Abstract
Several genome-wide association studies (GWAS) have identified a genetic polymorphism associated with the gene locus for interleukin 28B (IL28B), a type III interferon (IFN), as a major predictor of clinical outcome in hepatitis C. Antiviral effects of the type III IFN family have previously been shown against several viruses, including hepatitis C virus (HCV), and resemble the function of type I IFN including utilization of the intracellular JAK-STAT pathway. Effects unique to IL28B that would distinguish it from IFN-α are not well defined. By analyzing the transcriptomes of primary human hepatocytes (PHH) treated with IFN-α or IL28B, we sought to identify functional differences between IFN-α and IL28B to better understand the roles of these cytokines in the innate immune response. Although our data did not reveal distinct gene signatures, we detected striking kinetic differences between IFN-α and IL28B stimulation for interferon stimulated genes (ISGs). While gene induction was rapid and peaked at 8 h of stimulation with IFN-α in PHH, IL28B produced a slower, but more sustained increase in gene expression. We confirmed these findings in the human hepatoma cell line Huh7.5.1. Interestingly, in HCV infected cells, the rapid response after stimulation with IFN-α was blunted, and the induction pattern resembled that caused by IL28B. In conclusion, we describe the kinetics of gene induction as being fundamentally different for stimulations with either IFN-α or IL28B in hepatocytes suggesting distinct roles of these cytokines within the immune response. Furthermore, we demonstrate that the observed differences are substantially altered by infection with the hepatitis C virus.
Keywords: HCV, IFN-λ, lambda, hepatocyte, microarray
INTRODUCTION
In 2003, a novel family of antiviral cytokines was described and designated as type III interferons (1,2). Since their discovery, the three members comprising this family, interleukin (IL) 28A, also known as interferon (IFN) λ2, IL28B (IFN-λ3) and IL29 (IFN-λ1) have been extensively studied and have in many respects been found to resemble type I interferons. While type III interferons bind to a receptor complex that is distinct from the interferon-α receptor, type I and III interferons have been shown to share a common downstream signaling pathway via JAK-STAT, leading to the induction of interferon stimulated genes (ISGs), like OAS1 (3).
In contrast to the type I interferon receptor that is expressed ubiquitously on mammalian cells, the functional type III interferon receptor appears to be more tissue restricted with preferential expression on the surfaces of epithelial cells, including hepatocytes (4).
Chronic hepatitis C is estimated to affect 2 – 3% of the world’s population and is currently the leading cause of both hepatocellular carcinoma and liver transplantations in both the US and Europe (5,6). Peginterferon based therapies have been the mainstay of hepatitis C therapy for nearly a decade. However, they are fraught with an unfavorable side effect profile, long duration of treatment, a limited success rate and high costs (7). Furthermore, many patients have contraindications to treatment with peginterferon. The parameters that determine therapeutic success or failure in patients treated for hepatitis C are still incompletely understood.
A recent breakthrough in the field was the discovery that genetic variation in the proximity of the IL28B (otherwise known as IFN-λ3) gene is a major predictor of successful therapy with peginterferon and ribavirin (8–10). In addition, the detected IL28B associated single nucleotide polymorphisms (SNPs) have been shown to significantly determine spontaneous viral clearance (11, 13). Coupled with the extremely strong correlation between IL28B genotype, the previously known antiviral effect of the IL28B protein makes an important role of the gene product in the course of hepatitis C highly likely. However, the mechanism by which the genetic polymorphism impacts the outcome of viral infection is unknown; thus further studies to elucidate the mechanism are needed.
Recently, we have shown that IL28B inhibits HCV replication in a hepatoma cell line and that this action depends on an intact JAK-STAT pathway (3). Thus, IL28B’s action seems to resemble the effects of its close relatives IL28A and IL29 as well as IFN-α. While this study was important in order to establish that IL28B, like IL28A, IL29, and IFN is mainly signaling through the JAK-STAT pathway, it did not provide further insight regarding the unique contribution of IL28B in the response to HCV infection and therapy observed in the original GWA studies.
In an effort to identify functional differences between the effects of IL28B and IFN-α, we compared the transcriptomes of IL28B and IFN-α stimulated primary human hepatocytes at different doses and over different periods of time. We tested the hypothesis that there are distinct differences in the gene expression patterns of hepatocytes upon stimulation with IFN-α and IL28B, respectively, and that these differences could play a role in differential downstream antiviral action.
