Abstract
Two-component signal transduction systems of microbes are a primary means to respond to signals emanating from environmental and metabolic fluctuations as well as to signals coordinating the cell cycle with macromolecular syntheses, among a large variety of other essential roles. Signals are recognized by a sensor domain of a histidine kinase which serves to convert signal binding to an active transmissible phosphoryl group through a signal-induced ATP-dependent autophosphorylation reaction directed to histidine residue. The sensor kinase is specifically mated to a response regulator, to which it transfers the phosphoryl group that activates the response regulator’s function, most commonly gene repression or activation but also interaction with other regulatory proteins. Two-component systems have been genetically amplified to control a wide variety of cellular processes; for example, both Escherichia coli and Pseudomonas aeruginosa have 60 plus confirmed and putative two-component systems. Bacillus subtilis has 30 plus and Nostoc punctiformis over 100. As genetic amplification does not result in changes in the basic structural folds of the catalytic domains of the sensor kinase or response regulators, each sensor kinase must recognize its partner through subtle changes in residues at the interaction surface between the two proteins. Additionally, the response regulator must prepare itself for efficient activation by the phosphorylation event. In this short review, we discuss the contributions of the critical β4-α4 recognition loop in response regulators to their function. In particular, we focus on this region’s microsecond-millisecond timescale dynamics propensities and discuss how these motions play a major role in response regulator recognition and activation.
Keywords: β4-α4 recognition loop, millisecond dynamics, NMR, phosphorylation, response regulator
Introduction
The key to the exceptional adaptability of bacteria is their capacity to express only those genes for proteins and pathways which they need for survival and growth in the particular surroundings in which they find themselves. They do this by efficiently recognizing and responding to the composition of their environment by sensing environmental signals (1).
To elicit such responses, bacteria employ the services of two regulatory proteins in signal transduction modules referred to as two-component signal transduction systems (2, 3). These two-component systems are ubiquitous in bacteria and in their simplest form consist of a sensor protein kinase, most often located in the cytoplasmic membrane which senses a specific environmental circumstance, and a cytoplasmic response regulator protein which provokes the appropriate adaptive genetic response.
In the prototypical two-component system, signal binding to the sensor domain of the sensor kinase induces autophosphorylation in an ATP-dependent reaction at a conserved histidine residue (4). The kinase then transfers the phosphoryl group to a conserved aspartic acid residue in the N-terminal regulatory domain of the response regulator protein. Most frequently, phosphorylation of the response regulator activates its so-called ‘output’ C-terminal DNA binding capabilities and it subsequently engages in transcriptional control appropriate to its initiating signal (5).
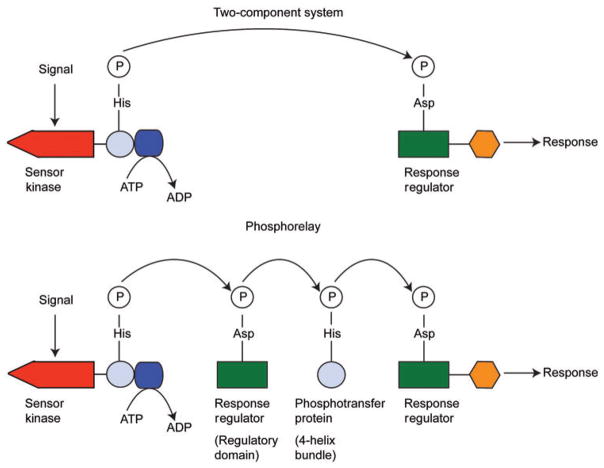
There is also a slightly more complex version of the two-component system called the phosphorelay (6). Here the sensor kinase first transfers the phosphoryl group to a response regulator which has only the regulatory domain but no output domain. The response regulator transfers the phosphoryl group to a histidine on a phosphotransferase protein, which then acts as the phosphodonor to a ‘conventional’ two-domain response regulator. This phosphorylated response regulator then acts as a transcription factor. In some phosphorelays, the sensor kinase, the ‘single-domain’ response regulator lacking the output domain, and the phosphotransfer domain are contained in a single protein (7). These are sometimes referred to as ‘hybrid kinases’ and they are the only type found in eukaryotic microbes. Schematic representations of the two-component system and the phosphorelay are shown in Figure 1.
