Curr Cardiol Rep. 2000;2:507–514. doi: 10.1007/s11886-000-0035-0. [DOI] [PubMed] [Google Scholar]
- 4.Makiyama T, Akao M, Tsuji K, Doi T, Ohno S, Takenaka K, Kobori A, Ninomiya T, Yoshida H, Takano M, Makita N, Yanagisawa F, Higashi Y, Takeyama Y, Kita T, Horie M. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol. 2005;46:2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 5.Gellens ME, George AL, Jr, Chen LQ, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci USA. 1992;89:554–558. doi: 10.1073/pnas.89.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang LL, Pfahnl AE, Sanyal S, Jiao Z, Allen J, Banach K, Fahrenbach J, Weiss D, Taylor WR, Zafari AM, Dudley SC., Jr Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ Res. 2007;101:1146–1154. doi: 10.1161/CIRCRESAHA.107.152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao G, Xie A, Huang SC, Zhou A, Zhang J, Herman AM, Ghassemzadeh S, Jeong EM, Kasturirangan S, Raicu M, Sobieski MA, Bhat G, Tatooles A, Benz EJ, Jr, Kamp TJ, Dudley SC., Jr Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–1131. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raven JF, Koromilas AE. PERK and PKR: old kinases learn new tricks. Cell Cycle. 2008;7:1146–1150. doi: 10.4161/cc.7.9.5811. [DOI] [PubMed] [Google Scholar]
- 9.Ito M, Nakagawa H, Okada T, Miyazaki S, Matsuo S. ER-stress caused by accumulated intracistanal granules activates autophagy through a different signal pathway from unfolded protein response in exocrine pancreas cells of rats exposed to fluoride. Arch Toxicol. 2009;83:151–159. doi: 10.1007/s00204-008-0341-7. [DOI] [PubMed] [Google Scholar]
- 10.Sanson M, Auge N, Vindis C, Muller C, Bando Y, Thiers JC, Marachet MA, Zarkovic K, Sawa Y, Salvayre R, Negre-Salvayre A. Oxidized low-density lipoproteins trigger endoplasmic reticulum stress in vascular cells: Prevention by oxygen-regulated protein 150 expression. Circ Res. 2009;104:328–336. doi: 10.1161/CIRCRESAHA.108.183749. [DOI] [PubMed] [Google Scholar]
- 11.Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44:453–459. doi: 10.1016/j.yjmcc.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanatous SB, Mammen PP, Rosenberg PB, Martin CM, White MD, Dimaio JM, Huang G, Muallem S, Garry DJ. Hypoxia reprograms calcium signaling and regulates myoglobin expression. Am J Physiol Cell Physiol. 2009;296:C393–C402. doi: 10.1152/ajpcell.00428.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zolk O, Solbach TF, Eschenhagen T, Weidemann A, Fromm MF. Activation of negative regulators of the hypoxia-inducible factor (HIF) pathway in human end-stage heart failure. Biochem Biophys Res Commun. 2008;376:315–320. doi: 10.1016/j.bbrc.2008.08.152. [DOI] [PubMed] [Google Scholar]
- 14.Wollert KC, Drexler H. The renin-angiotensin system and experimental heart failure. Cardiovasc Res. 1999;43:838–849. doi: 10.1016/s0008-6363(99)00145-5. [DOI] [PubMed] [Google Scholar]
- 15.Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest. 2005;115:500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anker SD, Chua TP, Ponikowski P, Harrington D, Swan JW, Kox WJ, Poole-Wilson PA, Coats AJ. Hormonal changes and catabolic/anabolic imbalance in chronic heart failure and their importance for cardiac cachexia. Circulation. 1997;96:526–534. doi: 10.1161/01.cir.96.2.526. [DOI] [PubMed] [Google Scholar]
- 17.Mao W, Fukuoka S, Iwai C, Liu J, Sharma VK, Sheu SS, Fu M, Liang CS. Cardiomyocyte apoptosis in autoimmune cardiomyopathy: mediated via endoplasmic reticulum stress and exaggerated by norepinephrine. Am J Physiol Heart Circ Physiol. 2007;293:H1636–H1645. doi: 10.1152/ajpheart.01377.2006. [DOI] [PubMed] [Google Scholar]