MATERIALS & METHODS
Cell cultures and viruses
The human hepatoma cell line Huh7.5.1 was grown at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (HyClone Laboratories, Inc., South Logan, UT). Viral RNA was made from plasmid Jc1FLAG2(p7-nsGluc2a), which represents an infectious recombinant HCV strain (based on Jc1), that harbors the Gaussia luciferase gene, and the RNA was transfected as previously described (14). Briefly, we used electroporation for the transfection of Jc1FLAG2(p7-nsGluc2a) RNA into Huh7.5.1 cells and collected the supernatants at day 3 post transfection. We then used these supernatants to inoculate naïve Huh7.5.1 cells with a tissue culture infectious dose 50 (TCID50) of 3.24 × 104/ml. TCID50 was determined using the method described by Lindenbach et al. with an antibody against HCV core protein (15). Infection was confirmed by quantitative polymerase chain reaction (qPCR) for viral RNA and Western blotting for HCV core protein, and infection with Jc1FLAG2(p7-nsGluc2a) by luciferase assay and qPCR for viral RNA. Primary human hepatocytes (PHH) were obtained after liver resections from three donors for colorectal metastasis from apparently healthy excess liver tissue portions that were not required for pathological diagnosis and therefore discarded if not used for research purposes. Liver resections were received from the University of Pittsburgh through the Liver Tissue and Procurement and Distribution System (LTPADS; Dr. Stephen C. Strom, Pittsburgh, PA) and have been isolated under the Institutional Review Board guidelines of the University of Pittsburgh as previously described (16). PHH were cultured in HMM (hepatocyte maintenance medium; Lonza, Walkersville, MD) supplemented with HMM Single Quots Kit, except that gentamycin was replaced by 100 U/ml penicillin and 100 μg/ml streptomycin (HyClone Laboratories, Inc., South Logan, UT). The hepatocytes in the microarray experiments were genotyped for an IL28B associated polymorphism at rs12979860 and we obtained the following results: C/C (1602), C/T (1638), T/T (1649).
Interferons
Peginterferon alfa-2b was obtained from Schering Corporation, Kenilworth, NJ. For the microarray studies in PHH, we used IL28B derived from an expression system as previously described (17). After the protein became commercially available, we used human IL28B from R&D Systems, Minneapolis, MN. We used 0.21 ng/ml peginterferon alfa-2b (equivalent to 6.91 × 10−6 nmol/ml or 15 IU/ml) and 1,000 ng/ml IL28B (4.97 × 10−2 nmol/ml) for the stimulation of PHH. Doses for the Huh7.5.1 cells were 0.43 ng/ml (1.38 × 10−5 nmol/ml or 30 IU/ml) IFN-α and 50 ng/ml (7.46 × 10−4 nmol/l) IL28B.
RNA extraction
For extraction of total RNA, cells were lysed and processed with the QIA shredder and RNeasy kits (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNA was reconstituted and stored in RNase free water. RNA concentrations were measured using the NanoDrop spectrophotometer (Thermo Scientific NanoDrop Products, Wilmington, DE). For the microarrays, RNA quality was additionally tested by the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA).
qRT-PCR
Total RNA was reverse transcribed to cDNA with random primers using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Invitrogen, Carlsbad, CA). Specific cDNA levels were quantified by real-time PCR using the Bio-Rad IQ5 machine (Bio-Rad Laboratories, Hercules, CA) and Thermo Scientific DyNAmo HS SYBR green qPCR kit (Thermo Fisher Scientific, Waltham, MA). GAPDH expression was used as an internal reference. Primers for the qPCR experiments are listed in Table 1.
Table 1.
Primer sequences for quantitative RT-PCR
| Target gene | Primer* | Nucleotide sequence (5′ to 3′) |
|---|---|---|
| GAPDH | F R |
ACCTTCCCCATGGTGTCTGA TCGCAACCCAACGCTACTCG |
| ACTB | F R |
CATGTACGTTGCTATCCAGG CTCCTTAATGTCACGCACGAT |
| MX1 | F R |
GTTTCCGAAGTGGACATCGCA GAAGGGCAACTCCTGACAGT |
| OAS1 | F R |
GATCTCAGAAATACCCCAGCCA AGCTACCTCGGAAGCACCTT |
| EIF2AK2 | F R |
TGGAAAGCGAACAAGGAGTAAG CCATCCCGTAGGTCTGTGAA |
| ISG15 | F R |
TCCTGGTGAGGAATAACAAGGG GTCAGCCAGAACAGGTCGTC |
| STAT1 | F R |
ATGTCTCAGTGGTACGAACTTCA TGTGCCAGGTACTGTCTGATT |
| IRF9 | F R |
GTGCTGGGATGATACAGCTAAG CAGGCGAGTCTTCCAGACAG |
| IFIT1 | F R |
TCAGGTCAAGGATAGTCTGGAG AGGTTGTGTATTCCCACACTGTA |
| IFI6 | F R |
GGTCTGCGATCCTGAATGGG TCACTATCGAGATACTTGTGGGT |
| RSAD2 | F R |
CAAGACCGGGGAGAATACCTG GCGAGAATGTCCAAATACTCACC |
| HERC5 | F R |
GATGGGCTGCTGTTTACTTTCG GAGTCACTCTATACCCAACAAGC |
| HERC6 | F R |
CCCTCAGTGGGCGTAATGTC AGAGCGATTGTCTCCAAATGTG |
| JFH-1 | F R |
CTGTCTTCACGCAGAAAGCG TCGCAACCCAACGCTACTCG |
F: forward, R: reverse
Microarrays
The Illumina BeadChip HT-12 was used for microarray analysis according to the manufacturer’s protocol (Illumina Inc., San Diego, CA) and to the methods described in Urban et al. (17). One single RNA sample was not available for further analysis and therefore, no microarray data are available (1602/IL28B/24 h).
