Abstract
The comparative efficacies of different conventional parasitological methods and nested PCR for diagnosis of bovine cryptosporidiosis in faecal samples were evaluated. Among the 100 samples collected from calves in and around Ludhiana Direct faecal smear staining technique revealed 25.0 % positivity for the oocysts of Cryptosporidium spp. with sensitivity and specificity of 68.12 and 92.98 %, respectively. Zinc sulphate solution floatation and saturated sugar solution floatation staining techniques showed sensitivity and specificity of 83.92 and 96.36; 81.03 and 98.14 %, respectively. Products of the primary PCR of Cryptosporidium spp. directed against small subunit (18S) ribosomal RNA when employed as template in nested PCR produced the amplicons of desired size (834 bp) in 47.0 % of the samples. Amplification of 834 bp fragment was also observed in positive control, while no amplification was observed in negative control. Results indicated PCR assays as highly sensitive and specific techniques for the screening of the samples for Cryptosporidium spp. but in developing countries and under field conditions where limited resources do not allow the application of PCR assays, concentration staining methods are recommended.
Keywords: Cryptosporidium spp, mZN staining, Diagnosis, Nested PCR
Introduction
Cryptosporidium spp. infection is regarded as one of the most common etiological agent of diarrhoea in humans and animals worldwide (Casemore et al. 1997). Infection of susceptible hosts follows ingestion of Cryptosporidium oocysts which excyst within the intestinal tract and release sporozoites capable of binding to and penetrating intestinal epithelial cells (Fayer et al. 1997). Bovine cryptosporidiosis is essentially a disease of newborn calves particularly of 1–3 weeks of age (Leek and Fayer 1984) but it has also been recorded in older animals above 2 years of age (Henriksen and Krogh 1985; Singh et al. 2006).
Cryptosporidiosis is generally diagnosed by microscopical detection of oocysts in faecal smears by conventional staining methods. However, immunofluorescence staining techniques, antigen detection by enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (Morgan et al. 1998; Kaushik et al. 2008) had been proved to be useful in sensitive and specific diagnosis of cryptosporidiosis. The nested PCR with the utilization of primers specific for the SSU-rRNA gene has been found to be highly sensitive as a diagnostic tool for cryptosporidiosis i.e. about one oocyst in 1 ml of faecal sample (Xiao et al. 1999). Modern improved PCR methods are also applied for sensitive detection of oocysts in faecal samples or environmental samples like water (Bakheit et al. 2008; Skotarczak 2009).
However, these diagnostic techniques differ significantly in the sensitivity and specificity for diagnosis of bovine cryptosporidiosis by faecal sample. Therefore, the current study was undertaken to determine the comparative efficacies of different conventional parasitological methods and nested PCR in faecal samples collected from neonatal dairy calves.
Materials and methods
Faecal samples (n = 100) were collected from neonatal dairy calves from in and around Ludhiana district and were used for diagnosis of cryptosporidiosis by various techniques
In direct faecal smear examination, a thin and transparent faecal smear was made with the help of ear bud, air dried, fixed in methanol for 3 min, air dried and stained by modified Ziehl-Neelsen (mZN) staining method as recommended by OIE (2008). In brief, after fixation, smears were stained with 1 % cold carbol–fuchsin solution for 15 min and rinsed thoroughly in tap water. Then decolorization was done in 1 % acid methanol for 15 s and again the smears were rinsed with tap water and then, the smears were counterstained with 0.4 % malachite green for 30 s. The smears were finally washed in tap water, air-dried and were examined microscopically.
In concentration methods, the faecal samples (1–2 g) were suspended in floatation medium (zinc sulphate solution, sp. gr. 1.18 and sugar solution, sp. gr. 1.18) for 20 min. After this, the meniscus was gently removed with a disposable pipette and dispensed gently on to a microscope slide to prepare faecal smears. Subsequently these were air dried, fixed in methanol and stained by mZN staining as described above.
