Abstract
The immune response to viral infections is determined by a complex interplay between the pathogen and the host. Innate immune cells are equipped with a set of cytosolic sensors to detect viral infections. Recognition by these sensors induces the production of type I interferons and the assembly of inflammasome complexes that activate caspase-1. Here, I discuss recent progress in our understanding of the central roles of NOD-like receptors (NLRs) and inflammasomes in the immune response during viral infections. This information will improve our understanding of host defence mechanisms against viruses and provide new avenues for interfering in the pathogenesis of infectious diseases.
To detect and respond rapidly to diverse groups of microorganisms, vertebrates have evolved multiple germline-encoded pattern-recognition receptors (PRRs) that detect conserved microbial and viral components known as pathogen-associated molecular patterns (PAMPs)1. These PAMPs represent molecules that are expressed by the microbes and are usually required for their survival. Bacterial PAMPs include flagellin, the bacterial cell-wall components lipopolysaccharide (LPS), lipoteichoic acid and peptidoglycan, and nucleic acid structures that are unique to bacteria. Viral PAMPs are composed mainly of unique nucleic acids such as double-stranded (ds)RNA, uncapped single-stranded (ss)RNA, and cytosolic DNA but also include several viral fusion glycoproteins (e.g. respiratory syncytial virus (RSV)-F protein and Ebola virus GP1 proteins). Four main classes of PRRs have been described: Toll-like receptors (TLRs), RIG-I (retinoic acid inducible gene-I)-like receptors (RLRs), nucleotide oligomerization and binding domain (NOD)-like receptors (NLRs) and the IFI200 family member absent in melanoma 2 (AIM2). Whereas TLRs sample the extracellular space and endosomes, NLRs, RLRs and AIM2 function as pathogen sensors in intracellular compartments 2.
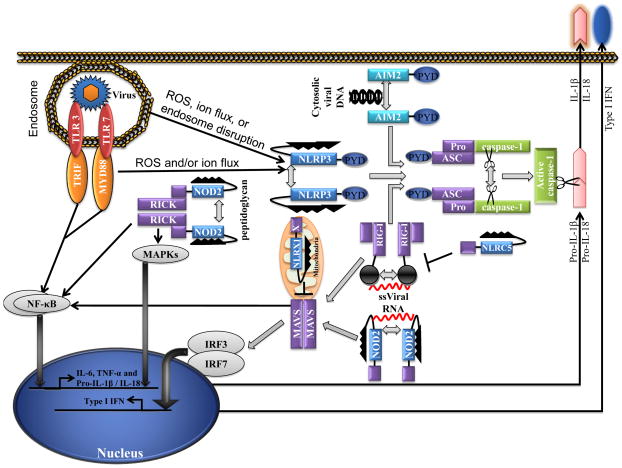
NLRs comprise a large receptor family characterized by the presence of a conserved NOD motif 3, 4. Notably, the domain structure of NLRs resembles that of a subset of plant disease-resistance (R) genes, which trigger the hypersensitive response and confer resistance to infection with fungal, viral, parasitic and insect pathogens 5. The general domain organization of NLRs includes: an amino-terminal effector binding region that consists of protein–protein interaction domains such as the caspase recruitment domain (CARD), pyrin domain (PYD) and baculovirus inhibitor repeat (BIR) domain; an intermediary NOD that is required for nucleotide binding and self-oligomerization; and an array of carboxy-terminal leucine-rich repeat (LRR) motifs presumed to detect conserved microbial patterns and to modulate NLR activity. Bioinformatics studies have shown that there are 22 NLR genes in the human genome and 34 NLR genes in the mouse genome 3, 4. The current model proposes that when a PAMP or DAMP is sensed by the C-terminal LRRs of an NLR, the molecule undergoes conformational rearrangements that trigger oligomerization through the NOD. In turn, NLRs expose the N-terminal effector domains to induce the recruitment and activation of CARD and PYD-containing effector molecules by promoting their proximity and oligomerization. As a result, NLRs are involved in the activation of diverse signalling pathways. For example, the NLR proteins NOD1 and NOD2 interact with the receptor-interacting serine-threonine protein kinase 2 (RIPK2; also known as RICK or RIP2) to induce nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) signalling. Furthermore, NOD1 and NOD2 have been shown to control the induction of autophagy by recruiting autophagy-related gene 16-like 1 (ATG16L1) to the plasma membrane 6. NOD2 also associates with mitochondrial antiviral signalling protein (MAVS; also known as VISA, CARDIF or IPS1) for the induction of type I interferons (IFNs) in response to viral infection 7. Moreover, the NLR family members NLRP1, NLRP3 and NLRC4 assemble large protein complexes known as inflammasomes, which are responsible for the activation of the inflammatory caspase caspase-1 and hence the production of interleukin-1β (IL-1β) and IL-18. Finally, NLRX1 and NLRC5 have been shown to inhibit NF-κB and type I IFN signalling pathways suggesting important roles for these negative regulators in homeostatic control of innate immunity 8, 9. Clearly, NLRs and inflammasomes participate in a diverse set of innate immune signalling pathways (FIG 1).
Figure 1. Intracellular sensors in innate immunity to viruses.
Viral pathogen-associated molecular patterns (PAMPs) activate nucleotide oligomerization and binding domain (NOD)-like receptors (NLRs) and inflammasomes to initiate signalling cascades that lead to the production of proinflammatory cytokines, thereby amplifying antiviral resistance. In the presence of viral PAMPS, NLRP3 and absent in melanoma 2 (AIM2) oligomerize and recruit ASC, the adapter protein engaged by inflammasomes through their pyrin (PYD) domains. The CARD domain within ASC binds the CARD domain of caspase-1, leading to caspase 1 activation and the production of interleukin-1β (IL-1β) and IL-18. Retinoic acid inducible gene-I (RIG-1) contains an RNA helicase domain and N-terminal CARD domains. The helicases senses the 5′ triphosphate moiety of RNA viruses and then signals via CARD–CARD interactions with the mitochondrial localized adapter molecule MAVS to the downstream IKK-related kinase, TBK1, which subsequently phosphorylates and activates IRF3 to turn on transcription of type I interferon (IFNα/β) genes. RIG-I also regulates IL-1β production transcriptionally and post-translationally following recognition of 5′-triphosphate dsRNA. Whereas RIG-I triggered transcription of pro-IL-1β is NF-κB-dependent and mediated by a MAVS, the caspase-1 inflammasome formation and IL-1β/IL-18 production by RIG-I involves ASC. The NLRs NOD2, NLRX1 and NLRC5 associate with MAVS. Whereas NOD2 mediates the induction of IFNα/β, NLRX1 and NLRC5 inhibit RIG-I–MAVS and thus negatively regulate type I IFN signalling. Both MAVS and caspase-1 signalling are blocked by influnenza NS1 protein.
Our understanding of the crucial roles that NLRs and inflammasomes have in mounting protective antiviral responses and the molecular mechanisms used by viral pathogens to evade them has progressed remarkably in the past few years. Depending on the nature of the infectious agent, particular NLRs and inflammasome complexes become activated to elicit tailored immune responses. Here, I review recent findings on the roles of the inflammasome regulators NLRP3, RIG-I and AIM2 and the NLRs NOD2, NLRX1 and NLRC5 in activating or inhibiting major antiviral signalling pathways and discuss the crosstalk that exists between these PRRs. Finally, I provide a brief overview of the mechanisms of viral evasion of these pathways.