Figure 1.
Schematic representations of the two-component system (top) and the phosphorelay (bottom).
As two-component systems represent the principal means by which bacteria protect themselves and respond to the environment, it is critical that precise interactions between the sensor kinase and its proper target response regulator are established. Flawed recognition between an activated sensor kinase and any inappropriate response regulators can lead to regulation of the wrong genes and the incorrect response to the current environmental circumstance (1).
Such precise interactions are not trivial for three main reasons (1). First, as continuing genomic studies attest, there are many such two-component systems in bacterial genomes. Second, though there is significant divergence in sequence and structure within the C-terminal DNA binding output domains of response regulators, their N-terminal regulatory domains are highly homologous from both a sequence and structure standpoint. Finally, the domains with which they interact are all quite similar four-helix bundles. Yet, despite all this similarity, each protein must only interact with its specific partner in order to activate unique genes. Ensuring such fidelity and accurate molecular recognition is quite complex. How can this be achieved?
It is also critical that the phosphorylation event occurs with some ease. At times of bacterial cell stress and the potential for significant morphological change, the frivolous use of cellular energy is particularly unwise (8). For this reason, the response regulator must prepare itself for efficiently accepting the incoming phosphoryl group and ensuring straightforward activation. How does it do this?
In this short review, we initially discuss the region(s) in response regulators which instigate kinase target specificity. Subsequently, and in more detail, we concentrate on the contributions of protein dynamics in preparing response regulators for activation. We focus solely on the N-terminal regulatory domain, as this is the domain which interacts with the four-helix bundle motif in the sensor kinase. The discussions here should serve as a complement to the many purely structural contributions involved in response regulator-target interactions.
Response regulator-sensor kinase structural considerations
Response regulator N-terminal domains have highly homologous structures (5, 9). They consist of an α-β structure with five α-helices arranged around a central five-stranded parallel β-sheet. The active-site aspartic acid is located at one end (the ‘top’) of the protein surrounded by the loops connecting the β-strands to the α-helices. The active site generally consists of three Asp residues including the phosphorylatable Asp at the end of strand β3. The other two Asp residues (or occasionally a Glu) are adjacent in the β1-α1 loop and help form an aspartic acid-binding pocket which initially accepts a divalent metal ion prior to phosphorylation (in order to offset negative charge in the pocket). The phosphorylation event causes subtle changes in structure and dynamics across the N-terminal regulatory domain, propagating an activation signal which releases the C-terminal output domain and allows for efficient DNA binding. The propagation of the signal across the regulatory domain has been described in terms of both dynamics and structural adjustments.
Histidine-containing four-helix bundles seem to be the main structural motif targeted by response regulators in sensor kinases (and phosphotransferases) in order to pick up the activating phosphoryl group (10–13).
Zapf and coworkers used information derived from a Spo0F-Spo0B co-crystal structure (13) to better define correlations between residues which comprise the interaction surface between the receiver domain of response regulators and the four-helix bundle motif of sensor kinases and phosphotransferases. They observed that the response regulator interaction surface involves, to some degree, all the β-α loops on the ‘top’ of the protein. They also noted that the N-terminal portion of the α1-helix is involved in the interaction. They proposed three general types of amino acids which play roles: (i) essential invariant ‘catalytic’ residues which are directly involved in the phosphotransfer mechanism, (ii) somewhat conserved ‘anchor’ residues which establish broad orientational contacts for catalysis, and (iii) variable ‘recognition’ residues which ensure that the correct two proteins come together. This patch of residues on the response regulator is found on the surface containing α1-helix, α5-helix, and the β4-α4 loop. Subsequent co-crystal structures and mutational analyses have confirmed the role of this surface in the interaction between the response regulator and its cognate four-helix bundle motif (11, 12, 14–16). Figure 2 shows co-crystal structures of representative response regulator/four-helix bundle complexes.
