Abstract
Four genetic polymorphisms located at the promoter (C-257T) and coding regions of CFH gene (exon 2 G257A, exon 14 A2089G and exon 19 G2881T) were investigated in 121 dengue patients (DENV-3) in order to assess the relationship between allele/haplotypes variants and clinical outcomes. A statistical value was found between the CFH-257T allele (TT/TC genotypes) and reduced susceptibility to severe dengue (SD). Statistical associations indicate that individuals bearing a T allele presented significantly higher protein levels in plasma. The –257T variant is located within a NF-κB binding site, suggesting that this variant might have effect on the ability of the CFH gene to respond to signals via the NF-κB pathway. The G257A allelic variant showed significant protection against severe dengue. When CFH haplotypes effect was considered, the ancestral CG/CG promoter-exon 2 SNP genotype showed significant risk to SD either in a general comparison (ancestral × all variant genotypes), as well as in individual genotypes comparison (ancestral × each variant genotype), where the most prevalent effect was observed in the CG/CG × CA/TG comparison. These findings support the involvement of –257T, 257A allele variants and haplotypes on severe dengue phenotype protection, related with high basal CFH expression.
1. Introduction
Dengue fever is one of the most relevant arthropod-borne diseases worldwide. Infection by any dengue virus serotypes can result in dengue fever (DF), a viral acute febrile disease. However, DF occasionally progresses to dengue hemorrhagic fever (DHF), a potentially life-threatening illness characterized by vascular leakage [1]. The dengue disease severity has been correlated with viral loads [2–4], circulating viral proteins [5,6] and exacerbated complement activity [7–11]. The main proposed mechanism leading to DHF development is that the presence of non-neutralizing antibodies against different dengue virus genotypes that facilitate the virus uptake into the cells expressing Fc receptors and enhancing the virus infectivity; thus leading to higher levels of circulating viral loads [12,13]. Several host genetic factors [2–4,10,12–19] are known to be involved with either predisposition or protection of some individuals against DHF. Among these factors, some complement system genes, such as C1q, C4 and MBL [10,11,20], have been shown as important elements of innate immunity response to the dengue progress.
The complement system has been found activated at high levels in DHF patients, as illustrated by the correlation between vasoactive anaphylatoxins (C3a and C5a) and C5b-9 and dengue severity [7,9,21,22]. Additional evidence suggests the involvement of the alternative complement pathway in the abnormal complement activation [7,11]. Soluble dengue NS1 has been shown to activate the complement in the absence of antibodies [8,9], whereas the complement factor H (CFH), a regulatory protein that inactivates the alternative pathway C3 convertase [23], has been found at low plasmatic levels in DHF patients [11]. Interestingly, other flaviviruses, such as West Nile, use the NS1 to bind to the factor H and evade the immune system [24].
CFH is an abundant protein essential to maintain the complement homeostasis and restrict the complement action to specific (infected) cell surfaces. The normal levels of factor H in human plasma vary significantly in the population and it is estimated that 63% of this variation is due to genetic polymorphisms [25]. CFH gene (1q32, locus ID: 3075) encodes for the CFH. The CFH promoter was characterized by Willians et al. [26] and the conserved transcription starting sites were described. To date, the single nucleotide polymorphism (SNP) NCBI database reported some SNPs located within the promoter and CFH gene [25].
We have sequenced a portion of the CFH gene promoter region and three exons (2, 14 and 19) in samples from a dengue patient cohort [27,28] to investigate potential gene polymorphisms, relating them with the factor H expression and dengue severity. It was found that the variant named C-257T (rs3753394), located in a putative NF-B responsive element biding site: GGATAT[T/C]ACC [29], is associated with higher factor H plasmatic levels and reduced susceptibility to develop severe dengue (SD). Also, G257A (Val/Ile, rs800292) locus and C-257T-G257A haplotypes were analyzed showing significant effects on the dengue phenotype. These findings support the involvement of –257T, 257A variant allele and haplotypes on the severe dengue phenotype protection, possible related with the high CFH expression in patients.