Inflammasome activation during viral infection
A breakthrough in the identification of the mechanisms controlling the activation of inflammatory caspases came from the identification and characterization of the inflammasome, a large (700 kDa) multi-protein complex that recruits inflammatory caspases and triggers their activation 10. Inflammasomes are often defined by the constituent PRR family member, which functions as a scaffold protein to bring caspase-1 molecules together and induce caspase-1 activation through proximity-induced auto-activation. Currently, three distinct inflammasomes have been shown to be involved in antiviral immunity: the NLRP3 inflammasome 11–15, the RIG-I inflammasome 16 and the AIM2 inflammasome 17–19. In these complexes, the bipartite, 22-kDa adaptor protein apoptosis-associated speck-like protein containing a CARD (ASC) provides a link between NLRP3, RIG-I or AIM2 and the pro-form of the inflammatory caspases through homotypic interactions with N-terminal PYDs and C-terminal CARDs (FIG 2). Activated caspase-1 cleaves pro-IL-1β and pro-IL-18 into biologically active IL-1β and IL-18, which upon secretion induce various biological effects associated with infection, inflammation and autoimmune processes 20, 21. Whereas most cytokines traffic through the Golgi complex before exocytosis, biologically active IL-1β and IL-18 are secreted through an unconventional and ill-defined mechanism following cleavage of their cytosolic precursors by caspase-1 22. IL-1β participates in the generation of systemic and local responses to infection, injury and immunological challenge by generating fever, activating lymphocytes and promoting leukocyte infiltration at sites of injury or infection20. IL-18 lacks the pyrogenic activity of IL-1β, but is mainly known as a factor that induces IFNγ production by activated T cells and natural killer (NK) cells, thereby contributing to T helper 1 (TH1) cell polarization 22, 23.
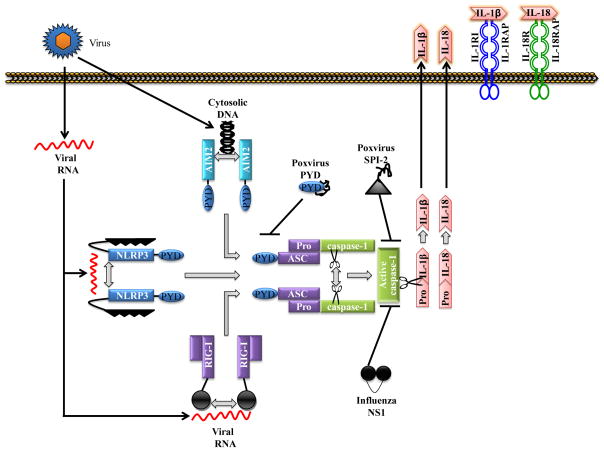
Figure 2. Inflammasome pathways.
The pattern recognition receptors (PRRs) nucleotide oligomerization and binding domain (NOD)-like receptor protein 3 (NLRP3), absent in melanoma 2 (AIM2) and retinoic acid inducible gene-I (RIG-I) assemble caspase-1-activating inflammasome complexes in response to microbial pathogen-associated molecular patterns (PAMPs). The NLRP3 inflammasome recognizes multiple PAMPs including viral RNA in combination with ATP, ion flux or reactive oxygen species. By contrast, AIM2 and RIG-I directly bind to dsDNA and ssRNA viral genomes, respectively. All three types of inflammasome induce caspase-1 activation and interleukin-1β (IL-1β) and IL-18 production. The adaptor protein apoptosis-associated speck-like protein containing a CARD (ASC) is essential in all inflammasome complexes to bridge the interaction between upstream PRRs and inflammatory caspases through its N-terminal pyrin domain (PYD) and C-terminal caspase-recruitment domain (CARD) motifs, respectively. To interfere with inflammasome signalling, the large genomes of poxviruses encode multiple inhibitors such as PYD and SPI-2 that inhibit ASC and caspase-1, respectively.
Many viruses activate caspase-1 and induce IL-1β and IL-18 production (Table 1). Recent studies with a mouse-adapted strain of influenza A/PR/8/34 (PR8) virus, these animals demonstrated impaired viral clearance at days 7 and 10 after infection and a 33% increase in mortality compared to wild-type controls in Il1r1−/− mice. In addition, neutrophil and lymphocyte recruitment to the infected respiratory tract of Il1r1−/− mice was reduced at day 7 after infection and comparable to the wild-type controls by day 10. In addition, there were greatly diminished IgM levels in both serum and lung at days 7 and 10 after infection 24. Similar to Il1r1−/−, Il18−/− mice also had increased mortality, enhanced virus growth, increased neutrophil numbers, and increased nitric oxide production over the first 3 days after respiratory challenge 25. However, with a less virulent influenza virus challenge, IL-18 deficiency was associated with decreased cytokine production by CD8+ T cells 26. In addition to influenza A virus, IL-1β-deficient mice are also susceptible to herpes simplex virus 1 encephalitis owing to increased viral load, decreased inflammation, including fewer infiltrating immune cells and decreased cytokine and chemokine levels 27. Ectromelia virus, an orthopoxvirus, activates caspase-1 in the brain tissue of infected mice and this is thought to have a role in mediating inflammation and cell death 28. Further, West Nile virus (WNV) infection causes the migration of resident skin dendritic cells, known as Langerhans cells, to the draining lymph nodes. The migration of Langerhans cells requires IL-1β, as treatment with an IL-1β-specific neutralizing antibody decreased Langerhans cell migration and decreased the total number of immune cells recruited to lymph nodes of infected mice 29. Hepatitis B virus (HBV) stimulates IL-18 production in human peripheral blood mononuclear cells by means of the histone-like domain of the virus’ core antigen (HBVcAg)30. Interestingly, HBV virions deficient in the pre-core antigen (HBVeAg) induced higher levels of IL-18, suggesting a role for HBVeAg in the inhibition of IL-18 signalling. RSV induces the activation of caspase-1 and production of IL-1β, as well as secretion of IL-1β from neonatal human monocytes, possibly implicating one or more inflammasomes in the response to RSV infection 31. The fact that a wide range of viruses and diverse viral components activate caspase-1, IL-1β and IL-18 shows the potential role of inflammasomes in the recognition of viruses and research in this direction may provide new insights into the activation mechanisms of inflammasomes and their downstream signalling pathways.
Table 1.
NLRs and inflammasomes involved in the recognition of virus infection
| Gene (alternate names) | Signalling pathways during viral infection | Viruses recognized | Viral PAMP recognition | Cytokines | References |
|---|---|---|---|---|---|
| NLRP3 | Inflammasome | Influenza virus, Sendai virus, adenovirus, EMCV | RNA | IL-1β and IL-18 | 11, 12, 15, 32, 33, 36, 37 |
| RIG-I | MAVS, Inflammasome | Vesicular stomatitis virus, hepatitis C virus, Japanese encephalitis virus, Rabies virus, measles virus, respiratory syncytial virus, influenza virus, Sendai virus | 5′-triphosphate ssRNA | IFNα/IFNβ, IL-1β and IL-18 | 52–64 |
| AIM2 | Inflammasome | Vaccinia virus, m cytomegalovirus | Cytosolic DNA | IL-1β and IL-18 | 17–19 |
| NOD2 (NLRC2) | MAVS OAS2 |
Respiratory syncytial virus, influenza A virus, parainfluenza virus-3, vesicular stomatitis virus | ssRNA | IFNα and IFNβ | 7 |
| NLRX1 | MAVS | Sendai virus, Sindbis virus | Exact sensing capacity is unknown | Negatively regulates IFNα and IFN β production | 9, 81 |
| NLRC5 | RIG-I, MDA5, NF-κB | Sendai virus, vesicular stomatitis virus, cytomegalovirus | PolyI:C | IFNα, IFNβ, CCL3, CCL5, IL-6, TNFα, IP10 | 82–86 |
NLRP3 inflammasome
The first evidence for the involvement of NLR inflammasomes in viral infection came from a study in which Sendai virus and influenza A virus were shown to stimulate caspase-1 activation and the production of IL-1β and IL-18 32. Subsequent studies have shown that influenza A virus can activate NLRP3 in various cell types in vitro, including mouse bone marrow-derived dendritic cells and macrophages, a human nasal airway epithelial cell line and the human monocyte cell line THP-1 11, 12. In addition to influenza A virus, infections with modified vaccinia virus Ankara 33 or encephalomyocarditis virus (EMCV) 16 have also been shown to activate the NLRP3 inflammasome. It should be noted, however, that a direct interaction between NLRP3 and viral RNA has not been established. Further, based on the requirement for NLRP3 in IL-1β production after infection with EMCV but not with VSV suggest existence of NLRP3-dependent or NLRP3-independent mechanisms 16.