Genomic DNA isolation
For conducting the PCR, whole-genomic DNA was isolated from faecal sample using QIAamp® DNA mini stool kit (QIAGEN, GmbH, Germany) following the manufacturer’s recommendations with minor modifications. In brief, approximately 200 mg of the faecal sample was mixed with 1.4 ml ASL buffer in 2.0 ml microcentrifuge tube. The homogenous suspension was heated in water bath at 80 °C for 5 min and then centrifuged for 1 min at 14,000×g to pellet stool particles. Supernatant (1.2 ml) was pipetted out in new 2 ml centrifuge tube, one inhibitEX tablet was added and then vortex. After 1 min incubation, the sample was centrifuged at 14,000×g for 3 min to pellet out inhibitors bound to inhibitEX. Supernatant (200 μl) was added to new 1.5 ml centrifuge tube containing 30 μl of proteinase K and after vortexing, 200 μl of AL buffer was added. This lysate was incubated at 70 °C for 10 min, 200 μl of ethanol was added, and the mixture was applied to QIAamp mini spin column and centrifuged at 8,000×g for 1 min. Thereafter, 2 washings were given with wash buffers and DNA was eluted in 150 μl of elution buffer and stored at −20 °C till use. Concentration of the extracted DNA from samples was measured in Nanodrop instrument. Genomic DNA of Cryptosporidium spp was isolated from faeces sample positive and revealing large number of oocysts in faecal smear examination and was utilized as positive control. Genomic DNA was also isolated from the faeces of infection-free, neonatal bovine calf and used as a negative control.
18S rRNA gene amplification
The PCR (primary as well as nested) was optimized to identify the small subunit (18S) ribosomal RNA gene as described by Paul et al. (2009). The sequences of the primers were as follows
For primary PCR
CRP-DIAG1 forward 5′-TTCTAGAGCTAATACATGCG-3′
CRP-DIAG1 reverse 5′-CATTTCCTTCGAAACAGG A-3′
For nested PCR
CRP-DIAG2 forward 5′-GGAAGGGTTGTATTTATTAGATAAAG-3′
CRP-DIAG2 reverse 5′-AAGGAGTAAGGAACAACCTCCA-3′
Two rounds of PCR in a final volume of 25 μl were carried out in a PCR thermal cycler (Eppendorf, Germany). In the primary PCR assay, the master solution consisted of 2.5 μl of 10X
PCR buffer (Bangalore Genei), 0.5 μl of 10 mM dNTP mix (Bangalore Genei), 2.0 μl of 25 mM MgCl2 (Bangalore Genei), 0.5 μl Taq DNA polymerase (Bangalore Genei), 0.5 μl each (20 pmol) of the external forward (CRP-DIAG1 forward) and external reverse (CRP-DIAG1 reverse) primers and 4.0 μl of template DNA isolated from faecal samples. The volume was made up to 25 μl with nuclease-free water. The cycling conditions were as: initial denaturation at 94 °C for 5 min, 34 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min, and the final extension was performed at 72 °C for 10 min. For nested PCR similar quantities of the PCR mixture constituents except 1.5 μl MgCl2 (25 mM) and 3 μl of template was used. Identical thermocyclic parameters were kept in nested PCR except annealing was done at 57 °C. The PCR product was checked for amplification by electrophoresis on a 1.5 % agarose gel and visualized using gel documentation system (Syngene, UK).
Specificity and sensitivity of parasitological techniques
The nested PCR with the utilization of primers specific for the SSU-rRNA gene as a diagnostic tool for cryptosporidiosis has been found to be highly sensitive i.e. about one oocyst in 1 ml of faecal sample (Xiao et al. 1999). Hence, PCR assay has been considered 100 % sensitive and used as a standard reference test for calculation of sensitivities and specificities of other techniques as per Morgan et al. (1998) and Paul et al. (2009) with the following formula:
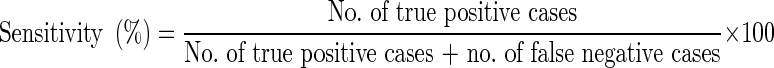 |
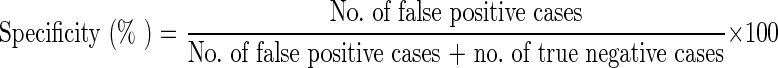 |
The faecal samples detected positive by PCR were considered as true positive. Number of false negative samples for a specific technique was obtained by subtracting the number of samples detected positive by the technique from those detected positive by PCR. False positive were the faecal samples which revealed oocyst like bodies in the mZN stained smears under microscope but failed to yield specific PCR amplification.