Figure 2. Co-crystal structures of the complexes.
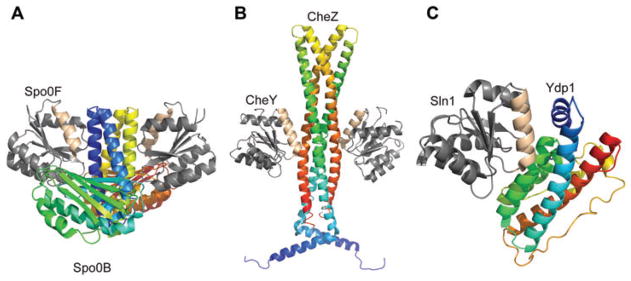
(A) Spo0F-Spo0B, (B) CheY-CheZ, and (C) Ydp1-Sln1. Spo0F, CheY, Sln1=response regulators; Spo0B=phosphotransferase; CheZ=phosphatase; Ydp1=domain of kinase. Interaction surfaces are highlighted in ‘wheat’ color.
In addition, a comparative homology modeling investigation examined the surface characteristics of two subfamilies of response regulators and found that the most variable hydrophobic regions were located around the α1-helix/α1-α5 interface and the β4-α4 loop (17). It was suggested that the surface variability in these regions, especially the α1-helix/α1-α 5 interface, contributed significantly to kinase target discrimination. Figure 3 shows hydrophobic surface homology models for the N-terminal regulatory domain of three members of the OmpR family of response regulators (OmpR, PhoB, and ArcA). In order to visualize subtle changes in surface side chain hydrophobicity, a color-coded hydrophobic scale is employed which highlights side chains based on a relative hydrophobicity scale derived from averaged physicochemical properties of the amino acid side chains. Hydrophobic rankings are assigned and color coded as a gradient, from high to low, as follows: ILV (red), GAF (orange), CM (yellow), ST (green), and WYPH (blue) (17). Note the high degree of variability, especially in the α1-helix/α1-α5 interface region.
Figure 3. Hydrophobic surfaces for homology models of the N-terminal regulatory domains of the response regulators.
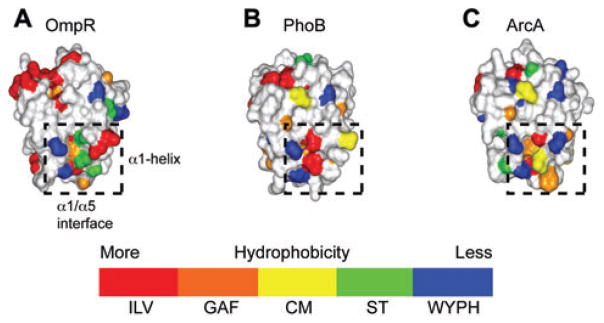
(A) OmpR, (B) PhoB, and (C) ArcA. The α1-helix/α1-α5 interface region is highlighted in a dashed box. The color-coded hydrophobic scale used is also shown (see text).
Overall, the structural, mutagenesis, and modeling studies support the notion that there is notable amino acid and, therefore, surface variability in the N-terminal α1-helix, β1-α1 loop, and the β4-α4 loop, and that these regions provide much of the specificity for a particular response regulator to target an appropriate four-helix bundle domain.
The motion of the β4-α4 recognition loop
It is clear that surface variability in the general α1-helix region plays a significant role in target kinase discrimination. We have analyzed the nuclear magnetic resonance (NMR) structures of 15 response regulator N-terminal domains and note, for the most part, that the α1-helix region is relatively stationary and only samples restricted conformational space. The implication is that protein dynamics in this region play a limited role in function. On the other hand, as discussed in detail below, the functionally important β4-α4 loop region exhibits significant motions on the microsecond-millisecond timescale. From this point onwards, we focus on the β4-α4 loop; to reflect its importance, we identify it as the β4-α4 recognition loop.