The requirement for NLRP3 inflammasome activation in vivo during virus infection is best characterized for influenza A virus. Two recent reports demonstrated a role for NLRP3 mediated immune responses to influenza A virus infection in vivo resulting in increased mortality 11, 15. Deletion of NLRP3, caspase-1 or the adaptor ASC resulted in decreased infiltration of neutrophils and monocytes to the lungs and decreased IL-1β, IL-18 and MIP2 levels in the BALF 11, 15. However, IL-6, TNF-α, KC, and IFN-α were also reported to be reduced in NLRP3 deficient mice by one group 15 and the other group reported differential roles for NLRP3 and ASC in the production of KC and TNF-α respectively 11 suggesting the existence of inflammasome-dependent paracrine and autocrine immune pathways. Similarly, a delay in virus clearance at day 7 post-infection 11 whereas no difference at day 6 post-infection 15 were reporter by these groups which might be due to the differences in the infectious doses (10–1000 PFU) used. Finally, the importance of NLRP3 in the resolution of inflammation and the proper repair of lung damage was reported as NLRP3−/− mice had increased amounts of collagen deposition in the lungs and increased necrosis 15. These activities might be mediated directly by IL-1β, as the repair of mechanically injured rat type II alveolar epithelial cell monolayers was promoted by adding IL-1β to in vitro cultures 34. IL-1β was shown to be the crucial component of edema fluid that stimulated repair in a lung epithelial cell line, which is consistent with reports that IL-1β can have a pro-fibrotic role in wound healing at particular stages of an infection 35. It should also be noted that both of these papers on NLRP3 observed some slight differences in the severity or magnitude of several outcomes between NLRP3 and ASC or Caspase-1, with NLRP3 at times having the more severe phenotype. This was observed for survival by one group 11 and for collagen deposition by the other 15 and it was suggested that theses results may indicate additional roles for NLRP3 outside of the inflammasome 11. Further, neither of these reports indicated any role for the NLRP3 inflammasome in the generation of the adaptive immune response. In line with above studies, Ichinohe et. al., 12 also demonstrated a role for the NLRP3 inflammasome in vitro as well as in the alveolar space in vivo, as BALF had reduced levels of IL-1β and IL-18 in NLRP3-, ASC- and Caspase-1-deficient mice 12. Further, at days 8 and 10 post-infection ASC−/− and Caspase-1−/− mice had higher viral titers and were more susceptible to infection with influenza A and had reduced neutrophil and monocytes infiltrates 11, 12, 15. However, this study did not report any difference in cytokines except for IFN-γ in whole lung homogenates for NLRP3−/−, and decreased CD4+ and CD8+ T cell responses and significantly reduced isotype-specific antibodies in ASC−/− or caspase-1−/− mice and not in NLRP3−/− animals 12. One possible explanation for the differences found in these reports is the timing of the experiments (days 6, 7, 8, 10 and 11 post-infection), and the infectious doses used (10 PFU-1000 PFU). Together, currently published studies unequivocally establish a role for NLRP3 inflammasome during influenza A infection 11, 15, 36 but whether or not NLRP3 is required for adaptive immunity require further validation.
Although a role of NLRP3 in sensing viruses is proposed, the precise molecular mechanism involved remains poorly understood. Several lines of evidence suggest that the NLRP3 inflammasome might monitor the presence of viral RNA and DNA in intracellular compartments. For example, transfection of human and mouse cell lines with ssRNA or dsRNA analogues such as polyinosinic–polycytidylic acid (poly(I:C) is sufficient to activate NLRP3 11. Similarly, purified dsRNA from rotavirus or brome mosaic virus and ssRNA of influenza A virus also activate NLRP3 11, 12, 32. The NLRP3 pathway has also been implicated in the detection of viral DNA from adenovirus in cell culture 37. Adenovirus vector capsids alone did not activate NLRP3. By contrast, replication-deficient adenovirus vectors comprising an intact genome activated the NLRP3 inflammasome. In vivo administration of poly(I:C) or purified influenza A virus ssRNA in mice also resulted in IL-1β secretion and inflammation through NLRP3 11, 13, 15.
The discovery that NLRP3 can detect a wide range of divergent PAMPs such as bacterial lipopolysaccharide (LPS), peptidoglycan and bacterial or viral nucleic acids, as well as endogenous danger signals such as monosodium urate (MSU), calcium pyrophosphate dihydrate (CPPD) and ATP, has led to the hypothesis that NLRP3 may sense a common downstream effect caused by these molecules, rather than the PAMPs themselves. Nicotinamide adenine dinucleotide phosphate (NADPH) oxidase-generated reactive oxygen species (ROS) have been suggested as likely secondary messengers activating the NLRP3 inflammasome 11, 38. This conclusion was based on pharmacological inhibition of the NADPH oxidase complex in THP-1 cells and peritoneal mouse macrophages, as well as small interfering RNA (siRNA)-mediated downregulation of the NADPH oxidase subunit p22phox 38. NLRP3 was also shown to physically interact with a ROS sensing protein TXNIP suggesting a role for NLRP3 inflammasome activation in response to ROS 39. Further, a role for ROS during influenza A infection was shown by treating cells in culture or mice infected with influenza A with the ROS inhibitors N-acetyl-L-cysteine (NAc) which resulted in significant decreases in IL-1β production 11. However, NLRP3 activation triggered by silica, MSU crystals or ATP-induced P2X7R activation seems to be normal in macrophages from mice lacking the NADPH oxidase subunit gp91phox 40. Moreover, the increased ROS production in superoxide dismutase-1 (SOD1)-deficient macrophages was recently found to inhibit rather than promote caspase-1 activation 41. The destabilization and rupture of phago-lysosomal compartments, which then release endogenous mediators into the cytosol was also suggested as the NLRP3 activation mechanism 42. It is possible that these potential mechanisms of NLRP3 activation may not be mutually exclusive 43 and additional studies are required for further validation.
Alternatively, TLRs have been proposed to activate the NLRP3 inflammasome directly 44. However, most reports indicate that TLR activation or ROS alone is insufficient for NLRP3 inflammasome activation and the production of significant amounts of IL-1β or IL-18 45, 46, and so a two-signal hypothesis for inflammasome activation has emerged in which pro-IL-1β and pro-IL-18 production is regulated independently of caspase-1 activation and only when both signals are received are significant amounts of active IL-1β or IL-18 produced. In the case of virus infection, dual signalling cues are common. For example, in influenza A virus infection, viral RNA triggers signalling through TLR3 or TLR7 47–49 and during virus entry into the host cell cytoplasm endosomes are compromised, resulting in the release of ions and ROS into the cytosol. Recently, the ability of influenza A virus to activate NLRP3 as a result of ion fluxes was found to be mediated by the M2 ion channel of the virus 36. After infection, virus-encoded M2 is expressed in the secretory compartment, including the trans-Golgi network. The ion-channel activity of M2 enabled the export of protons from acidified Golgi, and triggered activation of the NLRP3 inflammasome. In this case, M2 expression alone was sufficient to activate the NLRP3 inflammasome 36. Vaccinia virus was also shown to require TLR2-dependent production of pro-IL-1β and endocytic internalization of the virus for robust IL-1β production by the NLRP3 inflammasome 33. The fact that multiple pathogens and danger signals activate the NLRP3 inflammasome has probably necessitated the evolutionary pressures to develop a precise regulatory mechanism for production of active IL-1β and IL-18. The requirement for two activating signals, one signal to produce the pro-form of these two cytokines and a second signal to engage their processing by the inflammasome may allow for sensing a wide range of PAMPS and DAMPS and at the same time prevent unwanted inflammation by accidental activation of this pathway.