Protein Sample Preparation and Western Blotting were performed as previously described (31). The following antibodies were used: STAT1 (Tyr701) rabbit monoclonal, and STAT1 mouse monoclonal from Cell Signaling Technology (Beverly, MA), β(actin (A2228) mouse monoclonal from Sigma-Aldrich (Saint Louis, MO), and the secondary antibodies anti-mouse IgG (NA931) and anti-rabbit IgG (N933, GE Healthcare).
Statistics
Data analysis was carried out using a 2-tailed Student’s t-test with pooled variance. Data are expressed as mean +− SD of at least three sample replicates, unless stated otherwise. Microarray data was first analyzed as in (17). For the identification of relevant interferon stimulated genes in the PHH microarrays, we used gene induction by IL28B at 4, 8, and 24 h with a q-value (false discovery rate) ≤ 0.05. The hits were further analysed by means of the database and web-tool STRING (Search Tool for the Retrieval of Interacting Genes/Proteins), a meta-resource that aggregates most of the available information on protein-protein associations, scores and weights it and augments it with predicted interactions, as well as with the results of automatic literature-mining searches as described in Jensen et al. (35). We fitted cubic splines to the expression data in figure 3 to demonstrate trends in expression kinetics.
Figure 3. IFN-α and IL28B induction of 50 genes in PHH over 24 h suggests general trends.
The 50 genes significantly induced by IL28B were analyzed and gene expression at 0, 4, 8, and 24 h depicted for both IFN-α (15 IU/ml) and IL28B (1μg/ml) induction..
RESULTS
The transcriptomes of both IFN-α and IL28B stimulated PHH comprise typical ISGs
In order to identify differential effects of IFN-α and IL28B on the hepatocyte, we sought to determine gene expression patterns using an unbiased approach by transcriptomic microarrays in the currently best model for the isolated study of this cell type, PHH cultures. Interestingly, the magnitude of induction was generally much lower for most interferon stimulated genes in PHH compared to hepatoma cells. We stimulated PHH from three different donors with either IFN-α or IL28B over a 24 h timecourse, isolated RNA and determined gene induction by expression microarrays (Fig. 1). We evaluated data from three different donors as described in methods. After sorting the genes by relevant induction by IL28B, we compared to the expression of the same genes after IFN-α stimulation. Most of the genes with relevant induction represented known ISGs, which are by definition inducible by IFN-α, after stimulation with both IFN-α and IL28B (Fig. 2A), including OAS1, IFIT3, ISG15, and RSAD2 (Viperin). Many of the significantly induced genes are known to be coexpressed (Fig. 2B). Based on this observation, and in line with previous studies, we concluded that at least a majority of genes substantially induced by IL28B in PHH are shared with those induced by IFN-α, and are therefore typical ISGs. Hence, from these experiments we are not able to identify a unique expression signature of IL28B, that comprises distinct genes from those induced by IFN-α. These findings strongly suggest that IL28B and IFN-α have near complete overlap of signal transduction.
Figure 1. Gene expression upon stimulation with IFN-α and IL28B in primary human hepatocytes (PHH).
PHH from three different donors (1602, 1638, 1649) were isolated from liver resections, plated and cultured as described in methods. The PHH cultures were treated with IFN-α (15 IU/ml) and IL28B (1,000 ng/ml) for 4, 8, and 24 h. RNA was isolated and, after quality control and reverse transcription to cDNA, gene expression was detected by microarrays. The heatmap shows statistically significant gene induction for IL28B stimulation (false discovery rate, q ≤ 0.05). Gene expression for the 50 hit genes by IL28B was compared to the respective data after IFN-α induction. Intensity of red correlates with intensity of gene expression compared to the row Z-score.
Figure 2. STRING protein interaction view of significantly induced genes by IFN-α and IL28B in PHH.
The 50 genes significantly induced by IL28B were analysed with the database and web-tool STRING (Search Tool for Retrieval of Interacting Genes/Proteins), a meta-resource that integrates available information on protein-protein associations (http://string-db.org; 33). (A) Connecting lines between the protein symbols illustrate the many known interactions of the proteins (protein association network), most of the genes are known to be IFN-α induced genes (ISGs). (B) Coexpression view. As illustrated in the figure, many of these proteins are known to be coexpressed under certain conditions. The color intensity reflects the association score (0–1).