Nohaile and coworkers suggested that mutations in NtrC caused the response regulator to adopt a more ‘active’ (i.e., phosphorylated) conformation (18). This work was followed by studies in our group on the response regulator Spo0F, indicating that inactive (unphosphorylated) and active (phosphorylated) share functionally important fluctuations (19). In particular, it was proposed that in its inactive form, as it seeks its kinase target, Spo0F sampled its active conformation such that phosphorylation merely shifts this dynamic equilibrium. Excellent work by Kern and coworkers on NtrC corroborated this notion, and the NMR structure of transiently phosphorylated NtrC was also solved (20). In initial dynamics studies on unphosphorylated NtrC, microsecond motions were particularly evident in the β4-α4 recognition loop (21). Figure 4A shows the superposition of unphosphorylated NtrC (blue) and phosphorylated NtrC (orange/gold). Figure 4B shows microsecond-millisecond timescale motions superimposed on the unphosphorylated NtrC structure. It is evident that significant motions are occurring in the β4-α4 recognition loop (blue color in Figure 4B).
Figure 4. Dynamics of NtrC.
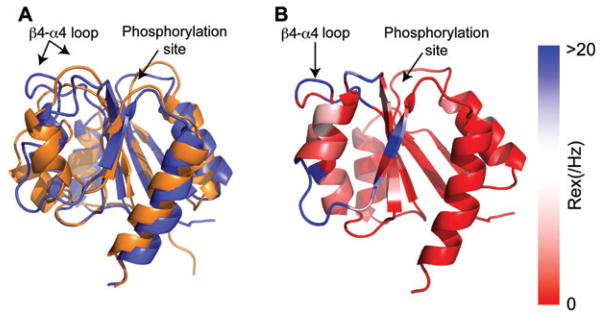
(A) Superimposition of the NMR structures of the N-terminal domain of unphosphorylated, inactive NtrC (blue, PDB 1DC7) and active NtrC (orange, PDB 1DC8) states are superimposed. (B) NMR 15N backbone Rex values (millisecond-microsecond motions) superimposed on unphosphorylated, inactive NtrC (PDB 1DC7). These data were adapted with permission from Ref (21). The NMR exchange term Rex is shown on a continuous color scale from red to blue. The color scale indicates the extent of conformational exchange between states that sense different chemical environments.
A comparison between the NMR structural data from our studies of unphosphorylated Spo0F (19, 22) and the NMR structural data of phosphorylated Spo0F provided by Gardino and coworkers (23) offers perhaps the best visualization of what the microsecond motions in the β4-α4 recognition loop are really doing (Figure 5).
Figure 5. Conformational families of Spo0F.
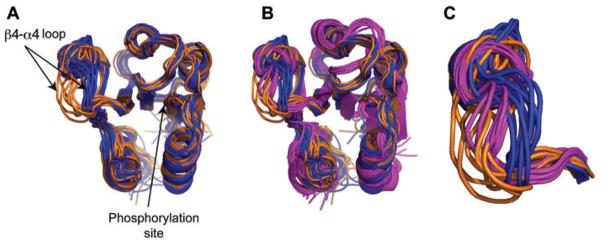
(A) Unphosphorylated-inactive Spo0F occupies two distinct conformational families (blue and orange, PDB code 1FSP) in the critical β4-α4 recognition loop. (B) One of these families structurally overlaps with the phosphorylated-active conformation of Spo0F (pink, PDB code 1PUX). (C) Magnified view of the β4-α4 recognition loop in its unphosphorylated-inactive form (blue and orange) and in its phosphorylated-active form (pink).
Figure 5A shows the two dominant NMR structural families populated by unphosphorylated Spo0F (blue and orange). Figure 5B shows these structures overlaid with the NMR structure of active BeF3-Spo0F (pink). The BeF3 moiety is used as a phosphorylation mimic and activates the response regulator. Figure 5C shows an expansion of the β4-α4 recognition loop. It is evident that in this region, the ‘orange’ structural family from inactive unphosphorylated Spo0F superimposes well on the structure of active BeF3-Spo0F (pink). This clearly shows that even in its inactive unphosphorylated form, Spo0F samples its active phosphorylated state and that this happens on the microsecond-millisecond timescale.
