Abstract
In Iran, theileriosis is normally diagnosed with traditional Giemsa staining method. This is not applicable for identification of the carrier animals. The aim of this study was to compare conventional Giemsa staining method with the PCR technique in the detection of Theileria organisms. In this study, examinations were performed on 150 blood samples from cattle without clinical signs. Sensitivity and specificity of 50 microscopic fields were compared with Theileria specific PCR. The degree of agreement between PCR and microscopic test was determined by Kappa (κ) values with 95 % confidence intervals. PCR showed that 42 samples were Theileria spp. positive, while routine microscopy showed erythrocytes harboring Theileria like structures in 11 blood samples. Examination of 50 microscopic fields showed 57 % sensitivity and 99 % specificity compared to 100 % sensitivity and specificity for PCR. The κ coefficient between PCR and Microscopy (50 fields) techniques indicated no level of agreement. Our results showed that the microscopic examination remains the convenient technique for day-to-day diagnosis of clinical cases in the laboratory but for the detection of carrier animal containing low parasitemia. Therefore, molecular methods such as PCR can be used as a safe method for identifying cattle persistently infected with Theileria spp.
Keywords: Theileria, Carrier cattle, Microscopic method, Giemsa staining, PCR
Introduction
Theileriosis is the main tick-borne diseases in tropical and sub tropical regions (Uilenberg 1981). Tropical theileriosis caused by Theileria annulata is an endemic and economically important disease of cattle especially in pure Holsteins and crossbreds (Hashemi-Fesharki 1988). Hyalomma, vectors of causative organism of tropical theileriosis, are most abundant ticks recorded throughout Iran (Rahbari et al. 2007). The disease is characterized by a marked rise in body temperature, reaching 40–41.5 °C, depression, lacrimation, nasal discharge, and swelling of the superficial lymph nodes and anemia (Gubbels et al. 2000). The recovered cows from acute or primary theileriosis, remain infected for a long period and even for the rest of their lives, so acting as reservoirs of infection for ticks and cause natural transmission of the disease (Cacci et al. 2000; Kirvar et al. 2000). Therefore, detection of Theileria in asymptomatic carrier cattle is important for the implementation of successful control programmes.
Conventional method for identification includes examination of blood smears using Giemsa staining, which is accompanied with some critical problems. Giemsa-stained blood smears can be indeed used as a suitable method to detect Theileria agents in the animals clinically suspected acute theileriosis, but it is not applicable for the determination of pre-symptomatic or carrier animals, where parasitemia is very low (Friedhoff and Bose 1994).
Several serological tests have been established for detection of subclinical infections in epidemiological studies. Unfortunately, because of false-positive and false negative results due to antigen cross reactivity and weakening of specific immune responses due to disappearance of antibodies in long-term, these tests are unreliable for detection of carrier cattle (Passos et al. 1998). Therefore, a sensitive and specific method as compared to serological tests and examination of Giemsa-stained blood smears for the diagnosis of piroplasms is required. Molecular methods, with a high degree of sensitivity and specificity, have been developed to identify various Theileria species in persistently infected cattle (Altay et al. 2008).
In Iran, theileriosis is normally diagnosed with the traditional Giemsa staining method, yet it seems not to be applicable for identifying of the carrier animals (Azizi et al. 2008). The aim of this study was to compare conventional Giemsa staining method with the PCR technique in the detection of Theileria organisms.
Materials and methods
Collection of blood samples
From June 2009 to October 2009, 30 farms in Isfahan province, central part of Iran, were selected for the study based on their history of outbreak of bovine theileriosis. Blood samples were collected from jugular vein of 150 Holstein and crossbred cattle. Five hundred micro liters of each collected blood samples was fixed with 1 ml 96 % ethanol in 1.5 ml sterile eppendorf tubes. Additionally, two thin blood smears by puncturing ear vein for each cattle were prepared. The blood smears were air dried, fixed in methanol, stained with Giemsa and analyzed for the presence of Theileria in the erythrocytes at 100× magnification. In each blood smear 50 fields were examined separately by a single observer. The presence of one piroplasm was considered as a positive blood smear. Microscopic examination was done in a manner that was blinded to the molecular results. All smears carefully examined to estimate the Percent Parasitized Erythrocytes (PPE).