IFN-α leads to a rapid but transient increase of gene expression in PHH whereas IL28B gene induction is slower and sustained
We next sought to determine the temporal nature of ISG induction by IFN-α and IL28B. When comparing the timecourses of induction, we found that many of the IFN-α responsive genes, like MX1, OAS1 and IFI6, were induced at 4 h, were further upregulated at 8 h and showed lower expression at 24 h compared to 8 h. While these genes were also found to be induced by IL28B, the kinetic profile was markedly different: The signal increased steadily over 24 h with the highest fold-induction value after the longest period of stimulation (Fig. 1, 3). The lack of evidence for specific IL28B induced genes and the observation that transcriptomes of IFN-α and IL28B largely overlap in PHH, made us consider alternative mechanisms as to how IFN-α and IL28B execute their distinct effects on hepatocytes. Indeed, we observed different kinetic profiles when analyzing induction of the same genes by IFN-α and IL28B. We therefore hypothesized that the two cytokines exert their differential effects on innate immunity temporally and therefore further investigated these differences. We analyzed a number of the ISGs induced on the microarrays, MX1 (Fig. 4A), OAS1 (Fig. 4B), RSAD2 (Viperin; Fig. 4C), STAT1 (Fig. 4D), and IRF9 (Fig. 4E), in qRT-PCR experiments from the same RNA extracts. While the magnitude of fold induction differed substantially between genes, the chosen doses led to graphs similar in range for IFN-α and IL28B. We were able to confirm induction of these ISGs by both IFN-α and IL28B on qPCR. As observed in the microarray experiments, we demonstrated that within 24 h for all but one sample there was maximum induction after 8 h of treatment with IFN-α whereas during the same period, IL28B led to a monotonic increase of expression. The one exception was a slight shift of the peak to 4h in the PHH from one of the donors in IRF9 induction (Fig. 4E).
Figure 4. Kinetic differences in induction of typical ISGs by IFN-α and IL28B in PHH determined by qRT-PCR.
IFN-α leads to a rapid but transient increase of gene expression in PHH whereas IL28B gene induction is slower and sustained. PHH were stimulated with IFN-α (0.21 ng/ml peginterferon alfa-2b, equivalent to 6.91 × 10−6 nmol/ml or 15 IU/ml) and IL28B (1,000 ng/ml) for 0, 4, 8, and 24 h, (A) MX1, (B) OAS1, (C) RSAD2, (D) STAT1, and (E) IRF9 expression were analyzed by qRT-PCR and normalized to GAPDH expression. The expression of the gene of interest at timepoint 0 was set to 1 and fold inductions determined for the other timepoints. The kinetics are demonstrated in two biological replicates (1638, 1649), largely confirming the trends as seen in the microarray experiments.
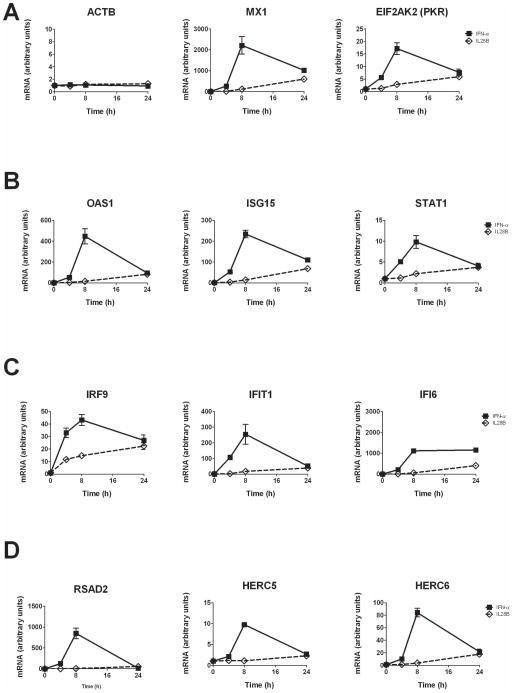
The typical patterns of ISG induction by IFN-α and IL28B over time are preserved in Huh7.5.1 cells
While regarded a model closely resembling the in vivo situation in many respects, PHH cultured in a monolayer have several limitations. They do not proliferate, rapidly lose cell function and are only viable for a limited number of days (16). Moreover, we found that when the hepatoma cell line Huh7.5.1 was used as a robust in vitro model for hepatocytes, the magnitude of ISG induction is much higher than in PHH and therefore permits detection of expression differences below a threshold value or below level of detection in PHH. Hence, we chose several of the top induced genes from the microarray experiments and further studied them in Huh7.5.1 cells (Fig. 5). After initial dose finding experiments, we submitted Huh7.5.1 cells to a timecourse over 48 h of stimulation with IFN-α and IL28B (data not shown and Fig. 5). We studied the expression patterns of the following genes in qPCR experiments: the housekeeping gene ACTB as a non-ISG control, MX1, EIF2AK2 (PKR; Fig. 5A); OAS1, ISG15, STAT1 (Fig. 5B); IRF9, IFIT1 (ISG56), IFI6 (Fig. 5C); RSAD2 (Viperin), HERC5, HERC6 (Fig. 5D). All ISGs, with one exception (IFI6) demonstrated a characteristic expression pattern over 48 h: IFN-α leads to a rapid increase of expression with a maximum after 8 h of incubation followed by a decline in the signal. In contrast, IL28B upregulates the same genes, showing a slower increase of induction that appears more sustained. On the other hand, we detected no changes in gene expression over time upon stimulation with either IFN-α or IL28B in expression of ACTB as a control gene.
Figure 5. Kinetic differences in the induction of ISGs by IFN-α and IL28B in hepatoma cells resembles characteristics seen in PHH and suggests the usefulness of Huh7.5.1 culture as a model system.