Results and discussion
Direct faecal smear staining (DFSS) technique revealed 25 animals as positive for the oocysts of Cryptosporidium spp. under 100× resolution by oil immersion which showed 25.0 % positivity in dairy calves from in and around Ludhiana district, Punjab. Whereas, Zinc sulphate solution floatation staining (ZSFS) and saturated sugar solution floatation staining (SSFS) techniques detected 36 and 38 samples positive for oocysts, respectively in the faecal samples. The oocysts stained bright pink to apple red on a pale green background in mZN staining (Figs. 1, 2).
Fig. 1.
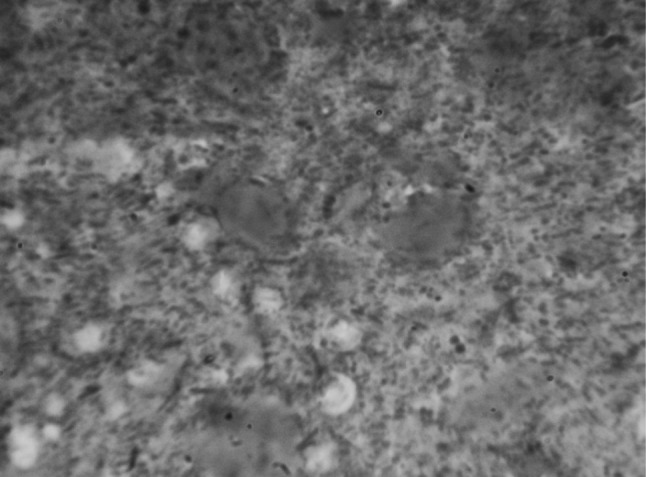
Cryptosporidium spp. oocysts in mZN staining at ×1000 magnification in DFSS method
Fig. 2.
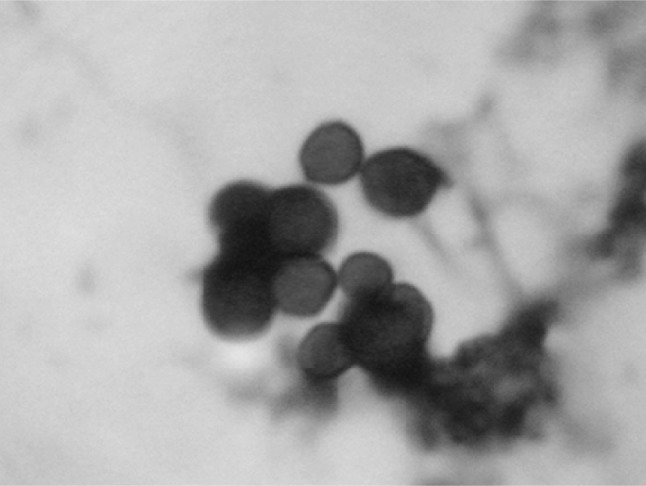
Cryptosporidium spp. oocysts in mZN staining at ×1000 magnification in concentration method
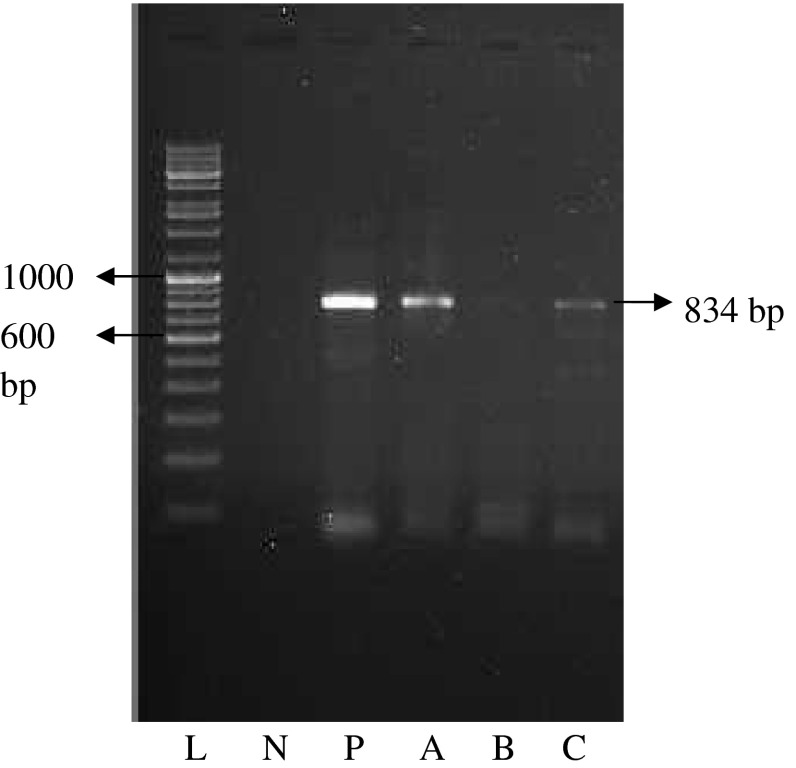
In order to assess the true status of Cryptosporidium spp. infection in dairy calves these samples were analyzed initially by primary PCR followed by nested PCR, to detect any amplification in the form of ethidium bromide-stained amplicons, after standardization of the assays. Of the total samples subjected to primary PCR, none were found positive for Cryptosporidium spp. infection as revealed by the absence of the amplification of a 1,317 bp product. PCR products obtained from the primary PCR of Cryptosporidium spp., when employed as template in nested PCR produced the amplicons of desired size (834 bp) in 47.0 % of the samples (Fig. 3). Amplification of 834 bp fragment was also observed in positive control, while no amplification was observed in negative control. This validates that nested PCR, when coupled with primary PCR, resulted in increased sensitivity of the assay, as the products from the primary PCR that were not visualized in the ethidium bromide-stained agarose gel electrophoresis, when subjected to nested PCR, could be detected.
Fig. 3.
Nested PCR product (834 bp) after PCR amplification of 18S SSU rRNA gene of Cryptosporidium spp. Lane L is ladder (medium range, Banglore Genei), Lane N and P are negative and positive control, respectively. A, B and C are denote samples
Out of the total samples detected positive in DFSS, SSFS and ZSFS; 4, 2 and 1, respectively were shown negative by nested PCR and therefore, were declared as false positive (Table 1). The sensitivity and specificity of DFSS was 68.12 and 92.98 %, respectively and was least as compared to the other techniques. Whereas, the sensitivity and specificity of SSFS and ZSFS technique was recorded as 83.92 and 96.36; 81.03 and 98.14 %, respectively (Table 1).