- 18.Szegezdi E, Duffy A, O'Mahoney ME, Logue SE, Mylotte LA, O'brien T, Samali A. ER stress contributes to ischemia-induced cardiomyocyte apoptosis. Biochem Biophys Res Commun. 2006;349:1406–1411. doi: 10.1016/j.bbrc.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol. 2006;291:H1411–H1420. doi: 10.1152/ajpheart.01378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M, Kitakaze M. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- 21.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson JA, Herron TJ, Jalife J, Kamp TJ. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rab A, Bartoszewski R, Jurkuvenaite A, Wakefield J, Collawn JF, Bebok Z. Endoplasmic reticulum stress and the unfolded protein response regulate genomic cystic fibrosis transmembrane conductance regulator expression. Am J Physiol Cell Physiol. 2007;292:C756–C766. doi: 10.1152/ajpcell.00391.2006. [DOI] [PubMed] [Google Scholar]
- 24.Raven JF, Baltzis D, Wang S, Mounir Z, Papadakis AI, Gao HQ, Koromilas AE. PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2alpha phosphorylation. J Biol Chem. 2008;283:3097–3108. doi: 10.1074/jbc.M709677200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Wang M, Fan XH, Chen JH, Guan YY, Tang YB. Upregulation of TRPM7 channels by angiotensin II triggers phenotypic switching of vascular smooth muscle cells of ascending aorta. Circ Res. 2012;111:1137–1146. doi: 10.1161/CIRCRESAHA.112.273755. [DOI] [PubMed] [Google Scholar]
- 26.Keskanokwong T, Lim HJ, Zhang P, Cheng J, Xu L, Lai D, Wang Y. Dynamic Kv4.3-CaMKII unit in heart: an intrinsic negative regulator for CaMKII activation. Eur Heart J. 2011;32:305–315. doi: 10.1093/eurheartj/ehq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao L, Li Y, Schultz HD, Wang WZ, Wang W, Finch M, Smith LM, Zucker IH. Downregulated Kv4.3 expression in the RVLM as a potential mechanism for sympathoexcitation in rats with chronic heart failure. Am J Physiol Heart Circ Physiol. 2010;298:H945–H955. doi: 10.1152/ajpheart.00145.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sridhar A, Nishijima Y, Terentyev D, Khan M, Terentyeva R, Hamlin RL, Nakayama T, Gyorke S, Cardounel AJ, Carnes CA. Chronic heart failure and the substrate for atrial fibrillation. Cardiovasc Res. 2009;84:227–236. doi: 10.1093/cvr/cvp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Tang K, Xie B, Li S, Rozanski GJ. Regulation of Kv4 channel expression in failing rat heart by the thioredoxin system. Am J Physiol Heart Circ Physiol. 2008;295:H416–H424. doi: 10.1152/ajpheart.91446.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose J, Armoundas AA, Tian Y, DiSilvestre D, Burysek M, Halperin V, O'Rourke B, Kass DA, Marban E, Tomaselli GF. Molecular correlates of altered expression of potassium currents in failing rabbit myocardium. Am J Physiol Heart Circ Physiol. 2005;288:H2077–H2087. doi: 10.1152/ajpheart.00526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaab S, Dixon J, Duc J, Ashen D, Nabauer M, Beuckelmann DJ, Steinbeck G, McKinnon D, Tomaselli GF. Molecular basis of transient outward potassium current downregulation in human heart failure: a decrease in Kv4.3 mRNA correlates with a reduction in current density. Circulation. 1998;98:1383–1393. doi: 10.1161/01.cir.98.14.1383. [DOI] [PubMed] [Google Scholar]
- 32.Patberg KW, Plotnikov AN, Quamina A, Gainullin RZ, Rybin A, Danilo P, Jr, Sun LS, Rosen MR. Cardiac memory is associated with decreased levels of the transcriptional factor CREB modulated by angiotensin II and calcium. Circ Res. 2003;93:472–478. doi: 10.1161/01.RES.0000088785.24381.2F. [DOI] [PubMed] [Google Scholar]
- 33.Hu D, Barajas-Martinez H, Medeiros-Domingo A, Crotti L, Veltmann C, Schimpf R, Urrutia J, Alday A, Casis O, Pfeiffer R, Burashnikov E, Caceres G, Tester DJ, Wolpert C, Borggrefe M, Schwartz P, Ackerman MJ, Antzelevitch C. A novel rare variant in SCN1Bb linked to Brugada syndrome and SIDS by combined modulation of Nav1.5 and Kv4.3 channel currents. Heart Rhythm. 2012;9:760–769. doi: 10.1016/j.hrthm.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.