Human hepatoma cells, Huh7.5.1, were plated and cultured as described in methods. One day after seeding, the cells were left untreated or incubated with IFN-α (0.43 ng/ml, equal to 30 IU/ml) or IL28B (50 ng/ml) for 4, 8, and 24 h. RNA was extracted and analyzed by qRT-PCR. (A) The non-ISG ACTB (β-Actin) was used as a negative control. Time courses for the ISGs MX1, EIF2AK2 (PKR), (B) OAS1, ISG15, STAT1, (C) IRF9, IFIT1, IFI6, (D) RSAD2 (Viperin), HERC5, HERC6 are shown. Gene expression at timepoint 0 was set to 1 and further timepoints are measured as fold induction in relation to timepoint 0. Error bars represent SD. Experiments were done in quadruplicates (n=4).
Differences in induction kinetics by IFN-α and IL28B are altered by HCV infection
We next investigated whether these general and reproducible patterns were preserved upon IFN-α or IL28B stimulation in the setting of infection with HCV. After all, genotypic variation related to IL28B profoundly determines therapeutic response in chronic hepatitis C by a yet unidentified mechanism pointing towards an important role of the cytokine in the course of the disease. We treated both uninfected and HCV-infected Huh7.5.1 (after established infection over several days) cells in parallel with IFN-α or IL28B over 48 h (Fig. 6A–E and Supplemental Figure 1A,B). While the overall pattern of IL28B induced gene expression in HCV infected Huh7.5.1 cells was similar compared to uninfected cells with a slow increase of signal over time, surprisingly, the typical IFN-α response was not preserved in infection: the pattern with early maximum followed by decline of the induction signal observed in uninfected Huh7.5.1 was blunted in the HCV infected cells, and the course of gene expression over time became similar for IFN-α and IL28B treated cells, reminiscent of a type III response. We assessed the effect of the cytokines on viral replication in the same samples over time and detected comparable antiviral effects (Supplemental Figure 1C) and no clear pattern of increased or decreased expression for a selected number of representative ISGs (Supplemental Figure 1D)
Figure 6. The pattern of gene induction by IFN-α is profoundly changed after infection with HCV.
Viral RNA was made from plasmid Jc1FLAG2(p7-nsGluc2a), an infectious recombinant HCV strain that harbors the Gaussia luciferase gene, and the RNA was transfected into Huh7.5.1 cells and supernatants collected at day 3 post transfection. We then left Huh7.5.1 cells either untreated or used the described infectious supernatants to inoculate naïve Huh7.5.1 cells. After ongoing viral replication in the system for 5 d, we continued with the gene induction experiments in both uninfected and infected Huh7.5.1 cells. Induction kinetics for several ISGs after stimulation with IFN-α (0.43 ng/ml, equal to 30 IU/ml) or IL28B (50 ng/ml) for 0, 4, 8, and 24 h in Huh7.5.1 in the absence and presence of HCV infection are shown. For (A) MX1, (B) OAS1, (C) EIF2AK2 (PKR), (D) RSAD2 (Viperin), and (E) USP18, gene induction is shown in uninfected Huh7.5.1 and compared to the kinetics in cells that were previously infected with Jc1Gluc. Error bars represent SD. Experiments were done in quadruplicate.
Differential kinetics in gene expression are not explained by alteration in STAT1 phosphorylation
Next, we investigated whether the observed RNA levels of ISGs were reflected in STAT1 phosphorylation patterns under the different conditions: we therefore studied phosphorylated STAT1 (pSTAT1) and STAT1 expression over time by Western Blot (Suppl. Fig. 2). After optimization of relevant timepoints (time course up to 48 h, data not shown) we saw an increase in the pSTAT1 signal with a maximum within 6 h of stimulation followed by a decline. There were no apparent differences in the phosphorylation pattern observed in IFN-α or IL28B stimulated uninfected or infected cells. We conclude that STAT 1 phosphorylation patterns do not explain the distinct kinetic ISG mRNA patterns and that the basis for these distinct patterns potentially lie outside of the IFN signal transduction pathway.
DISCUSSION
In order to better understand the role and function of IL28B, we sought to characterize differences in gene induction by IFN-α and IL28B. While our comprehensive transcriptomic analysis in ex vivo cultured PHH did not primarily reveal distinct genes induced by IL28B, there were significant kinetic differences when looking at the expression profiles of the studied typical ISGs. For many of these genes, the response to IFN-α was a fast and sharp increase followed by a decline in expression within 24 h, whereas the IL28B response was slower but more sustained. This led us to confirm and further investigate these patterns in a more stable cell culture model that is permissive for robust HCV infection, human hepatoma cells. In infected cells, we obtained a similar temporal response to IL28B and IFN-α. Therefore, interestingly, the kinetic difference we have found previously was substantially altered after infection with HCV.