RIG-I inflammasome
RIG-I is a member of the RLR family. RIG-I has been shown to induce type I IFN signaling through the mitochondrial adaptor protein MAVS, followed by TRAF3/6, IKKε and IRF3/7 signaling 50, 51. Several studies have examined the role of RIG-I during RNA virus infections in vitro, including Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), Sendai virus, hepatitis C virus (HCV), Japanese encephalitis virus (JEV), influenza A virus, rabies virus, measles virus, and RSV 52–59 (Table 1). In vivo, RIG-I–deficient mice have only been studied in the context of JEV infection and were found to be highly susceptible to infection with this virus 53. However, due to the difficulty in generating RIG-I−/− mice many other viruses have yet to be examined in vivo. Another member of the RLR family is MDA5. This RNA sensing cytosolic PRR has also been shown to recognize RNA virus infection and some debate has occurred over the last few years concerning exactly what RNA species RIG-I and MDA5 recognize. Recent work identified full-length virus genomes as the major RIG-I agonist in cells infected with influenza virus and Sendai virus 60. These studies also suggested that nongenomic viral transcripts, short replication intermediates and cleaved self-RNA do not contribute substantially to type I IFN induction in cells infected with negative-strand RNA viruses. Rather, ssRNA viral genomes bearing 5′-triphosphates constitute the natural RIG-I agonists that trigger cell-intrinsic innate immune responses during viral infection. In case of MDA5, the RNA species appears to be specific dsRNA intermediates produced during positive-strand RNA virus infections, particularly by members of the picornavirus family like encephalomyocarditis virus 53, 61 but also by flaviviruses like West Nile virus 62 and Dengue virus 63.
Until recently, RIG-I was thought to be solely involved in activation of IFNα/β genes through recruitment of the adaptor MAVS and the transcription factors IRF3 and IRF7 2. Two recent reports, however, show that RIG-I can also trigger IL-1β production and/or IL-18 production through the caspase-1 and -3 activation 16, 64. RIG-I activation by transfected 5′-triphosphate RNA or VSV infection resulted in the production of pro-IL-1β through a MAVS–CARD9–NF-κB signalling pathway. In response to the same stimuli, RIG-I also directly activated the inflammasome pathway by recruiting caspase-1 through the adaptor ASC. This pathway is not universal for all RLRs as MDA5 does not interact with ASC. These findings indicate that RIG-I might be able to mediate crosstalk between the type I IFN and inflammasome pathways. With regard to this crosstalk, RIG-I may also be able to single handedly mediate the dual signaling discussed above for the NLRP3 inflammasome as RIG-I mediated MAVS signaling can induce NF-κB activation and pro-IL1β production as well as form an inflammasome and activate caspase-1. However, this possibility will require additional research.
AIM2 inflammasome
AIM2 belongs to a family of proteins that includes at least six members in mice (IFI202, IFI203, IFI204, IFI205, Pyhin1 and AIM2) and four members in humans (IFI16, MNDA, IFIX and AIM2). These IFI200 family members (also known as HIN200 proteins) are IFN-inducible proteins with a 200-amino acid repeat at the C-terminus known as the HIN domain and an N-terminal PYD. Four groups recently identified AIM2 as a cytosolic dsDNA sensor that binds cytosolic dsDNA with its C-terminal HIN domain 17–19, 65–67. Similar to NLRP3, the AIM2 N-terminal PYD motif is capable of recruiting ASC and caspase-1. Cytosolic dsDNA from both viruses and bacteria can serve as ligands to activate the AIM2 inflammasome 19, 37. These observations were recently confirmed using AIM2-deficient mice 65, 68. Macrophages from AIM2 deficient mice were found to be defective in caspase-1 activation and induction of pyroptotic cell death in response to liposome-delivered cytosolic DNA. Furthermore, AIM2 was also shown to be required for the activation of caspase-1 in response to infection with vaccinia virus and mouse cytomegalovirus (mCMV) 65, 68 (Table 1). These cell culture experiments were confirmed by testing AIM2−/− mice with mCMV 65. AIM2−/− mice were defective in IL-18 and IFN-γ production and their NK cells especially produced less IFN-γ upon restimulation compared to littermate controls 65. Because activation of the AIM2 inflammasome not only leads to the generation of pro-inflammatory cytokines, but also to the elimination of infected cells through pyroptosis, AIM2 is likely to be important for the efficient clearance of viral infections. Indeed, AIM2−/− mice had higher mCMV titers than control mice likely as a result of decreased pyroptosis and NK cell activity 65, although the contribution of both these factors remains to be examined independently.
Thus, studies in AIM2-deficient macrophages provided conclusive genetic evidence for a crucial role of the AIM2 inflammasome in recognition of cytosolic DNA and infection with vaccinia virus and mCMV. Further, AIM2 was suggested to have a crucial role in mediating the immune response against additional viruses that produce cytosolic DNA during their lifecycle (such as adenovirus and perhaps ssRNA retroviruses, which reverse transcribe their RNA into double-stranded DNA in the cytoplasm). Intriguingly, this same paper reported that the dsDNA virus herpes simplex virus-1 (HSV-1) did not require AIM2 for inflammasome activation 65. These results may indicate that AIM2 is unable to recognize certain DNA viruses or that some viruses have evolved the ability to inhibit AIM2 signaling or block AIM2 mediated recognition of their viral genomic DNA. Additional research will be needed to determine if all families of DNA viruses are recognized and what factors control AIM2 mediated recognition and signaling.
NLRs in type I IFN responses
Although most NLRs are thought to be involved in either NF-κB and MAPK signalling or inflammasome activation and IL-1β and IL-18 production, recent studies have revealed a new and unexpected role for NLRs in type I IFN signalling. Type I IFNs were originally described as antiviral agents and comprise multiple IFN-α molecules (13 subtypes in humans) and IFN-β 69, 70. IFN-β expression is required to exert positive-feedback control over the induction of IFN-α genes 71 and to transduce intracellular signals through the common receptor IFNAR1–IFNAR2, leading to further enhancement of type I IFN gene induction through a complex of STAT1/2 and IRF9 (also called Interferon-stimulated gene factor 3 (ISGF3)) 71. Thus, type I IFNs are key components of the antiviral innate immune system and might be responsible for driving broad cellular antimicrobial activity in general. Below is a brief description of the new roles of NLRs in type I IFN responses.