Table 1.
Number of samples detected positive out of 100 samples screened in different diagnostic techniques
| Different diagnostic techniques | DFSS | ZSFS | SSF | nPCR |
|---|---|---|---|---|
| Samples detected positive | 25 (4) | 36 (1) | 38 (2) | 47 |
| Specificity (%) | 92.98 | 98.14 | 96.36 | 100 |
| Sensitivity (%) | 68.12 | 81.03 | 83.92 | 100 |
Figures in the parentheses denote the false positive cases
Direct demonstration of the oocysts in the faeces is considered to be the gold standard in the conventional parasitology and has been made customary since earlier for the diagnosis of gastrointestinal protozoal disease (Paul et al. 2009). Among the various conventional parasitological techniques viz. DFSS, ZSFS and SSFS techniques utilized in the current study higher sensitivity and specificity was recorded in methods employing concentration of oocysts before modified Ziehl-Neelsen staining. Similar results of better efficacies of concentration techniques had been reported earlier (Current et al. 1983; Barwick et al. 2000; Kar et al. 2011; Paul et al. 2009). Thus, the efficacy of staining methods can be increased by utilizing the procedures involving concentration of oocysts and could conveniently be used as a routine diagnostic tool for detection of Cryptosporidium infection.
The present study utilized both conventional parasitological techniques as well as PCR assays for determining the comparative efficacies of these techniques as diagnostic tools for the diagnosis of cryptosporidiosis. After evaluation of different diagnostic techniques for the detection of Cryptosporidium spp. infection, PCR was found to be most efficient as compared to the other diagnostic techniques utilized. Results achieved are congruous with the findings of Current et al. (1983), MacPherson and McQueen (1993), Webster et al. (1996), Morgan et al. (1998), Xiao et al. (2006), Paul et al. (2009) and Kar et al. (2011).
It can thus be concluded that PCR assays being highly sensitive and specific should be used for the screening of the samples for Cryptosporidium spp. However, in developing countries and under field conditions where limited resources do not allow the application of PCR assays, concentration staining methods are recommended.
Acknowledgments
Authors are highly thankful to Director of Research, Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana, India for providing all necessary facilities.