Recently, several constitutively active Spo0F mutants (L66A, I90A, and H101A) have been studied (24–26). I90 is in the β4-α4 recognition loop, H101 in β5-strand, and L66 in the middle of α3-helix. These mutant proteins are more readily phosphorylated by histidine kinases, resulting in hypersporulation phenotypes. It was suggested that these point mutants possess structural characteristics of the activated protein, particularly in the important β4-α4 loop region as they mimicked activated Spo0F (24). The high-resolution NMR structures of I90A, H101A, and L66A Spo0F did indeed show that in each case, the β4-α4 recognition loop adopts a conformation more closely approximating the activated form of Spo0F than the inactive form (26). As an example, Figure 6 shows the NMR structure of the β4-α4 recognition loop from H101A Spo0F (yellow), overlaid with the unphosphorylated/inactive (blue and orange) and activated structures of Spo0F (pink).
Figure 6.
The NMR structure of the β4-α4 recognition loop from H101A Spo0F (yellow, PDB code 2JVI), overlaid with the unphos-horylated-inactive (blue and orange) and activated structures of Spo0F (pink).
Certainly, we know that the dynamics of this region play a role in target identification and activation. With this in mind, molecular dynamics simulations were carried out in order to report on changes in the conformational space being sampled in the β4-α4 recognition loop for both wild-type Spo0F and the hypersporulating mutants (26). These data, for wild-type and L66A Spo0F (as a representative example), are shown in Figure 7.
Figure 7. Molecular dynamics simulation data.
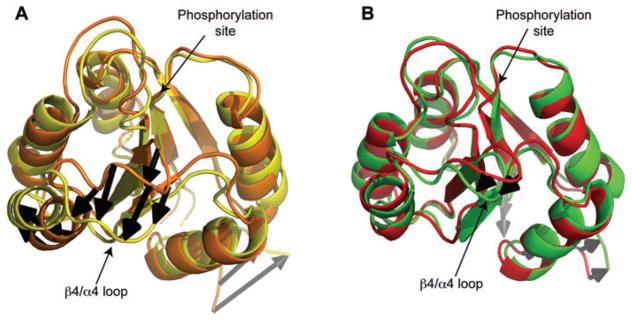
(A) Wild-type Spo0F and (B) L66A Spo0F (PDB code 2JVK). In (A), the two structures shown (orange and yellow) represent the structures from the wild-type Spo0F molecular dynamics ensemble which have the greatest root mean square deviations (RMSDs) with respect to one another. In (B), the two structures shown (red and green) represent the structures from the L66A Spo0F molecular dynamics ensemble which have the greatest RMSDs with respect to one another. The large difference in the β4-α4 recognition loop region (indicated by black arrows) for wild-type Spo0F (A) suggests significant motional propensity in this region. In the case of L66A Spo0F (B), this motional propensity is restricted.
The molecular dynamics simulation data for wild-type Spo0F (Figure 7A) show that the β4-α4 recognition loop has significant propensity for motion. In fact, this region displays the largest motional propensity in the protein by far (neglecting the termini). However, the molecular dynamics data for the L66A mutant shows that dynamic propensity and amplitude is appreciably reduced in the β4-α4 recognition loop region (26). The same is seen for each mutant.
These data show that in the β4-α4 recognition loop region, the mutants appear ‘dynamically trapped’ in a conformation which promotes function. These studies support the earlier work of Volkman and coworkers (21), and again demonstrate the role of β4-α4 recognition loop motions in the activity of Spo0F.
Covariance and the β4-α4 recognition loop
Interestingly, and rather surprisingly, the β4-α4 recognition loop was identified by covariance analyses as a co-evolving region in the general response regulator-sensor kinase interaction surface (15, 27, 28). This result is unexpected if the motional propensity of this loop region is not considered. As discussed below, evaluation of an interaction between two static structures does not provide insight into how the β4-α4 recognition loop could be in the correct orientation to be involved in the response regulator-sensor kinase interface. However, the inherent dynamics of this structural element provide an attractive explanation (28).