NOD2
The NLR family member NOD2 is known to sense bacterial molecules that are produced during the synthesis and/or degradation of peptidoglycan, allowing recruitment of downstream adaptor molecule RIPK2 through homotypic CARD interactions. This CARD-containing serine-threonine kinase directly binds and promotes K63-linked polyubiquitylation of the regulator inhibitor of NF-κB kinase-γ (IKKγ) and activation of the kinase TAK172, 73, which are prerequisite for activation of the IKK complex. IKK activation results in degradation of the NF-κB inhibitor IκBα and the subsequent translocation of NF-κB to the nucleus, where transcription of NF-κB-dependent target genes, such as those encoding IL-6 and TNF occurs. In addition to activation of the NF-κB pathway, NOD2 stimulation results in activation of the MAPKs p38, ERK and JNK 74, 75. The CARD-containing adaptor protein CARD9 was found to be important for the activation of p38 and JNK downstream of NOD2, although it was dispensable for NF-κB activation 76. Nevertheless, the NF-κB and MAPK pathways are thought to cooperate to upregulate the expression of proinflammatory molecules such as IL-1β, TNF and IL-6 that stimulate both innate and adaptive immune responses. In addition, NOD2 has been shown to induce a type I interferon response to the intracellular bacterium mycobacterium tuberculosis (MTB) 77. In this context it was shown that recognition of unique peptidoglycans from MTB by NOD2 resulted in activation of a RIPK2 and IRF5 mediated signaling cascade with type I IFN production.
Recent studies have examined the role of NOD2 in response to ssRNA using a transfection assay designed to detect activation of the transcription factor IRF3, which is involved in the production of type I IFNs 7. Both synthetic ssRNA and viral ssRNA genomes were found to activate IRF3 in a NOD2 and MAVS-dependent manner 7. The important physiological function of NOD2 in antiviral defence was evident from decreased type I IFN responses, and increased RSV pathogenesis, lung disease and virus susceptibility of Nod2-deficient mice. Similarly, macrophages and mice lacking NOD2 had reduced IRF3 phosphorylation and diminished production of type I interferons in response to influenza A and parainfluenza viruses 7. Furthermore, NOD2−/− cells were deficient in their ability to inhibit VSV replication 7. This work suggested that NOD2 functions as a viral PRR that is important for the production of type I IFNs in response to RNA viruses and suggests that, similar to RIG-I, NOD2 can trigger multiple signalling pathways in response to cytoplasmic pathogens. Furthermore, RSV infection was shown to result in NOD2 relocalization to mitochondria, where it binds to MAVS 7. It is therefore likely that NOD2 binding to peptidoglycan induces recruitment and activation of RIPK2 signalling, whereas binding of NOD2 to ssRNA results in mitochondrial relocalization and activation of MAVS-dependent pathways (FIG 1). It should be noted, however, that NOD2 was not specifically shown to bind ssRNA and only to be required for ssRNA induction of type I interferons. However, how NOD2 is capable of recognizing the presence of both bacterial peptidoglycan and viral ssRNA and the biochemical mechanism that links MAVS to IRF3 activation during NOD2 signaling require further analysis.
Another novel role for NOD2 during virus infection was recently implicated when NOD2 was shown to interact with 2′-5′-oligoadenylate synthetase type 2 (OAS2) 78. This interaction was shown to positively regulate the enzymatic activity of OAS2 in response to poly I:C, which resulted in increased degradation of RNAs by RNAse L. OAS2 is required for the activation of RNAse L which degrades viral and cellular RNA and inhibits virus replication 79. At the same time, RNAse L provides a feedback mechanism for type I IFN production by generating new PAMPS for RIG-I and MDA5 80. These reports further implicate NOD2 in the efficient induction of type I IFNs in response to viral infection.
NLRX1
NLRX1 is the only NLR known so far to localize to mitochondria. Indeed, this NLR family member contains a mitochondrial targeting sequence in its N-terminus 8, 9, 81. Overexpression of NLRX1 resulted in inhibition of RIG-I signalling induced by synthetic RIG-I ligands and viruses such as Sendai virus or Sindbis virus. Depletion of NLRX1 using specific siRNA amplified virus-induced type I IFN production and decreased viral replication. However, the precise subcellular localization of NLRX1 in mitochondria remains to be determined. In one study, NLRX1 was found in close proximity to the antiviral RLR adaptor protein MAVS, which is known to be associated with the mitochondrial outer membrane. Physical association between MAVS and NLRX1 prevented viral RNA-induced type I IFN responses and NF-κB activation, suggesting that NLRX1 is a negative regulator of RLR signalling 9. A second study contested the localization of NLRX1 to the outer mitochondrial membrane and showed that NLRX1 fully translocates to the mitochondrial matrix 8. Accordingly, NLRX1 was found to amplify NF-κB and JNK signalling by augmenting ROS production in mitochondria in response to poly I:C treatment 81. Although the mechanism of NLRX1 function is unclear and its role in antiviral signaling is under debate, the mitochondrial localization of NLRX1 seems to be essential for regulating its activity. Further studies will be required to resolve the precise localization of NLRX1 in mitochondria, and whether NLRX1 exerts a pro- or anti-inflammatory effect or if the role of NLRX1 is context dependant. The generation of NLRX1-deficient mice may prove instrumental in determining the physiological role of this unique NLR family member.
NLRC5
Similar to NLRX1, NLRC5 is an atypical NLR family member in that it contains a C-terminal effector domain that is predicted to adopt a death-domain (DD) fold without obvious homology to the CARD and PYD motifs found in most NLRs 82. Furthermore, its LRR domain is the longest among NLRs and comprises over 1000 residues. It is thought to organize into a helical structure rather than the ‘horseshoe’ fold that is typical for most LRR domains 83. One study recently reported that the protein is primordially expressed in hematopoietic tissues and cells of the myeloid and lymphoid lineages 83, whereas another study suggested a ubiquitous expression pattern with lowest expression levels found in lymphoid tissue and leukocytes 84.
NLRC5 was recently shown to be specifically upregulated by IFNγ and was implicated in modulating IFN-dependent gene transcription 83–86. This suggests an important role in antiviral immune responses, although the precise molecular mechanism remains enigmatic. Knockdown of endogenous NLRC5 in THP-1 cells and in primary human dermal fibroblasts impaired type I IFN responses as shown by decreased IFN-β and CCL5 release, and decreased type I IFN-β and IP-10 mRNA induction, after Sendai virus infection and poly(I:C) stimulation 83. In agreement, NLRC5 knockdown diminished the induction of IFN-α in human foreskin fibroblast cells stimulated with poly(I:C) or infected with CMV 84. In contrast to these two reports, it was recently suggested that NLRC5 is a negative regulator of NF-κB and type I IFN signalling during viral infection 87. These results were comparable to another recent study which showed that siRNA knock down of NLRP5 in RAW264.7 mouse macrophage cell line resulted in increased NF-κB and IFN signaling and IL-6, IL-1β, and TNF-α production 85. Further, an inhibitory role for NLRC5 in MHC I and CD40 expression was also demonstrated. However, another study outlined NLRC5 as a positive regulator of MHC I expression along with other genes involved in antigen presentation 86. Although above studies are contradictory, the ability of IFN-γ in upregulating NLRC5 expression is consistent. Thus, the generation of NLRC5-deficient mice will undoubtedly contribute to resolving these apparent discrepancies and to characterizing the physiological role of this NLR in antiviral immune signalling.
NLRs and inflammasomes recognize specific nonself-motifs in viral products, the viral replication program (a cytosolic phase of genome amplification and/or mRNA metabolism), viral protein expression or the ion-channel activity of viruses. After virus infection, multiple signalling pathways are activated simultaneously or sequentially to eliminate the invading pathogen and to prevent the establishment of a persistent infection. This activation of distinct and shared signalling pathways leads to pathway crosstalk as described above and eventually determines the specific immune response directed at clearing the virus. As the host has evolved factors and strategies for detecting and responding to viral infection, viruses have developed countermeasures to inhibit these processes. Owing to genomic constraints, many pathogenic viruses focus their disruptive efforts on host targets that are key players in the antiviral response. Thus, common host molecules that connect virus recognition to downstream effectors and inflammatory cytokine expression are usually targeted by viral immune evasion proteins. An overview of the mechanisms by which viruses block NLRs and inflammasomes is provided below.
