Abstract
Objective(s): Breast cancer is the most common type of cancer among women worldwide. This study investigated the expression and clinical significance of activating transcription factor 3 (ATF3) in human breast cancer and its relationship with the clinical outcome of breast cancer.
Materials and Methods : ATF3 expressions were detected in 114 primary breast cancer tissues and 114 adjacent normal tissues using immunohistochemistry (IHC) assay. Categorical variables were statistically compared by chi-square or Fisher’s exact test. Survival curves were evaluated using the Kaplan-Meier method and comparisons of survival rates were tested using a Log-rank test.
Results : IHC analysis showed that the positive expression of ATF3 protein was detected in breast cancer tissue with a positive ratio of 76.3%, and the positive ATF3 expression in adjacent normal breast tissue was 13.2%, which is lower than that in breast cancer tissue samples (P<0.01). Furthermore, ATF3 expression showed significant correlation with TNM stage, invasion, lymph node metastasis and number of metastatic lymph nodes (P=0.038, P=0.029, P=0.026, and P=0.039 respectively), and did not correlate with patients’ age and tumor size (P>0.05). A significant difference in overall survival rate was found between the patients with positive expression of ATF3 protein and those with negative expression (P=0.041).
Conclusion : Increased ATF3 expression participate in the tumorigenesis, invasion and metastasis of breast cancer, and ATF3 may be useful as a new prognostic indicator for breast cancer patients.
Key Words: Activating transcription factor 3 (ATF3), Breast cancer, Clinical significance, Immunohistochemistry, Prognostic indicator
Introduction
Breast cancer is the most common type of cancer among women worldwide, and the incidence of breast cancer is increasing in the developing world (1). Risk factors for breast cancer include repro-ductive factors associated with prolonged exposure to endogenous estrogens, genetic factors, lifestyle factors, and others (1, 2). Activating transcription factor 3 (ATF3) is a member of the ATF/CREB family of transcription factors, which shares the basic region leucine zipper DNA binding motif and binds to the ATF/CRE consensus sequence TGACGTCA(3-5). Overwhelming evidence indicates that ATF3 gene expression may be upregulated by a variety of stress signals during their development (6, 7). However, until recently, there have been no studies investigating the clinical significance of ATF3 in human breast cancer.
In this study, we collected 114 cases of breast cancer and 114 cases of adjacent non-tumor breast tissues, and explored the expression of ATF3 protein to determine whether or not ATF3 expression influences breast cancer malignancy and clinical outcome.
Materials and Methods
Patients
A total of 114 patients who underwent potentially curative resection for breast cancer at the Department of General Surgery, the 2nd Affiliated Hospital of Soochow University from January 2009 to December 2011 were eligible for this study, ranging in age from 24 to 88 years (median age 53.5 years) (Table 1). There are 15 intraductal carcinoma, 42 infiltrative ductal carcinoma, 44 infiltrative lobular carcinoma, and 13 other infiltrative carcinoma.
Table 1.
Relationship between the expression of ATF3 and the clinical pathological characteristics in breast cancer tissues
| Characteristic | n | Expression of ATF3 | P - value | |
|---|---|---|---|---|
| Positive (%) | Negative (%) | |||
| Age (yr) | 0.871 | |||
| ≤50 | 48 | 36 (75.0) | 12 (25.0) | |
| >50 | 66 | 51 (77.3) | 15 (22.7) | |
| Primary tumor size (cm) | 0.206 | |||
| ≤2 | 18 | 9 (50.0) | 9 (50.0) | |
| >2, ≤5 | 90 | 72 (80.0) | 18 (20.0) | |
| >5 | 6 | 6 (100.0) | 0 (0.0) | |
| TNM stage* | 0.038 | |||
| I-II | 46 | 28 (60.9) | 18 (39.1) | |
| III-IV | 68 | 59 (86.8) | 9 (13.2) | |
| Invasion | 0.029 | |||
| No | 15 | 3 (20.0) | 12 (80.0) | |
| Yes | 99 | 82 (82.8) | 17 (17.2) | |
| Lymph node metastasis | 0.026 | |||
| No | 28 | 11 (39.3) | 17 (60.7) | |
| Yes | 86 | 75 (87.2) | 11 (12.8) | |
| Number of metastatic lymph nodes | 0.039 | |||
| 0 | 28 | 11 (39.3) | 17 (60.7) | |
| 1-3 | 29 | 15 (51.7) | 14 (48.3) | |
| 4-9 | 37 | 30 (81.1) | 10 (18.9) | |
| ≥10 | 20 | 19 (95) | 1 (5) | |
ATF3: activating transcription factor 3; *According to the 7th AJCC (American Joint Committee on Cancer) TNM classification for breast cancer
Eligibility criteria for this study included: 1) histologically proven primary breast cancer, 2) no history of mastectomy or other malignancy, 3) a lack of noncurative surgical factors except for distant metastasis (such as liver, lung, brain, or bone-marrow metastasis) and supraclavicular lymph node metastasis, 4) axillary lymph node dissection (ALND) performed, and 5) no patients died during the initial hospital stay or for 1 month after surgery, 6) no neoadjuvant therapy (including chemotherapy, radiotherapy, and hormone therapy).