2. Materials and methods
2.1. Patients and clinical definitions
Patients with dengue-related symptoms were screened from three hospitals in the city of Recife, Brazil. After a full explanation of the proposed study, only patients who consented to participate were enrolled (study reviewed and approved by the ethics committee of the Brazilian Ministry of Health, under the number 4909; Process 25000.119007/2002-03; and also reviewed by the Johns Hopkins IRB according to the protocol JHM-IRB-3: 03-08-27-01). Total blood from enrolled patients was processed and submitted to laboratorial exams that included: hematocrit, hemogram, white blood cell count, differential leucocyte count, platelet count, serum albumin, serum aspartate transaminase – AST, serum alanine transaminase – ALT (performed by insured laboratories), ELISA for IgM and IgG evaluation [28]. Dengue virus (serotype 3) was detected by reverse transcriptase–polymerase chain reaction (RT-PCR) according to Lanciotti et al. (1992) [30] and the virus isolation in C6/36 cell line was identified by immunofluorescence test [31] with serotype-specific and anti-dengue monoclonal antibodies (Bio-Manguinhos, Fundação Oswaldo Cruz, Brazil). After clinical and laboratorial evaluations, a subset of 121 well-characterized dengue cases was selected for genotyping the CFH promoter (Table 1). All personal information and clinical/laboratorial results were handled confidentially and integrated into a customized digital database that includes the research results, and the inventories of cryopreserved samples of Peripheral blood mononuclear cells (PBMCs), plasma and serum [32].
Table 1.
Dengue patient stratification according to sex, serological history, clinical classification and age.
| Sex | Type of infection* | Course of infection | Age | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Male (n = 61) | Female (n = 60) | Primary (n = 46) | Secondary (n = 70) | DF (n = 34) | DS (n = 87) | Years |
| 9 | 10 | 8 | 9 | 10 | 9 | 0–15 |
| 4 | 4 | 3 | 4 | - | 8 | 16–20 |
| 8 | 5 | 10 | 3 | 3 | 10 | 21–25 |
| 6 | 10 | 6 | 9 | 5 | 11 | 26–30 |
| 9 | 5 | 7 | 9 | 2 | 12 | 31–35 |
| 5 | 7 | 6 | 7 | 3 | 9 | 36–40 |
| 8 | 3 | 2 | 8 | 3 | 8 | 41–45 |
| 6 | 3 | 1 | 8 | 3 | 6 | 46–50 |
| 6 | 13 | 3 | 13 | 5 | 14 | 51–76 |
Five patients did not have information about type of infection.
Dengue cases were classified following the World Health Organization (WHO) guidelines [33]. According to these guidelines, DF is characterized by a high fever accompanied by at least two of the associated symptoms: severe headache, retro-orbital pain, myalgia, arthralgia, and rash. DHF is defined by the same clinical manifestations as for DF, but with the addition of hemorrhagic manifestations including: positive tourniquet test, thrombocytopenia (platelet count <100,000/mm3), hemoconcentration or other sign of plasma leakage. Dengue virus was identified by immunofluorescence test, with specific-serotype, anti-dengue monoclonal antibodies (Bio-Manguinhos, Fundação Oswaldo Cruz, Brazil). In addition, we applied one more classification, namely, “dengue fever complicated” (DFC), which refers to a subset of DF patients who develop thrombocytopenia but do not completely fulfill the WHO criteria for DHF [28]. For statistical analysis, data for DHF and DFC patients, both with thrombocytopenia phenotype manifestation, were analyzed together as the “severe dengue” group. The definition about acute and convalescent phase was made base on [11]. Convalescent samples were considered a set of samples from the same individuals collected 1–9 months after the onset of symptoms.
The HLA diversity of our population was compared with two other major HLA databases from Sao Paulo and Minas and showed great correlation indicating that the cohort in this study is representative of the genetic mixture present in Brazil. The allele and genotype frequencies from dengue patients and healthy control were not statistically different [34].
2.2. Peripheral blood mononuclear cells (PBMCs) Isolation
PBMCs were isolated from whole blood by centrifugation (931g, 30 min) on a Ficoll-PaqueTM PLUS gradient (Amersham Biosciences, Uppsala, Sweden). Mononuclear cells were collected from the interface and washed in cold phosphate-buffered saline. After centrifugation (335g, 15 min) the supernatant was discarded and the pellet was washed in ACK (Ammonium–Chloride–Potassium) lysing buffer (Gibco BRL, Gaithersburg, Md.) to remove residual red blood cells. PBMCs were re-suspended with supplemented culture medium (RPMI 1640 medium containing 10% FBS, 100 U/ml penicillin/streptomycin, 2 mM L-glutamine and 10% DMSO) and cryopreserved.