Virus immune evasion of NLRs and inflammasomes
In the evolutionary arms race between a pathogen and their host, the host has selected for various mechanisms to detect the presence of viruses and closely regulates the production of master cytokines such as IL-1β and IL-18 by the inflammasome. In response, viruses have undergone selective pressure to evade the inflammasome. Several viruses are known to encode proteins that interfere with inflammasome signalling (Table 2). Inflammasome signalling is interrupted at the level of the adaptor protein ASC or caspase-1 itself, as both these molecules are downstream of multiple NLRs, as well as AIM2 and RIG-1 inflammasomes.
Table 2.
Virus encoded inhibitors and the inflammasome targets
| Inhibitor classes | Virus | gene | Cellular target | References |
|---|---|---|---|---|
| PYD homologues | Myxoma virus Shope Fibroma virus |
M13L-PYD S013L |
Inhibits inflammasome formation by binding ASC; inhibits subsequent caspase-1, IL-1β and IL-18 activation | 89–91 |
| SPI-2 homologues | Cowpox virus Rabbit pox virus Myxoma virus Vaccinia virus |
CrmA CrmA Serp2 SPI-2 |
Protease inhibitor that directly blocks caspase- 1, IL-1β and IL-18 activation | 92–99 |
| NS1 | Influenza A virus | NS1 | Inhibits caspase-1 and IL-1β activation; also inhibits RIG-I | 100–102 |
The large genomes of poxviruses encode multiple inhibitors that interfere with innate and adaptive immunity 88. Bioinformatics studies show that many poxviruses encode a putative PYD-containing protein that is hypothesized to interact with ASC; however, the myxoma virus M13L-PYD and Shope Fibroma virus S013L proteins are the only examples that have been studied in detail 89–91. M13L-PYD is required for the pathogenesis of myxoma virus and deletion of this protein results in severe virus attenuation in vivo, as evidenced by a decrease in viremia due to inefficient replication in lymphocytes and leukocytes and increased inflammation at the initial site of infection, which lead to more rapid resolution of disease. Infection of cells in vitro with myxoma virus lacking M13L-PYD leads to increased activation of caspase-1 and thereby increased levels of IL-1β and IL-18, which indicates that M13L-PYD has an immune-suppressive activity 91. M13L-PYD and S013L associate with ASC through direct PYD–PYD interactions. Expression of either M13L-PYD or S013L alone inhibits caspase-1 activation and IL-1β maturation downstream of the NLRP3 inflammasome in cell culture 90, 91. Poxvirus PYD-containing proteins may therefore inhibit inflammasome activation at the level of the adaptor protein ASC and prevent PRRs from activating caspase-1 in virus-infected cells. In addition, ASC might have several caspase-1-independent immune functions, such as T cell and lymph node cell activation. It is therefore possible that poxvirus PYD-containing proteins block inflammation by inhibiting these processes as well.
Poxviruses also encode various serpin-like protease inhibitors that inhibit caspase-1. For example, CrmA (also known as SPI-2) of cowpox virus is a well-known caspase-1 inhibitor. Deletion of CrmA decreases the number and severity of pox lesions on the allantoic membrane when the virus is grown in chicken eggs 92, 93. Infecting the respiratory tract of mice with cowpox virus or rabbit poxvirus mutants that lack CrmA results in decreased inflammation and viral loads compared with wild-type virus 94, 95. After intradermal inoculation with cowpox virus, a more rapid virus clearance and more robust inflammatory response is observed with virus lacking CrmA 96. It is interesting to note that no differences in IL-1β levels were observed in the lungs of C57BL/6 mice infected with wild-type cowpox virus versus mutant virus lacking CrmA 96 even though the studies in vitro indicate that CrmA can inhibit IL-1β maturation and secretion.
Other poxviruses also encode SPI-2 homologues, such as Serp2 in myxoma virus 97. Deletion of myxoma virus Serp2 results in severe attenuation of the virus in rabbits 98. Some vaccinia viruses also encode a SPI-2 protein. However, deletion of vaccinia virus SPI-2 had no effect on IL-1β-induced fever responses seen in infected mice and vaccinia virus SPI-2 mutants were not attenuated 94, 95. Instead, fever reduction and attenuated weight loss in vaccinia virus-infected mice were dependent on vaccinia virus-encoded IL-1β scavenger receptor (vIL-1βR) 94. Several poxviruses also encode an IL-18-binding protein that blocks IL-18-induced signalling 99. It is becoming clear that poxviruses evolved a variety of inflammasome inhibitors and molecules interfering with downstream signalling of the inflammasome products IL-1β and IL-18. However, viral inflammasome inhibitors are not limited to poxviruses. Influenza A/PR/8/34 H1N1 virus NS1 (PR/8 NS1) protein prevents caspase-1 activation and IL-1β maturation 100. The N-terminus of PR/8 NS1 was also suggested to interfere with inflammasome activation, although the details remain unclear. Influenza A/PR/8/34 H1N1 virus lacking the N-terminus of NS1 is attenuated in cell culture and induces higher levels of IL-1β and cell death. Interestingly, inhibition of the dsRNA kinase PKR also decreases IL-1β maturation in PR/8 NS1-mutant influenza virus. Although these results do not provide a clear inhibitory mechanism for PR/8 NS1, they do show the importance of this protein for virus replication. However, the ability of NS1 to block caspase-1 activation seems to be strain specific as NS1 from highly pathogenic H5N1 bird flu activates caspases and induces apoptosis 101. One possible explanation for the ability of H5N1 virus to grow despite activating caspases is that it downregulates the NLRP3 and IL-1β expression levels by 24h post infection 102 and therefore H5N1 virus may not require inhibition of caspase-1 to suppress inflammation. By contrast, PR/8 influenza virus has been shown to upregulate inflammasome components 11 and may therefore require NS1-mediated inhibition of caspase-1 to prevent inflammation and to replicate successfully.
It is likely that additional viral evasion pathways exist that target inflammasome components. Therapeutic interventions that are designed to target the virus–host interface of such immune evasion strategies to restore the functions of PRRs and their signalling pathways will offer the potential to restore innate immune induction and inflammatory programs that initiate immunity for the control of virus infection.
Concluding remarks
Host proteins involved in the recognition of virus infection have only recently been identified and the initial characterization of their roles in this intricate system is just now starting to be understood. Identification of the NLR and RLR family of sensors within the past several years has provided critically important and necessary information concerning cellular recognition of RNA viruses. It is evident from the information presented here that NLRs and inflammasomes are necessary for the innate inflammatory control of virus infection, as well as healing responses. In addition, the presence of multiple viruses that encode inflammasome inhibitory proteins is consistent with the importance of this pathway for inhibiting virus replication. However, our knowledge of inflammasome biology is still in its infancy and many questions remain to be answered. Specifically, how are these pathways regulated? Is there crosstalk with other PRRs recognizing the same pathogen? What is the physiological significance of each of these systems and which viral triggers are responsible for their activation? As research continues into the role of NLRs and inflammasomes in antiviral immunity it is likely that the list of PRRs involved and viruses that activate and suppress NLR and inflammasome signalling will continue to grow.
Online Summary.
Inflammasomes and nucleotide oligomerization and binding domain (NOD)-like receptors (NLRs) mediate the recognition of viruses by detecting evolutionary conserved motifs in their genomic RNA or DNA. In addition to viral RNA, NLRP3 senses a myriad of pathogen-associated molecular patterns (PAMPs) through incompletely characterized mechanisms, such as ion flux caused by viral ion channels or multiple coordinated signals.