DNA extraction
The DNA was extracted using a DNA isolation kit (MBST, Iran) according to the manufacturer’s instructions. Briefly, ca. 5 mm3 big pieces of fixed blood samples were air dried and subsequently lysed in 180 μl lysis buffer and the proteins were degraded using 20 μl proteinase K for 10 min at 55 °C. After addition of 360 μl Binding buffer and incubation for 10 min at 70 °C, 270 μl ethanol (96 %) was added to the solution and after vortexing, the complete volume was transferred to the MBST-column. The column was first centrifuged, and then washed twice with 500 μl washing-buffer. Finally, DNA was eluted from the carrier using 100 μl Elution buffer. The amount of extracted DNA and its purity was measured by OD260 and the ratio of OD260–OD280 respectively. In addition the extracted DNA was analyzed on agarose gel before use.
PCR
For assessment of the Theileria infection in samples, primers Tbs-S (5′CACAGGGAGGTAGTGACAAG3′) and Tbs-A (5′AAGAATTTCACCTCTGACAG3′) were used, which amplify simultaneously an approximately 426–430 bp fragment of the 18SrRNA gene for members of genera Theileria. Approximately 100 ng DNA was used for the PCR analysis. The PCR was performed in 100 μl total volume including one time PCR buffer, 2.5 U Taq DNA Polymerase (Cinnagen, Iran), 2 μl of each primer (Tbs-S/Tbs-A, 20 μM, Cinnagen, Iran), 200 μM of each dATP, dTTP, dCTP and dGTP (Fermentas, EU) and 1.5 mM MgCl2 in automated Thermocycler (MWG, Germany) with the following program: 5 min incubation at 95 °C to denature double strand DNA, 38 cycles of 45 s at 94 °C (denaturing step), 45 s at 55 °C (annealing step) and 45 s at 72 °C (extension step). Finally, PCR was completed with the additional extension step at 72 °C for 10 min. The PCR products were analyzed on 2 % agarose gel in 0.5 times Tris–Borate–EDTA (TBE) buffer and visualized using ethidium bromide and UV-illuminator. A molecular mass ladder (100 bp) and positive and negative controls were used for each batch run. Each sample was spiked with positive control Theileria spp. DNA to detect any inhibition of the PCR that might lead to false- negative results.
Statistical analysis
The degree of agreement between PCR and the microscopical tests was determined by Kappa (κ) values with 95 % confidence intervals. The PCR as the reference test was used to calculate the relative sensitivity and relative specificity of the microscopical tests.
Results
The DNA was extracted from blood samples and analyzed by PCR using primers derived from the 18S rRNA gene. The nucleotide sequence of 18S rRNA gene is highly conserved in Theileria spp. and the primers Tbs-S/Tbs-A can amplify the corresponding fragments of the gene in all known Theileria species.
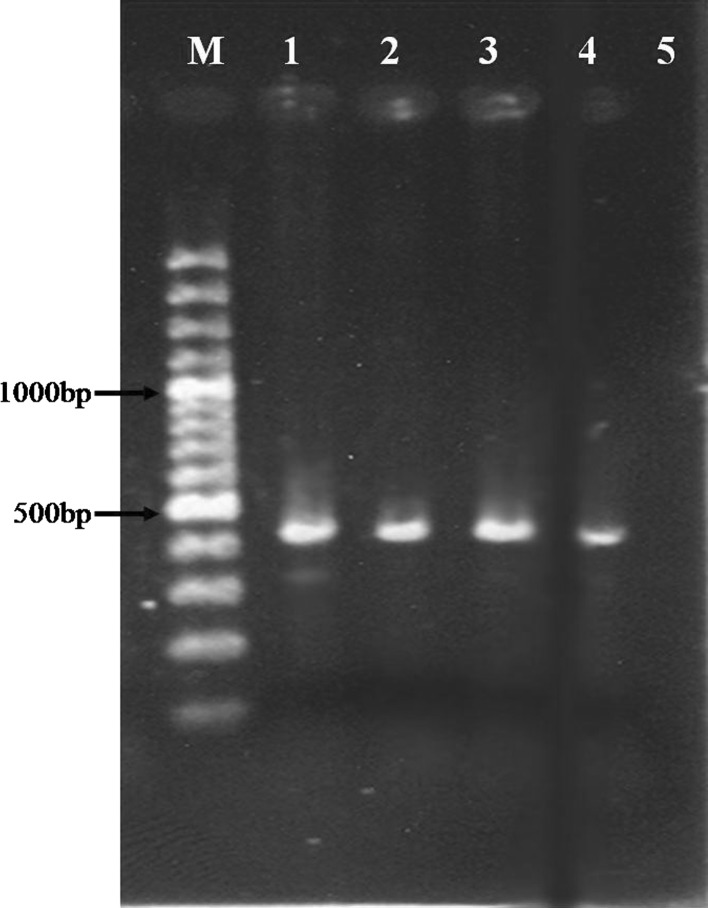
The PCR analysis of the DNA isolated from blood samples showed that 42 out of the total 150 blood samples were Theileria spp. positive and revealed an expected PCR product of 426–430 bp in length (Fig. 1).