Immunohistochemistry
Tissue specimens of primary breast cancer samples and adjacent normal tissue samples from 114 patients were fixed in 10% formalin and paraffin-embedded according to standard procedures. Tissue sections of 4 μm were prepared, and endogenous peroxidase activity was blocked by incubation in 0.5% H2O2 for 10 min. Microwave antigen retrieval was performed in 0.1 mol/l citrate buffer (pH 6.0) at 750 W for 10 min. To reduce nonspecific binding, the slides were incubated with 1% normal goat serum for 30 min at room temperature. After washing three times with phosphate buffered saline (PBS), the slices were incubated with mouse anti-human ATF3 antibody (1:50; Abcam) at 4°C overnight. After centrifuging three times with PBS, the specimens were incubated with mouse secondary antibody (1:200; DAKO, Canada) at room temperature for 1 hr. Finally, the centrifuged slices were stained with diaminobenzidine (DAB) as a chromogen. PBS-only stained sample was used as a background control.
The grade of staining was determined independ-ently by two experienced pathologists. Staining intensity (0 to 3: weakly intense to strongly intense) and the proportion of stained cells (0 to 3: no cells stained to more than 50% cells stained) were determined as described. A cumulative score of ≥3 was considered to be positive expression.
Follow-up
After curative surgery, all patients were followed every half a year or until death. The median follow-up time was 31 months (range, 13-48 months). The follow-up of all patients who were included in this study was completed in January 2013.
Statistical analysis
All statistical analyses were performed with statistical analysis program package SPSS version 18.0 (SPSS Inc., Chicago, USA). Categorical variables were statistically compared with chi-square or Fisher’s exact test. Survival curves were evaluated using Kaplan-Meier method and comparisons of survival rates were tested using a Log-rank test. A P value of less than 0.05 was considered statistically significant.
Results
E xpression of ATF3 in breast cancer and adjacent normal breast tissues
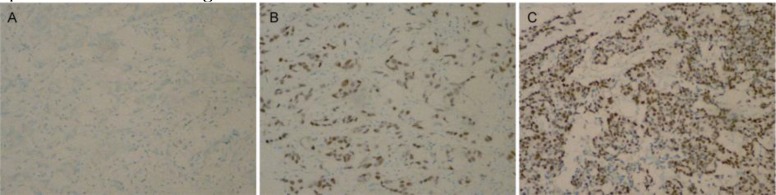
ATF3 expression was undetectable in the negative control samples (Figure 1A) and ATF3 protein was weakly expressed mostly in the adjacent normal breast tissues (Figure 1B), however, in breast cancer cells, there was a remarkable increase (Figure 1C). ATF3 was observed in the nuclei of breast cancer cells and typically appeared as buffy granulo-staining (Figure 1). The positive expression rate of ATF3 was 76.3% (87/114) in breast cancer tissues, whereas the rate was 13.2% (15/114) in adjacent normal breast tissues. The difference in ATF3 expression between breast cancer and adjacent normal breast tissues was statistically significant (P=0.009).
Figure 1.
Detection of ATF3 expression in breast cancer tissues and adjacent normal tissues by immunohistochemistry assay (DAB, ×40). A: Negative control; B: Adjacent normal tissues sample; C: Breast cancer tissues sample
Relation ship between the expression of ATF3 and the clinical pathological characteristics in breast cancer tissues
Positive expression rate of ATF3 was significantly correlated with TNM stage (P=0.038), invasion (P=0.029), lymph node metastasis (P=0.026), and number of metastatic lymph nodes (P=0.039), but not with patients’ age and primary tumor size (P>0.05, Table 1). As TNM stage and number of metastatic lymph nodes increased, the positive expression rate of ATF3 was higher.