Inflammasomes have a crucial role in the immune response to viral infections. NLR proteins were previously thought to be the only mediators of inflammasome activation. However, recent research has shown that members of additional cytoplasmic pattern recognition receptor (PRR) families such as retinoic acid inducible gene_I (RIG-I) and absent in melanoma 2 (AIM2) can engage the inflammasome in virus-infected cells.
NOD2, which was originally reported to activate mitogen-activated protein kinase and nuclear factor-κB signalling through the kinase receptor interacting serine-threonine protein kinase 2 (RIPK2) in response to bacterial peptidoglycan, has been shown to also signal through mitochondrial antiviral signalling protein (MAVS) and induce the production of type I interferons (IFNs) in response to viral ssRNA.
NLR proteins also have regulatory functions during virus infection. NLRX1 negatively regulates type I IFN production mediate by the RIG-I–MAVS–IFN response factor 3 (IRF3) signalling cascade.
Crosstalk between innate immune signalling pathways has become increasingly more evident. RIG-I was originally discovered as a mediator of type I IFN production but is now recognized as a potential inflammasome activator.
Several viruses encode inhibitors of inflammasome and MAVS signalling pathways, thus suggesting a key role for these pathways in controlling virus replication and the requirement for viruses to interfere with their activation in order to successfully infect a host.
Acknowledgments
I acknowledge the very large number of researchers who have contributed to this field whose work was not cited or was cited through others’ review articles because of space limitations. This work is supported by National Institute of Health Grants AR056296 and AI088177, a NIAMS Centers of Excellence for Influenza Research and Surveillance (CEIRS) grant and the American Lebanese Syrian Associated Charities (ALSAC).
Glossary
- Pattern-recognition receptors (PRRs)
Receptors that bind to molecular patterns found in pathogens, but not mammalian cells. Examples include the mannose receptor, which binds to terminally mannosylated and polymannosylated compounds, and Toll-like receptors, which are activated by various microbial products, such as bacterial lipopolysaccharides, hypomethylated DNA, flagellin and double-stranded RNA
- Pathogen-associated molecular patterns (PAMPs)
A molecular pattern that is found in pathogens but not mammalian cells. Examples include terminally mannosylated and polymannosylated compounds, which bind the mannose receptor, and various microbial products, such as bacterial lipopolysaccharides, hypomethylated DNA, flagellin and double-stranded RNA, which bind Toll-like receptors
- NOD-like receptors (NLRs)
A large family of cytosolic PRRs that resembles a subset of plant disease-resistance (R) genes, which are involved in the hypersensitive response against virulent plant pathogens. There are 23 NLR genes in the human genome and at least 34 in the mouse genome. NLRs are involved in innate immune sensing and in the regulation of inflammatory and cell death responses
- Nucleotide-binding oligomerization-domain (NOD)
Members of the NLR family and plant disease-resistance (R) gene products contain a central domain known as a NOD
- Hypersensitive response
A form of programmed cell death. The hypersensitive response is a marker of disease resistance in plants and is characterized by rapid localized pathogen-induced cell death and restriction of further pathogen growth
- Autophagy
An evolutionarily conserved process in which acidic double-membrane vacuoles sequester intracellular contents (such as damaged organelles and macromolecules) and target them for degradation, through fusion to secondary lysosomes
- Inflammasome
A large multiprotein complex containing certain NLR, RLR and IFI200 proteins, the adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) and pro-caspase 1. The assembly of the inflammasome leads to the activation of caspase 1, which cleaves pro-IL-1β and pro-IL-18 to generate the active pro-inflammatory cytokines
- Caspases
Caspases are evolutionarily conserved cysteine proteases that are synthesized as inactive zymogens. Upon proteolytic activation, they initiate or execute cellular programs, leading to inflammation or cell death. Caspases are categorized as either proinflammatory or proapoptotic, depending on their participation in these cellular programs. The inflammatory caspases are caspase-1, caspase-11 and caspase-12 in mouse and caspase-1, caspase-4, and caspase-5 in human
- Danger signal
Signal released by injured or damaged tissues that trigger an innate immune response
- Pyroptosis
A form of cell death associated with antimicrobial responses during inflammation and dependent on the activation of an inflammasome and inflammatory caspases such as caspase-1
- Small interfering RNA
Synthetic double-stranded RNA molecules of 19–23 nucleotides, which are used to ‘knockdown’ (silence the expression of) a specific gene. This is known as RNA interference and is mediated by the sequence-specific degradation of mRNA
- K63-linked polyubiquitylation
Ubiquitin is a highly conserved 76kDa protein that is ligated to many proteins in both monomeric and polymeric forms. The two most common ubiquitin polymer chains are linked through an isopeptide bond between glycine-76 and either lysine-48 (K-48) or lysine-63 (K-63). K-63-linked polyubiquitination results in signals related to intracellular trafficking, cell signalling, ribosomal biogenesis and DNA damage repair. There are also reports that K63-linked polyubiquitin is involved in proteasome-independent proteolysis through autophagy
Footnotes
Competing interests statement
The author declares no competing financial interests.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–7. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–93. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 4.Inohara N, Nunez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20:6473–81. doi: 10.1038/sj.onc.1204787. References 3 and 4 report the discovery of the NLR family. [DOI] [PubMed] [Google Scholar]
- 5.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–33. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 6.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 7.Sabbah A, et al. Activation of innate immune antiviral responses by Nod2. Nature Immunol. 2009;10:1073–80. doi: 10.1038/ni.1782. This paper describe the role of NOD2 in antiviral responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnoult D, et al. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122:3161–8. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–7. doi: 10.1038/nature06501. References 8 and 9 show that NLRX1 is targeted to mitochondria. [DOI] [PubMed] [Google Scholar]
- 10.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–26. doi: 10.1016/s1097-2765(02)00599-3. This paper describes the inflammasome. [DOI] [PubMed] [Google Scholar]
- 11.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–65. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–8. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 14.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–6. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 15.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–75. doi: 10.1016/j.immuni.2009.02.006. References 11–15 describe activation of the NLRP3 inflammasome by RNA and viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poeck H, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nature Immunol. 2009;11:63–9. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 17.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 18.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. References 17–19 describe AIM2 as a cytosolic DNA sensor and its role in the activation of caspase-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–147. [PubMed] [Google Scholar]
- 21.Dinarello CA. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann N Y Acad Sci. 1998;856:1–11. doi: 10.1111/j.1749-6632.1998.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 22.Ghayur T, et al. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–23. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 23.Okamura H, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz J, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Liu B, et al. Interleukin-18 improves the early defence system against influenza virus infection by augmenting natural killer cell-mediated cytotoxicity. J Gen Virol. 2004;85:423–428. doi: 10.1099/vir.0.19596-0. [DOI] [PubMed] [Google Scholar]
- 26.Denton AE, Doherty PC, Turner SJ, La Gruta NL. IL-18, but not IL-12, is required for optimal cytokine production by influenza virus-specific CD8+ T cells. Eur J Immunol. 2007;37:368–375. doi: 10.1002/eji.200636766. [DOI] [PubMed] [Google Scholar]
- 27.Sergerie Y, Rivest S, Boivin G. Tumor necrosis factor-alpha and interleukin-1 beta play a critical role in the resistance against lethal herpes simplex virus encephalitis. J Infect Dis. 2007;196:853–860. doi: 10.1086/520094. [DOI] [PubMed] [Google Scholar]
- 28.Krzyzowska M, Cymerys J, Winnicka A, Niemialtowski M. Involvement of Fas and FasL in Ectromelia virus-induced apoptosis in mouse brain. Virus Res. 2006;115:141–149. doi: 10.1016/j.virusres.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Byrne SN, Halliday GM, Johnston LJ, King NJ. Interleukin-1beta but not tumor necrosis factor is involved in West Nile virus-induced Langerhans cell migration from the skin in C57BL/6 mice. J Invest Dermatol. 2001;117:702–709. doi: 10.1046/j.0022-202x.2001.01454.x. [DOI] [PubMed] [Google Scholar]