Fig. 1.
Isolated DNA from blood was analysed by PCR. Theileria spp. positive control (lane 1), DNA was amplified with Tbs-S/Tbs-A primer set resulting in PCR product of 426–430 bp in length (lanes 2–4), negative control (lane 5) and M (Marker 100 bp)
Interestingly, the Giemsa staining analysis of blood smears showed different results dependent upon the chosen number of examined microscopic fields by 100× magnification (Fig. 2). In 11 out of 150 blood samples, Theileria like structures could be identified when the examination was performed in 50 microscopic fields. Ten of samples (among 11 samples) showed also PCR positive (Table 1).
Fig. 2.
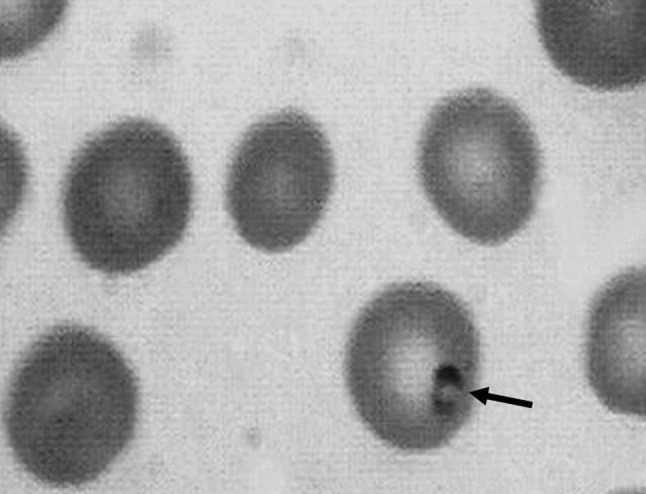
Blood smear was analyzed by Giemsa staining. Theileria like structure is detectable in one of the erythrocytes (arrow)
Table 1.
Comparison of results of PCR assay and microscopic examination for Theileria spp. in 150 cattle blood samples
| PCR assay results | Microscopy results | |
|---|---|---|
| 50 fields | ||
| + | – | |
| + | 10 | 32 |
| – | 1 | 107 |
The percentage of erythrocytes harboring Theileria like structures varied in the positive blood samples from 10−3 to 5 × 10−1 %. The examination of 50 microscopic fields showed 57 % sensitivity and 99 % specificity compared to 100 % sensitivity and specificity for PCR (Table 2).
Table 2.
Sensitivity and specificity of microscopical methods compared to 100 % sensitivity and specificity of PCR for detection of Theileria spp. in carrier cattle
| Method | No. of samples examined | No. of positives detected | Sensitivitya (%) | Specificityb (%) |
|---|---|---|---|---|
| PCR | 150 | 42 | 100 | 100 |
| Microscopy (50 fields) | 150 | 11 | 57 | 99 |
aCalculated as follows: [number of true positives/(number of true positives + number of false negatives)] × 100
bCalculated as follows: [number of true negatives/(number of true negatives + number of false positives)] × 100
The κ coefficient between PCR and Microscopy (50 fields) indicated no level of agreement (−1.06).
Discussion
The Giemsa stained of blood smear is the most used method for the identification and characterisation of these piroplasms in Iran. This method accompanied with some technical problems cause false morphological diagnosis, and in some cases needs special diagnostic knowledge (Shayan and Rahbari 2005). PCR is now routinely used around the world for investigations of Theileria infections, particularly to determine carrier animals (Altay et al. 2005; Shayan and Rahbari 2005). The forward and reverse primers used in the present study was previously designed as a Theileria genus specific primer based on hyper variable region V4 of 18S rRNA gene sequences for the detection of this parasite in the carrier animals (Shayan and Rahbari 2005). Previously, the prevalence of Theileria infections in cattle was determined in central region of Iran using PCR (Noaman 2012) but sensitivity and specificity of microscopic technique in comparison with PCR is not evaluated yet.
Our results indicated no level of agreement between the microscopic examination and PCR. The examination of 50 microscopic fields showed 57 % sensitivity and 99 % specificity compared to 100 % sensitivity and specificity of PCR. With sole use of this method, 11 blood samples were identified as Theileria positive, from which 1 sample was false positive. This means that within 150 blood samples, 32 PCR positive samples were recognized as false-negative. In Iran, the most of the veterinary laboratories examine 50 microscopic fields for detection of parasites in blood smears. Although this microscopic screening is specific, this approach often lacks the desired sensitivity. The detection of Theileria microscopically requires high parasitemia, good smear preparation, proper staining and a well-trained microscopist (in spite of the fact that the technique is cheaper and easier to perform). However, microscopic examination remains the convenient technique for day-to-day diagnosis of clinical cases in the laboratory but due to the very low amount (10−3–5 × 10−1) of infected erythrocyte is not able to detect carrier cattle (Salih et al. 2007).