We compared IFN-α and IL28B induced genes in PHH over 24 h in expression microarrays. When we sorted the genes by relevance in the IL28B induced samples, we found known IFN-α stimulated genes. This concurs with the initial reports describing type III IFNs, in which the authors detected intracellular signal transduction via the same JAK-STAT pathway that transduces the type I IFN signal (1,2). Several publications analyze microarrays in PHH treated with IFN-α: In 2002 Radaeva et al. published gene expression in PHH detected on an Affymetrix chip comprising 12,000 known genes and found 44 upregulated > 2-fold, including MX1, the OAS family and PKR, and nine genes downregulated by greater than 50% (18). He et al. compared transcriptomes of PHH after stimulation with IFN-α and IFN-γ at 6 h and 18 h (19). There were more IFN-α upregulated genes at 6 h than at 18 h whereas the opposite was true for IFN-γ. In HCV replicon cells, Marcello et al. compared the effects of IFN-α and IL29 (IFN-λ1) and found several different patterns of gene regulation over 24 h (20). However, the replicon model is limited to intracellular stages of the viral life cycle and may not be directly comparable to our experiments. Another group found different kinetics of RSAD2 and IFI44 induction between IFN-α and IL28B stimulation in PHH, as well as overlapping and distinct genes when comparing transcriptomes (21). Based on our findings we speculate that genes previously described as being induced by solely one cytokine are ascribable to false negatives in the comparison samples since the magnitude of induction appears to be considerably lower in PHH compared to cell lines and thus some genes have fallen below the threshold of significance. Interestingly, Doyle et al. treated HepG2 cells with IL-29 or IFN-α for 1, 6, and 24 h (4). The gene expression profiles upon stimulation with both cytokines were very similar with induction of typical ISGs, like PKR and MX1 at 6 h and declining signals thereafter. Therefore, in contrast to what we describe for IL28B, the kinetics for IL29 induced genes may rather resemble IFN-α induction.
PHH are exquisitely sensitive to perturbations, lose cell functions and viability rapidly in culture and do not proliferate (16). Moreover, their use is limited by availability and quality of surgical specimens and the complex isolation procedure. It is important to note that due to the nature of hepatocyte extraction, there is a risk of contamination with other cell types in the PHH cultures. While on daily microscopic examination we were unable to detect any other cell types but hepatocytes in the cultures used for our studies, we cannot fully exclude that some of the effects that we see may be due to non-hepatocyte cells present in the cultures. Huh7.5.1 cells therefore appear as a valid model system to analyze kinetics. Based on our data from two lines of investigation, we conclude that IFN-α and IL28B exert their differential effects through temporal expression differences. We further hypothesize that these differences may play a critical role in the antiviral immune response and that IFN-α and IL28B may complement each other’s effects. Cell type dependent expression of IL28RA and production of type III IFNs may facilitate tissue-specific regulation of antiviral effects, therefore allowing for fine-tuned responses that are able to match the organ and cell specificity of viruses. There is accumulating evidence that the type III IFNs function as first line of antiviral defense in epithelial tissues like the gut, human liver, and lung, e.g. upon infection with influenza virus, whereas lack of local receptor expression, for instance in the CNS, makes use of the system in other tissues unlikely (22–26).
Viruses have developed sophisticated mechanisms to counteract effects of IFN (27). HCV is notorious for its ability to evade host antiviral effector functions: mitochondrial antiviral-signaling protein (MAVS, also known as IPS-1) for example, is inactivated by the viral protease NS3/4, likely to diminish IFN induction. Further, HCV inhibits JAK-STAT signaling through a variety of mechanisms (28–32).
In this study, we observed that HCV’s modulation of the type I ISG response to resemble a type III response does not appear to be mediated by classical IFN signaling. Rather, they suggest that HCV may be acting through other, nonclassical mechanisms to sustain ISG expression, including induction of alternate transduction pathways or posttranscriptional regulation of ISGs. Blunting a rapid IFN response may be beneficial to survival of the virus. Once infection has stabilized, viral products may set up an effective anti-inflammatory armamentarium that is able to suppress downstream effects of IFN-α. This is compatible with the clinical reality that signs of active inflammation early in the disease are predictive of viral clearance in hepatitis C (33,34). This response may be important in many viral infections to rapidly induce innate antiviral action and protect the host from systemic spread of viruses.
We selected differing doses of IFNα and IL28B for the PHH experiments because our pilot studies indicated that higher concentrations of IL28B were required to produce comparable levels of peak ISG induction. These data are supported by our findings in Figure 3. This apparent dose disparity may be attributable to the two cytokine’s differential affinity for their cognate receptors. We also performed experiments using much higher doses of IFN-α and lower doses of IL28B and found no difference in the induction kinetics of ISGs (not shown), suggesting that the two IFNs retain their fundamental induction patterns over a wide range of concentrations.