- 30.Manigold T, et al. Hepatitis B core antigen is a potent inductor of interleukin-18 in peripheral blood mononuclear cells of healthy controls and patients with hepatitis B infection. J Med Virol. 2003;71:31–40. doi: 10.1002/jmv.10445. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi R, et al. Respiratory syncytial virus infection of human alveolar epithelial cells enhances interferon regulatory factor 1 and interleukin-1beta-converting enzyme gene expression but does not cause apoptosis. J Virol. 1998;72:4498–4502. doi: 10.1128/jvi.72.5.4498-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanneganti TD, et al. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006 doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 33.Delaloye J, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Geiser T, Jarreau PH, Atabai K, Matthay MA. Interleukin-1beta augments in vitro alveolar epithelial repair. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1184–1190. doi: 10.1152/ajplung.2000.279.6.L1184. [DOI] [PubMed] [Google Scholar]
- 35.Perkins GD, Gao F, Thickett DR. In vivo and in vitro effects of salbutamol on alveolar epithelial repair in acute lung injury. Thorax. 2008;63:215–220. doi: 10.1136/thx.2007.080382. [DOI] [PubMed] [Google Scholar]
- 36.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nature Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 38.Dostert C, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nature Rev Immunol. 10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 40.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nature Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 42.Horwood NJ, et al. Interleukin 18 inhibits osteoclast formation via T cell production of granulocyte macrophage colony-stimulating factor. J Clin Invest. 1998;101:595–603. doi: 10.1172/JCI1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz N, Kurrer M, Bachmann MF, Kopf M. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J Virol. 2005;79:6441–6448. doi: 10.1128/JVI.79.10.6441-6448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imamura M, et al. Contribution of TIR domain-containing adapter inducing IFN-beta-mediated IL-18 release to LPS-induced liver injury in mice. J Hepatol. 2009;51:333–341. doi: 10.1016/j.jhep.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 45.Maitra R, et al. Endosomal damage and TLR2 mediated inflammasome activation by alkane particles in the generation of aseptic osteolysis. Mol Immunol. 2009;47:175–184. doi: 10.1016/j.molimm.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nature Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 47.Guillot L, et al. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 48.Heer AK, et al. TLR signaling fine-tunes anti-influenza B cell responses without regulating effector T cell responses. J Immunol. 2007;178:2182–2191. doi: 10.4049/jimmunol.178.4.2182. [DOI] [PubMed] [Google Scholar]
- 49.Koyama S, et al. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179:4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 50.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 51.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 53.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, et al. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fredericksen BL, Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melchjorsen J, et al. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol. 2005;79:12944–12951. doi: 10.1128/JVI.79.20.12944-12951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mikkelsen SS, et al. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1. J Biol Chem. 2009;284:10774–10782. doi: 10.1074/jbc.M807272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 59.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 60.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. This paper describes ss viral genomes bearing 5′ triphosphate ssRNA as the ligands for RIG-1. [DOI] [PubMed] [Google Scholar]
- 61.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rintahaka J, Wiik D, Kovanen PE, Alenius H, Matikainen S. Cytosolic antiviral RNA recognition pathway activates caspases 1 and 3. J Immunol. 2008;180:1749–1757. doi: 10.4049/jimmunol.180.3.1749. [DOI] [PubMed] [Google Scholar]
- 65.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 68.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. References 65–68 describe AIM2 deficient mice and confirm the role of AIM2 in vivo in cytosolic DNA sensing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taniguchi T, Fujii-Kuriyama Y, Muramatsu M. Molecular cloning of human interferon cDNA. Proc Natl Acad Sci U S A. 1980;77:4003–4006. doi: 10.1073/pnas.77.7.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 71.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 73.Windheim M, Lang C, Peggie M, Plater LA, Cohen P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J. 2007;404:179–190. doi: 10.1042/BJ20061704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Girardin SE, et al. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park JH, et al. RICK/RIP2 Mediates Innate Immune Responses Induced through Nod1 and Nod2 but Not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 76.Hsu YM, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 77.Pandey AK, et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dugan JW, et al. Nucleotide oligomerization domain-2 interacts with 2′-5′-oligoadenylate synthetase type 2 and enhances RNase-L function in THP-1 cells. Mol Immunol. 2009;47:560–566. doi: 10.1016/j.molimm.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hovanessian AG. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2′-5′oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev. 2007;18:351–361. doi: 10.1016/j.cytogfr.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 80.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tattoli I, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 83.Neerincx A, et al. A role for the human NLR family member NLRC5 in antiviral responses. J Biol Chem. 2010 Jun 10; doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuenzel S, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 85.Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- 86.Meissner TB, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cui J, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 141:483–496. doi: 10.1016/j.cell.2010.03.040. References 83–87 report the role of NLRC5 in antiviral responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seet BT, et al. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 89.Benedict CA, Ware CF. Poxviruses aren’t stuPYD. Immunity. 2005;23:553–555. doi: 10.1016/j.immuni.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 90.Dorfleutner A, et al. A Shope Fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007;35:685–694. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnston JB, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 92.Komiyama T, et al. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 93.Ray CA, et al. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 94.Kettle S, et al. Vaccinia virus serpin B13R (SPI-2) inhibits interleukin-1beta-converting enzyme and protects virus-infected cells from TNF- and Fas-mediated apoptosis, but does not prevent IL-1beta-induced fever. J Gen Virol. 1997;78 (Pt 3):677–685. doi: 10.1099/0022-1317-78-3-677. [DOI] [PubMed] [Google Scholar]
- 95.Kettle S, Blake NW, Law KM, Smith GL. Vaccinia virus serpins B13R (SPI-2) and B22R (SPI-1) encode M(r) 38.5 and 40K, intracellular polypeptides that do not affect virus virulence in a murine intranasal model. Virology. 1995;206:136–147. doi: 10.1016/s0042-6822(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 96.MacNeill AL, Moldawer LL, Moyer RW. The role of the cowpox virus crmA gene during intratracheal and intradermal infection of C57BL/6 mice. Virology. 2009;384:151–160. doi: 10.1016/j.virol.2008.10.041. [DOI] [PubMed] [Google Scholar]
- 97.Petit F, et al. Characterization of a myxoma virus-encoded serpin-like protein with activity against interleukin-1 beta-converting enzyme. J Virol. 1996;70:5860–5866. doi: 10.1128/jvi.70.9.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Messud-Petit F, et al. Serp2, an inhibitor of the interleukin-1beta-converting enzyme, is critical in the pathobiology of myxoma virus. J Virol. 1998;72:7830–7839. doi: 10.1128/jvi.72.10.7830-7839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith VP, Bryant NA, Alcami A. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. J Gen Virol. 2000;81:1223–1230. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- 100.Stasakova J, et al. Influenza A mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1beta and 18. J Gen Virol. 2005;86:185–195. doi: 10.1099/vir.0.80422-0. [DOI] [PubMed] [Google Scholar]
- 101.Lam WY, et al. Avian influenza virus A/HK/483/97(H5N1) NS1 protein induces apoptosis in human airway epithelial cells. J Virol. 2008;82:2741–2751. doi: 10.1128/JVI.01712-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cilloniz C, et al. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 2009;5:e1000604. doi: 10.1371/journal.ppat.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]