Conclusion
The present study showed that the microscopic examination could fulfill the desired results for the diagnosis of acute Theileriosis but for the detection of carrier animal with low parasitemia, the PCR is preferable to microscopy with 50 fields. Our results showed that for the detection of cattle infected persistently with Theileria spp., the PCR can be used as a safe method and is recommended for epidemiological studies.
Acknowledgments
This work was supported by Isfahan research center for agriculture and natural resources.
References
- Altay K, Dumanli N, Holman PJ, Aktas M. Detection of Theileriaovis infected sheep by nested PCR. Vet Parasitol. 2005;127:99–104. doi: 10.1016/j.vetpar.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Altay K, Aydın MF, Dumanli N, Aktas M. Molecular detection of Theileria and Babesia infections in cattle. Vet Parasitol. 2008;158:295–301. doi: 10.1016/j.vetpar.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Azizi H, Shiran B, Farzaneh Dehkordi A, Salehi F, Taghadosi C. Detection of Theileria annulata by PCR and its comparison with smear method in native carrier cows. Biotechnology. 2008;7:574–577. doi: 10.3923/biotech.2008.574.577. [DOI] [Google Scholar]
- Cacci S, Cammà C, Onuma M, Severini C. The b-tubulin gene of Babesia and Theileria parasites is an informative marker for species discrimination. Int J Parasitol. 2000;30:1181–1185. doi: 10.1016/S0020-7519(00)00105-3. [DOI] [PubMed] [Google Scholar]
- Friedhoff K, Bose R. Recent developments in diagnostics of some tick-borne diseases. In: Uilenberg G, Permin A, Hansen JW, editors. Use of applicable biotechnological methods for diagnosing haemoparasites. Food and Agriculture Organisation of the United Nations (FAO), Rome, Italy: Proceedings of the expert consultation, Merida, Mexico; 1994. pp. 46–57. [Google Scholar]
- Gubbels JM, Katzer F, Hide G, Jongejan F, Shiels B. Generation of a mosaic pattern of diversity in the major merozoites piroplasm surface antigen of Theileria annulata. Mol Biochem Parasitol. 2000;110:23–32. doi: 10.1016/S0166-6851(00)00253-X. [DOI] [PubMed] [Google Scholar]
- Hashemi-Fesharki R. Control of Theileria annulata in Iran. Parasitol Today. 1988;4:36–40. doi: 10.1016/0169-4758(88)90062-2. [DOI] [PubMed] [Google Scholar]
- Kirvar E, Ilhan T, Katzer F, Hooshmand-Rad E, Zweygarth C, Gerstenberg PP, Brown CG. Detection of Theileria annulata in cattle and vector ticks by PCR. Parasitol. 2000;120(3):245–254. doi: 10.1017/S0031182099005466. [DOI] [PubMed] [Google Scholar]
- Noaman V. A molecular study on Theileria and Babesia in cattle from Isfahan province, Central Iran. J Parasit Dis. 2012 doi: 10.1007/s12639-012-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos LMF, Bell-Sakyi L, Brown CGD. Immunochemical characterization of in vitro culture-derived antigens of Babesia bovis and Babesia bigemina. Vet Parasitol. 1998;76:239–249. doi: 10.1016/S0304-4017(98)00095-8. [DOI] [PubMed] [Google Scholar]
- Rahbari S, Nabian S, Shayan P. Primary report on distribution of tick fauna in Iran. Parasitol Res. 2007;101(Suppl 2):S175–S177. doi: 10.1007/s00436-007-0692-7. [DOI] [PubMed] [Google Scholar]
- Salih DA, Hassan SM, El Hussein AM. Comparisons among two serological tests and microscopic examination for the detection of Theileria annulata cattle in Northern Sudan. Prev Vet Med. 2007;81:323–326. doi: 10.1016/j.prevetmed.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Shayan P, Rahbari S. Simultaneous differentiation between Theileria sp. and Babesia sp. on stained blood smear using PCR. Parasitol Res. 2005;97:281–286. doi: 10.1007/s00436-005-1434-3. [DOI] [PubMed] [Google Scholar]
- Uilenberg G. Theileria species of domestic livestock. In: Irvin AD, Cunningham MP, Young AS, editors. Advances in the Control of Theileriosis. The Hague: Martinus Nijhoff; 1981. pp. 4–37. [Google Scholar]