References
- Bakheit MA, Torra D, Palomino LA, Thekisoe OM, Mbati PA, Ongerth J, Karanis P. Sensitive and specific detection of Cryptosporidium species in PCR-negative samples by loop-mediated isothermal DNA amplification and confirmation of generated LAMP products by sequencing. Vet Parasitol. 2008;158:11–22. doi: 10.1016/j.vetpar.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Barwick RS, Mohammed HO, White AB, Bryan RT. Detection of Cryptosporidium parvum and Cryptosporidium muris in soil samples. Biol Fertil Soils. 2000;31:385–390. doi: 10.1007/s003749900185. [DOI] [Google Scholar]
- Casemore DP, Wright SE, Coop RL. Cryptosporidiosis-human and animal epidemiology. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton: CRC; 1997. pp. 65–92. [Google Scholar]
- Current WL, Reese NC, Ernst JV, Bailey WS, Heyman MB, Weinstein WM. Human cryptosporidiosis in immunocompetent and immunodeficient persons. Studies on outbreak and experimental transmission. N Engl J Med. 1983;308:1252–1258. doi: 10.1056/NEJM198305263082102. [DOI] [PubMed] [Google Scholar]
- Fayer R, Speer CA, Dubey JP. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca raton: CRC; 1997. pp. 1–42. [Google Scholar]
- Henriksen SA, Krogh HV. Bovine cryptosporidiosis in Denmark. Prevalence, age, distribution and seasonal variation. Nord Vet Med. 1985;37:34–41. [PubMed] [Google Scholar]
- Kar S, Gawlowska S, Daugschies A, Bangoura B. Quantitative comparison of different purification and detection methods for Cryptosporidium parvum oocysts. Vet Parasitol. 2011;177:366–370. doi: 10.1016/j.vetpar.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Kaushik K, Khurana S, Wanchu A, Malla N. Evaluation of staining techniques, antigen detection and nested PCR for the diagnosis of cryptosporidiosis in HIV seropositive and seronegative patients. Acta Trop. 2008;107:1–7. doi: 10.1016/j.actatropica.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Leek RG, Fayer R. Prevalence of Cryptosporidium infections and their relation to diarrhea in calves on 12 dairy farms in Glicia (NW Spain) Vet Parasitol. 1984;106:1–10. [Google Scholar]
- MacPherson DW, McQueen R. Cryptosporidiosis: multi-attribute evaluation of six diagnostic methods. J Clin Microbiol. 1993;31:198–202. doi: 10.1128/jcm.31.2.198-202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan UM, Pallant L, Dwyer D, Forbes DA, Rich G, Thompson RCA. Comparison of PCR and microscopy for the detection of Cryptosporidium parvum in human faecal samples: clinical trial. J Clin Microbiol. 1998;36:995–998. doi: 10.1128/jcm.36.4.995-998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE (2008) Cryptosporidiosis. Terr Man. Chapter 2.9.4
- Paul S, Chandra D, Tewari AK, Banerjee PS, Ray DD, Boral R, Rao JR. Comparative evaluation and economic assessment of coprological diagnostic methods and PCR for detection of Cryptosporidium spp. in bovines. Vet Parasitol. 2009;164:291–295. doi: 10.1016/j.vetpar.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Singh BB, Sharma R, Kumar H, Banga HS, Aulakh RS, Gill JPS, Sharma JK. Prevalence of Cryptosporidium parvum infection in Punjab (India) and its association with diarrhea in neonatal dairy calves. Vet Parasitol. 2006;140:162–165. doi: 10.1016/j.vetpar.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Skotarczak B. Methods for parasitic protozoan detection in the environmental samples. Parasite. 2009;16:183–190. doi: 10.1051/parasite/2009163183. [DOI] [PubMed] [Google Scholar]
- Webster KA, Smith HV, Giles M, Dawson L, Robertson LJ. Detection of Cryptosporidium oocysts in faeces: comparison of conventional coproscopical methods and polymerase chain reaction. Vet Parasitol. 1996;61:5–13. doi: 10.1016/0304-4017(95)00811-X. [DOI] [PubMed] [Google Scholar]
- Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Monsali RJ, Fayer R, Lal AA. Phylogenetic analysis of Cryptosporidium parasites based on the SSU rRNA gene locus. App Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Alderisio KA, Jiang J. Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl Environ Microbiol. 2006;72(9):5942–5947. doi: 10.1128/AEM.00927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]