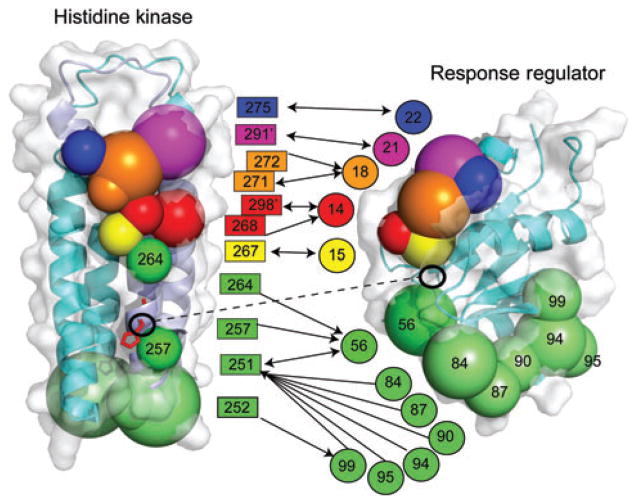
Six clusters of residues were identified between response regulators and sensor kinases which have co-evolved to ensure precise interactions by superimposing these clusters onto the structures of the histidine kinase domain of a sensor kinase HK853 (Thermotoga maritime) and Spo0F (Bacillus subtilis). This is shown in Figure 8. These interacting clusters are color coded to clarify which clusters co-vary.
Figure 8. Coupling of residues which co-vary across the sensor kinase-response regulator interaction surface.
The residues comprise six color-coded clusters. These are superimposed on the sensor kinase HK853 (T. maritime, PDB code 2C2A) on the left and the response regulator Spo0F (B. subtilis) on the right. The aspartate and histidine residues which are involved in the phosphotransfer are highlighted.
The first five clusters include HK853 residues T267, A268, A271, Y272, T275, F291, and Q298 and Spo0F residues G14, I15, L18, E21, and V22. The clusters are shown in yellow, red, orange, blue, and purple in Figure 8. These clusters define an interaction between the α1-helix of the histidine kinase and the α1-helix of the response regulator – as we expect. Indeed, this is the precise mode seen in the Spo0B-Spo0F structure (1, 13). Therefore, clusters 1–5 are the positions/residues interacting in the co-crystal which were predicted to be responsible for specificity. These residues co-vary to maintain the spatial arrangement of the active site.
The residues identified by covariance in cluster 6 (green in Figure 8) are more difficult to explain on the basis of the purely structural data. Specifically, residues in a subcluster (Y84, L87, I90, K94, E95 and L99) on the response regulator appear to be located some distance from the protein-protein interface. Accordingly, it seems impossible, at first sight, for these residues to form a direct interaction without undergoing significant conformational change. Not coincidentally, these residues are located in the β4-α4 recognition loop/helix α4 region, which is known to be both involved in kinase specificity and also to possess functional dynamic character, as discussed above. It appears that the covariance method captured specificity parameters not apparent from the static Spo0B-Spo0F structure. However, these recognition parameters were found in alanine scanning mutagenesis studies (14).
These residues, though not in suitable orientations in the X-ray complex, have the ability to ‘swing in’ to the interface such that they play a critical role in dictating the correct arrangement of the phosphotransfer active site. As discussed above, there is overwhelming evidence showing that the β4-α4 recognition loop/α4-helix region in response regulators is dynamic. Residues in this area may shift in order to inhabit functional positions at the sensor kinase-response regulator interface. Even a small population of such conformations at the interface has the capacity to influence specificity. This is an appealing mechanism for fine-tuning specificity. Variable dynamic characteristics among response regulators in this region may assist in precise targeting. It is possible that covariance clusters 1–5 contribute a more static and robust targeting process, whereas cluster 6 offers additional, delicate, contributions to the specificity (28).