Relation ship between ATF3 expression and survival
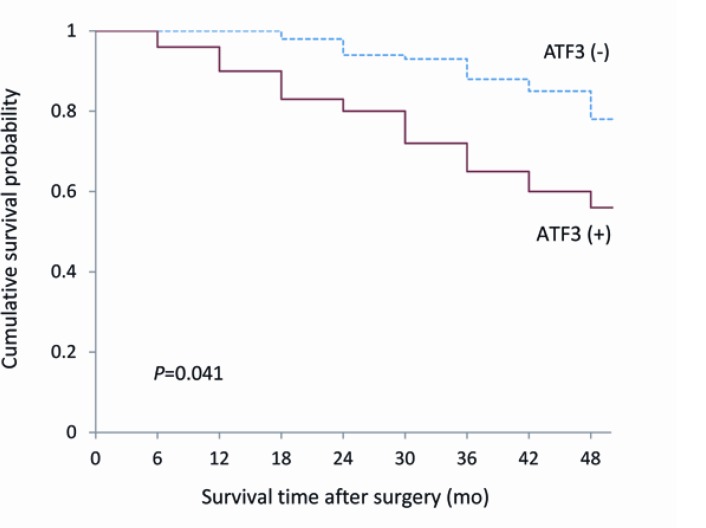
Among 114 breast cancer patients, 70 patients are alive, 31 patients died of distant metastases and 13 others died of local regional recurrence. As for the Kaplan-Meier survival analysis (Figure 2), a signi-ficant difference in overall survival rate was found between the patients with positive expression of ATF3 protein and those with negative expression (P=0.041).
Figure 2.
Kaplan-Meier survival curve for ATF3 expression (+) and ATF3 expression (–) breast cancer patients. P=0.041, Log-rank test
Discussion
ATF3 belongs to the ATF/cyclic AMP response element binding (CREB) family of transcription factors, and most normal cells have very weak or absent ATF3 expression under steady-state conditions (8-10). ATF3 is activated in most human cancers either directly or indirectly, which highlights its critical biological function as a tumor suppressor gene. However, significant upregulated ATF3 expression can be observed when cell stress is induced, which implies that ATF3 is a universal “adaptive response gene” (11, 12). In normal tissues, ATF3 may promote cell proliferation and cell apoptosis. In contrast, it has been identified in neoplasms as either an oncogene or as tumor suppressor, depending on tumor entity and grade. For example, ATF3 can mediate pro-apoptotic effects in human mammary epithelial cells. However, it may promote cell survival, motility, and invasiveness in breast cancer cells. Transgenic mice that over-express ATF3 in basal epithelial cells develop dysplastic lesions, epidermal hyperplasia, and oral squamous cell carcinoma (4, 13). Also in favor of oncogenicity, the tumor suppressor gene Drg-1 mediates its anti-metastatic properties through ATF3 downregulation in prostate cancer (4, 14, 15). Up to now, to the best of our knowledge, the relationship and molecular mechanisms of ATF3 in the progression and metastasis of breast cancer remain unclear. It is now necessary to elucidate the relationship between ATF3 and breast cancer based on the results of previous studies. We found that the ATF3 expression was markedly increased in breast cancer tissues compared to adjacent normal breast tissues (P<0.01), and ATF3 expression correlated with TNM stage, invasion, lymph node metastasis and number of metastatic lymph nodes (P=0.038, P=0.029, P=0.026, and P=0.039, respectively). Moreover, as TNM stage and number of metastatic lymph nodes increased, the positive expression rate of ATF3 was higher. This indicated that ATF3 might be involved in the tumorigenesis, invasion and metastasis of breast cancer, inferring that ATF3 might be a new tumor marker for breast cancer patients. A significant difference in overall survival rate was also found between the patients with positive expression of ATF3 protein and those with negative expression (P=0.041), suggesting that ATF3 may be a new prognostic indicator for breast cancer patients. In view of these data, ATF3 may play an oncogenic role in human breast tumorigenesis and development. However, our study has given limited data about role of ATF3 in breast cancer, and the diagnostic, predictive and prognostic value of ATF3 and its mechanism needs to be investigated thoroughly. Further studies on a large scale should be performed, and they might support the idea that ATF3 is useful as either a biomarker or therapeutic target of breast cancer, since breast cancer has been recognized as a heterogeneous disease (16).