Mechanisms that could potentially explain outcome differences upon HCV infection in patients with different IL28B related genotypes include: alterations in IL28B expression, mRNA splicing, half-life, or cytokine-receptor affinity or specificity (13). The literature on IL28A and IL28B expression levels is controversial, with some studies detecting slightly higher expression levels of the cytokines in the clearance associated genotype, e.g. in peripheral blood mononuclear cells (PBMCs) (9,10), and others being unable to find any such correlations in PBMCs and liver tissue, respectively (8,17). An interesting recent contribution describes a novel gene that may be an additional member of the type III IFN family and contains a functional SNP related to both the IL28B genotype and outcome in hepatitis C(35): Depending on genotype, a protein is either made or not made. Strikingly, the favorable IL28B genotype as well as viral clearance are associated with non-production of the protein. The functional role of this novel gene, named IFNL4 (IFN-λ4), and the mechanism of action of its protein are unknown. Another recent study detected the same polymorphism, but found a correlation with IL28B expression (37).
In summary, we demonstrate functional differences of IFN-α and IL28B by showing distinct kinetic induction patterns for many typical ISGs in hepatocytes. Furthermore, we demonstrate that the kinetic patterns of gene induction are altered during HCV infection. Given their common downstream signaling but temporal differences in gene induction, we speculate that IFN-α and IL28B play complementary roles with a massive, early IFN-α response and slower but more sustained gene induction by IL28B..
Supplementary Material
Infection with HCV alters the gene induction pattern of IFN-α. Viral RNA was made from plasmid Jc1FLAG2(p7-nsGluc2a), an infectious recombinant HCV strain that harbors a luciferase gene, and the RNA was transfected into Huh7.5.1 cells and supernatants collected at day 3 post transfection. We then left Huh7.5.1 cells either untreated or used the described infectious supernatants to inoculate naïve Huh7.5.1 cells. After ongoing viral replication in the system for 5 d, we continued with the IFN induction experiments in both uninfected and infected Huh7.5.1 cells. Induction kinetics for several ISGs after stimulation with IFN-α (0.43 ng/ml, equal to 30 IU/ml) or IL28B (50 ng/ml) for 0, 4, 8, and 24 h in Huh7.5.1 in the absence and presence of HCV infection are shown. For (A) STAT1, and (B) IRF9, gene induction is shown in uninfected Huh7.5.1 cells and compared to the kinetics in cells that were previously infected with Jc1Gluc. (C) HCV replication under treatment with IFN-α and IL28B is inhibited. In general, gene expression at timepoint 0 was set to 1 and further timepoints measured as fold induction in relation to timepoint 0. (D) Gene expression in uninfected, untreated controls and infected, untreated samples is compared for MX1, OAS1, EIF2AK2, USP18, STAT1, and IRF9 in the absence of cytokine stimulation. The ratio of expression in infected samples to expression in those not infected is shown. Error bars represent SD. Experiments were done in quadruplicate.
In order to further investigate whether the differences in kinetics were also apparent in the IFN signaling pathway, Huh7.5.1 cells were incubated with IFN-α (0.43 ng/ml, equal to 30 IU/ml) or IL28B (50 ng/ml) for 0, 4, 8, and 24 h in Huh7.5.1 in the absence and presence of HCV. Protein was extracted from the samples and phosphorylated STAT1 (pSTAT1) as well as total STAT1 were detected by immunoblot, β-actin served as a control.
Acknowledgments
Financial support: This work was supported by the NIH, U19 grant AI082630 (RTC, GML), DA033541 (RTC), by the Deutsche Forschungsgemeinschaft (German Research Foundation, DFG) research fellowship Ji 145/1-1 (NJ), and by the NIH Liver Tissue Procurement and Distribution System.
The authors thank F. V. Chisari (The Scripps Research Institute, La Jolla, CA) for Huh7.5.1 cells, T. Wakita (National Institute for Infectious Diseases, Tokyo, Japan) and Charles M. Rice (Rockefeller University, New York, NY) for the infectious HCV strains JFH1 and Jc1Gluc. Normal human hepatocytes were obtained through the Liver Tissue Cell Distribution System (S. C. Strom University of Pittsburgh, Pittsburgh, PA) which was funded by NIH Contract #N01-DK-7-004/HHSN26l7200700004C. We thank David Goldstein (Duke University, NC) for facilitating the microarray experiments; T. F. Baumert (INSERM U748, Strasbourg, France) for helpful suggestions.