Outlook and summary
There have been several intriguing studies on the precise mechanism of response regulator activation upon phosphorylation (25, 26, 29–31). These studies have centered on the propagation of the activating signal across the N-terminal regulatory domain. When the response regulator becomes phosphorylated, that information is relayed through and across the protein to other functionally relevant regions. For example, the state of phosphorylation must be communicated to the interface between the N-terminal domain and the C-terminal DNA-binding domain, such that structural adjustments can be made in order to allow the C-terminal domain to better carry out its DNA binding role. These messages are efficiently sent across the protein by intra-protein communication networks (29). A variety of studies using molecular dynamics, mutagenesis, NMR relaxation, and NMR chemical shift mapping suggest that transfer of information, and the setting up of the network, appears to involve coherent millisecond motions and transient non-native hydrogen bonds (29, 31). Interestingly, these studies find vital roles for residues at the top of α4, in the β4-α4 loop region. Certainly, further NMR dynamics and computational studies on a wider variety of response regulators would greatly enhance and refine the current models for activation.
Overall, several studies suggest that the motional properties of the β4-α4 recognition loop are critical in driving the interaction of a response regulator with its specific target. The motions have evolved to provide assistance in kinase target recognition and in preparing the response regulator to readily accept the activating phosphoryl group. Without these specific motions, the two-component system would not function correctly.
Acknowledgments
The authors thank Dr. Doro Kern (Brandeis) for permission to use modified NtrC figures (Figure 4) and Dr. Doug Kojetin (Scripps) for providing the images for Figure 3. This work was supported by National Institutes of Health grants to John Cavanagh (GM55769) and James A. Hoch (GM19416).
Biographies

John Cavanagh is a William Neal Reynolds Distinguished Professor of Structural Biochemistry at North Carolina State University. He received a PhD degree from the University of Cambridge, UK and was an NIH postdoctoral fellow in the group of Dr. Mark Rance at The Scripps Research Institute, La Jolla, CA, USA. Subsequently, he held positions as a Senior Research Associate at The Scripps Research Institute and Director of NMR Structural Biology/Associate Professor of Biomedical Sciences at the Wadsworth Center (New York State Department of Health). His early research efforts were in the design of novel NMR methods to study biomolecules. Now he focuses on the structure, dynamics, and function of bacterial signal transduction systems and the development of small molecules to overcome bacterial resistance.

James A. Hoch is a Professor of Molecular and Experimental Medicine at The Scripps Research Institute, La Jolla, CA, USA. He received a PhD degree from the University of Illinois, Urbana, IL and was an NIH postdoctoral fellow at the Centre de Génétique Moléculaire, Gif-sur-Yvette, France. His research has focused on the structure and function of bacterial signal transduction systems and more recently, on the development of statistical physics techniques to identify mechanisms of protein interaction between signaling molecules.

Benjamin Bobay began studying signal transduction as a student at Purdue University, West Lafayette, IN, USA. He received his PhD in Molecular and Structural Biochemistry in 2004 at North Carolina State University, Raleigh, NC, USA, followed by a post-doc with Drs. John Cavanagh and Michael Goshe at North Carolina State University where he studied the induced protein conformational changes as a result of ligand interactions through NMR and mass spectrometry. He is currently a Research Assistant Professor at North Carolina State University. His research program is focused on bacterial response proteins and the role of calcium signaling in diabetes and dementia.