Conclusion
Our data indicated ATF3 might be involved in the tumorigenesis, invasion and metastasis of breast cancer, and increased ATF3 expression may lead to poor prognosis, which suggests that ATF3 may play an oncogenic role in human breast tumorigenesis and development, and is useful as either a biomarker or therapeutic target of breast cancer.
References
- 1.Chávarri-Guerra Y, Villarreal-Garza C, Liedke PE, Knaul F, Mohar A, Finkelstein DM, et al. Breast cancer in Mexico: a growing challenge to health and the health system. Lancet Oncol. 2012;13:e335–e343. doi: 10.1016/S1470-2045(12)70246-2. [DOI] [PubMed] [Google Scholar]
- 2.Moody LC, Wen X, McKnight T, Chao C. Indications for sentinel lymph node biopsy in multifocal and multicentric breast cancer. Surgery . 2012;152:389–396. doi: 10.1016/j.surg.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Yan L, Della Coletta L, Powell KL, Shen J, Thames H, Aldaz CM, et al. Activation of the canonical Wnt/beta-catenin pathway in ATF3-induced mammary tumors. PLoS One . 2011;6:e16515. doi: 10.1371/journal.pone.0016515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin X, Wolford CC, Chang YS, McConoughey SJ, Ramsey SA, Aderem A, et al. ATF3, an adaptive-response gene, enhances TGF{beta} signaling and cancer-initiating cell features in breast cancer cells. J Cell Sci . 2010;123:3558–3565. doi: 10.1242/jcs.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patacsil D, Tran AT, Cho YS, Suy S, Saenz F, Malyukova I, et al. Gamma-tocotrienol induced apoptosis is associated with unfolded protein response in human breast cancer cells. J Nutr Biochem. 2012;23:93–100. doi: 10.1016/j.jnutbio.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ameri K, Hammond EM, Culmsee C, Raida M, Katschinski DM, Wenger RH, et al. Induction of activating transcription factor 3 by anoxia is independent of p53 and the hypoxic HIF signalling pathway. Oncogene . 2007;26:284–289. doi: 10.1038/sj.onc.1209781. [DOI] [PubMed] [Google Scholar]
- 7.Wang A, Arantes S, Yan L, Kiguchi K, McArthur MJ, Sahin A, et al. The transcription factor ATF3 acts as an oncogene in mouse mammary tumorigenesis. BMC Cancer. 2008;8:268. doi: 10.1186/1471-2407-8-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackl C, Lang SA, Moser C, Mori A, Fichtner-Feigl S, Hellerbrand C, et al. Activating transcription factor-3 (ATF3) functions as a tumor suppressor in colon cancer and is up-regulated upon heat-shock protein 90 (Hsp90) inhibition. BMC Cancer . 2010;10:668. doi: 10.1186/1471-2407-10-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem J . 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chromik AM, Hahn SA, Daigeler A, Flier A, Bulut D, May C, et al. Gene expression analysis of cell death induction by taurolidine in different malignant cell lines. BMC Cancer . 2010;10:595. doi: 10.1186/1471-2407-10-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai M, Jin M, Nishimura J, Dewa Y, Saegusa Y, Matsumoto S, et al. Hepatocarcinogenic susceptibility of fenofibrate and its possible mechanism of carcinogenicity in a two-stage hepatocarcinogenesis model of rasH2 mice. Toxicol Pathol . 2008;36:950–957. doi: 10.1177/0192623308327118. [DOI] [PubMed] [Google Scholar]
- 12.Kawai M, Saegusa Y, Jin M, Dewa Y, Nishimura J, Harada T, et al. Mechanistic study on hepatocarcinogenesis of piperonyl butoxide in mice. Toxicol Pathol. 2009;37:761–769. doi: 10.1177/0192623309344087. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Nguyen BC, Dziunycz P, Chang S, Brooks Y, Lefort K, et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature . 2010;465:368–372. doi: 10.1038/nature08996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene . 2008;27:2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- 15.Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) . 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taherian A, Mazoochi T. Different expression of extracellular signal-regulated kinases (ERK) 1/2 and phospho-Erk proteins in MBA-MB-231 and MCF-7 Cells after chemotherapy with doxorubicin or docetaxel. Iran J Basic Med Sci . 2012;15:669–677. [PMC free article] [PubMed] [Google Scholar]