Abbreviations
- GWA(S)
genome-wide association (study/studies)
- IL
interleukin
- IL28B
interleukin-28B
- IFN
interferon
- HCV
hepatitis C virus
- JAK-STAT
Janus kinase – Signal Transducer and Activator of Transcription
- PHH
primary human hepatocytes
- ISG
Interferon Stimulated Gene
- Huh
Human hepatoma
- SNP
single nucleotide polymorphism
- mRNA
messenger ribonucleic acid
- qPCR
quantitative polymerase chain reaction
- RT-PCR
reverse transcription PCR
- SD
standard deviation
References
- 1.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003 Jan;4(1):63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 2.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003 Jan;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Jilg N, Shao RX, Lin W, Fusco DN, Zhao H, Goto K, et al. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol. 2011 Aug;55(2):289–98. doi: 10.1016/j.jhep.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006 Oct;44(4):896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 5.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007 May 7;13(17):2436–41. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011 Feb;17(2):107–15. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 7.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Jr, Häussinger D, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002 Sep 26;347(13):975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 8.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009 Sep 17;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 9.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009 Oct;41(10):1100–4. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009 Oct;41(10):1105–9. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 11.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009 Oct 8;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010 Apr;138(4):1338–45. 1345.e1–7. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 13.Afdhal NH, McHutchison JG, Zeuzem S, Mangia A, Pawlotsky JM, Murray JS, Shianna KV, et al. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology. 2011 Jan;53(1):336–45. doi: 10.1002/hep.24052. [DOI] [PubMed] [Google Scholar]
- 14.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008 Dec;48(6):1843–50. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005 Jul 22;309(5734):623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 16.Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- 17.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010 Dec;52(6):1888–96. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radaeva S, Jaruga B, Hong F, Kim WH, Fan S, Cai H, Strom S, et al. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology. 2002 Apr;122(4):1020–34. doi: 10.1053/gast.2002.32388. [DOI] [PubMed] [Google Scholar]
- 19.He XS, Nanda S, Ji X, Calderon-Rodriguez GM, Greenberg HB, Liang TJ. Differential transcriptional responses to interferon-alpha and interferon-gamma in primary human hepatocytes. J Interferon Cytokine Res. 2010 May;30(5):311–20. doi: 10.1089/jir.2009.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006 Dec;131(6):1887–98. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 21.Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012 Apr;142(4):978–88. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jewell NA, Cline T, Mertz SE, Smirnov SV, Flaño E, Schindler C, Grieves JL, et al. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol. 2010 Nov;84(21):11515–22. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008 Mar 14;4(3):e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pott J, Mahlakõiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7944–9. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010 Jun;84(11):5670–7. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008 Sep 12;4(9):e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang BX, Fish EN. The yin and yang of viruses and interferons. Trends Immunol. 2012 Apr;33(4):190–7. doi: 10.1016/j.it.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009 Feb;83(3):1299–311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, Holtzman MJ, et al. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol. 2006 Sep;80(18):9226–35. doi: 10.1128/JVI.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard JT, Schmidt EV, Chung RT. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005 Apr;128(4):1034–41. doi: 10.1053/j.gastro.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Kumthip K, Chusri P, Jilg N, Zhao L, Fusco DN, Zhao H, Goto K, et al. Hepatitis C virus NS5A disrupts STAT1 phosphorylation and suppresses type I interferon signaling. J Virol. 2012 Aug;86(16):8581–91. doi: 10.1128/JVI.00533-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999 Jul 2;285(5424):107–10. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 33.Gerlach JT, Diepolder HM, Zachoval R, Gruener NH, Jung MC, Ulsenheimer A, Schraut WW, et al. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology. 2003 Jul;125(1):80–8. doi: 10.1016/s0016-5085(03)00668-1. [DOI] [PubMed] [Google Scholar]
- 34.Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, et al. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology. 2012 Feb;55(2):384–94. doi: 10.1002/hep.24678. [DOI] [PubMed] [Google Scholar]
- 35.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T. STRING 8 - a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009 Jan;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013 Feb;45(2):164–71. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FH, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCVclearance prediction. J Exp Med. 2013 Jun;3(6):210. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Infection with HCV alters the gene induction pattern of IFN-α. Viral RNA was made from plasmid Jc1FLAG2(p7-nsGluc2a), an infectious recombinant HCV strain that harbors a luciferase gene, and the RNA was transfected into Huh7.5.1 cells and supernatants collected at day 3 post transfection. We then left Huh7.5.1 cells either untreated or used the described infectious supernatants to inoculate naïve Huh7.5.1 cells. After ongoing viral replication in the system for 5 d, we continued with the IFN induction experiments in both uninfected and infected Huh7.5.1 cells. Induction kinetics for several ISGs after stimulation with IFN-α (0.43 ng/ml, equal to 30 IU/ml) or IL28B (50 ng/ml) for 0, 4, 8, and 24 h in Huh7.5.1 in the absence and presence of HCV infection are shown. For (A) STAT1, and (B) IRF9, gene induction is shown in uninfected Huh7.5.1 cells and compared to the kinetics in cells that were previously infected with Jc1Gluc. (C) HCV replication under treatment with IFN-α and IL28B is inhibited. In general, gene expression at timepoint 0 was set to 1 and further timepoints measured as fold induction in relation to timepoint 0. (D) Gene expression in uninfected, untreated controls and infected, untreated samples is compared for MX1, OAS1, EIF2AK2, USP18, STAT1, and IRF9 in the absence of cytokine stimulation. The ratio of expression in infected samples to expression in those not infected is shown. Error bars represent SD. Experiments were done in quadruplicate.
In order to further investigate whether the differences in kinetics were also apparent in the IFN signaling pathway, Huh7.5.1 cells were incubated with IFN-α (0.43 ng/ml, equal to 30 IU/ml) or IL28B (50 ng/ml) for 0, 4, 8, and 24 h in Huh7.5.1 in the absence and presence of HCV. Protein was extracted from the samples and phosphorylated STAT1 (pSTAT1) as well as total STAT1 were detected by immunoblot, β-actin served as a control.