References
- 1.Hoch JA, Varughese KI. Keeping signals straight in phosphorelay signal transduction. J Bacteriol. 2001;183:4941–9. doi: 10.1128/JB.183.17.4941-4949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 4.Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–90. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–34. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–52. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 7.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–70. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 8.Buckler DR, Anand GS, Stock AM. Response-regulator phosphorylation and activation: a two-way street? Trends Microbiol. 2000;8:153–6. doi: 10.1016/s0966-842x(00)01707-8. [DOI] [PubMed] [Google Scholar]
- 9.Volz K. Structural and functional conservation in response regulators. In: Hoch JA, Silvahy TJ, editors. Two-component signal transduction. Washington DC: American Society for Microbiology, ASM Press; 1995. pp. 53–64. [Google Scholar]
- 10.Ohta N, Newton A. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J Bacteriol. 2003;185:4424–31. doi: 10.1128/JB.185.15.4424-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Q, Porter SW, West AH. The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems. Structure. 2003;11:1569–81. doi: 10.1016/j.str.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002;9:570–5. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 13.Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–62. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 14.Tzeng YL, Hoch JA. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–12. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 15.Skerker JM, Perchuk BS, Siryaporn A, Lubin E, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–54. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capra EJ, Perchuk BS, Lubin EA, Ashenberg O, Skerker JM, Laub MT. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 2010;6:e1001220. doi: 10.1371/journal.pgen.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojetin DJ, Sullivan DM, Thompson RJ, Cavanagh J. Classification of response regulators based on their surface properties. Methods Enzymol. 2007;422:141–69. doi: 10.1016/S0076-6879(06)22007-X. [DOI] [PubMed] [Google Scholar]
- 18.Nohaile M, Kern D, Wemmer D, Stedman K, Kustu S. Structural and functional analyses of activating amino acid substitutions in the receiver domain of NtrC: evidence for an activating surface. J Mol Biol. 1997;273:299–316. doi: 10.1006/jmbi.1997.1296. [DOI] [PubMed] [Google Scholar]
- 19.Feher VA, Cavanagh J. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature. 1999;400:289–93. doi: 10.1038/22357. [DOI] [PubMed] [Google Scholar]
- 20.Kern D, Volkman BF, Luginbühl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–8. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 21.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–33. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 22.Feher VA, Zapf JW, Hoch JA, Whiteley JM, McIntosh LP, Rance M, Skelton NJ, Dahlquist FW, Cavanagh J. High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition. Biochemistry. 1997;36:10015–25. doi: 10.1021/bi970816l. [DOI] [PubMed] [Google Scholar]
- 23.Gardino AK, Volkman BF, Cho HS, Lee SY, Wemmer DE, Kern D. The NMR solution structure of BeF(3)(−)-activated Spo0F reveals the conformational switch in a phosphorelay system. J Mol Biol. 2003;331:245–54. doi: 10.1016/s0022-2836(03)00733-2. [DOI] [PubMed] [Google Scholar]
- 24.Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;33:389–95. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin PD, Bobay BG, Regel EJ, Thompson RJ, Hoch JA, Cavanagh J. predominantly buried residues in the response regulator spo0f influence specific sensor kinase recognition. febs lett. 2007;581:1425–9. doi: 10.1016/j.febslet.2007.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobay BG, Thompson RJ, Hoch JA, Cavanagh J. Long range dynamic effects of point-mutations trap a response regulator in an active conformation. FEBS Lett. 2010;584:4203–7. doi: 10.1016/j.febslet.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White RA, Szurmant H, Hoch JA, Hwa T. Features of protein-protein interactions in two-component signaling deduced from genomic libraries. Methods Enzymol. 2007;422:75–101. doi: 10.1016/S0076-6879(06)22004-4. [DOI] [PubMed] [Google Scholar]
- 28.Szurmant H, Bobay BG, White RA, Sullivan DM, Thompson RJ, Hwa T, Hoch JA, Cavanagh J. Co-evolving motions at protein-protein interfaces of two-component signaling systems identified by covariance analysis. Biochemistry. 2008;47:7782–4. doi: 10.1021/bi8009604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feher VA, Tzeng YL, Hoch JA, Cavanagh J. Identification of communication networks in Spo0F: a model for phosphorylation induced conformational change and implications for activation of multiple domain bacterial response regulators. FEBS Lett. 1998;425:1–6. doi: 10.1016/s0014-5793(98)00182-3. [DOI] [PubMed] [Google Scholar]
- 30.Lei M, Velos J, Gardino A, Kivenson A, Karplus M, Kern D. Segmented transition pathway of the signaling protein nitrogen regulatory protein C. J Mol Biol. 2009;392:823–36. doi: 10.1016/j.jmb.2009.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardino AK, Villali J, Kivenson A, Lei M, Liu CF, Steindel P, Eisenmesser EZ, Labeikovsky W, Wolf-Watz M, Clarkson MW, Kern D. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–18. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]