2.3. CFH genotyping
The CFH promoter region of 93 healthy Brazilian blood donors and 121 dengue-positive selected were sequenced for determination of the genotype and allele frequencies as described below. Genomic DNA was extracted from PBMCs of the patients by using the Wizard DNA extraction kit (Promega, Madison, MA) following the manufacturer’s protocol. Dengue patients CFH promoter genotyping was performed by direct sequencing. The following primers (fw 5′-CAAGCACTGCATTCTTGGCA-3′ and rev 5′-GCTAGGGAAATTCTCCGTTG- 3′) were used for PCR amplification of the CFH promoter region, between –443 and –195 CFH promoter position, where two polymorphic locus are known: one at –257 position (C/T rs3753394) [35] and another at –196 position (C/G rs35046519); however, the second allele variant was not found in our population. The experimental protocol was as follows: 38.5 μl H2O, 5 μl PCR buffer (Invitrogen), 2 μl 50 mM MgCl2, 1 μl 10 mM dNTPs, 1 μl 10 mM of each primer, 1 μl DNA template (1 μg/μl) and 0.5 μl Taq Polimerase (5 U/μl, Invitrogen) to 50 μ. The measurements have started, performed neatly, as shown in figure. The amplification reaction was performed with the GeneAmp® PCR system 9700 using the following cycling profile: 96 °C/3 min, 35 cycles of: 94 °C/30 s, annealing 50 °C/30 s, extension 72 °C/30 s), and 72 °C/10 min final extension. PCR products of 248 pb were purified from agarose gel after electrophoresis by using the MinElute™ PCR purification Kit (Qiagen) and sequenced. Complete DNA sequences were obtained from the PCR product encompassing the –337 to –195 region (142 pb DNA length) were obtained using the Big Dye Terminator sequencing kit (Applied Biosystems) and the reactions were run on a ABI 3100 Genetic Analyzer (Applied Biosystems). Sequences were handled by using the SeqScape 1.0 software (Applied Biosystem).
TaqMan Applied Genotyping Assay (ID C___2530387_10) was used only to genotyping the C-257T locus in control population following the recommended protocol. To the exon 2 rs800292, exon 14 rs3753396 exon 19 and rs1065489 SNPs the control and dengue cohort were genotyped by DNA sequencing as described before, using fw 5′-GGAGGATGACCACCCCTTTTTG-3′ and rev 5′-CCAA ACATATCCAGAAGGCACC-3′; fw 5′-AGCAAGTACAATCATGTGGT CC-3′ and rev 5′-GTAATAAGGAGGGGAAGAAAGCTGG-3′; fw 5′-TCCGATAGACAGACAGACACCAG-3′ and rev 5′-AGCTATAATTTCCCA CAGCAGTCC-3′ primers, respectively.
2.4. Factor H plasma determination
Sandwich ELISA was used to measure plasma level of factor H during both acute and convalescent phases as previously described in [11]. In that work, CFH measurements were performed in healthy subjects, therefore it was not carried out here. Briefly, Nunc microplates were coated with goat serum anti-human Factor H (Calbiochem) overnight at 4 °C. After washing, the plates were blocked with 5% (w/v) BSA (Sigma) for 1 h at 37 °C and then washed and incubated 2 h at 37 °C with either the plasma from dengue patients or purified human factor H (Calbiochem). After washing, the plates were incubated for 1 h at 37 °C with mouse monoclonal antibody anti-human factor H (Abcam) followed by washing and one hour incubation at 37 °C with goat anti-mouse IgG-HRP (Jackson ImmunoResearch). The reaction was developed with TMB substrate reagent set (BD Biosciences) and read at 450 nm in a microplate reader (Safari2, Tecan).
2.5. Statistical analysis
The risk analysis and plotting were carried out using the opensource R statistical package version 2.5.0, and PRISM version 4.0a. For association between genotype category and diagnostic class, Fisher’s statistical test was performed, using the R function fisher test, which was also used to find the associated OR, using a conditional maximum-likelihood method. Estimation of gametic phase (haplotypes) from multi-locus diploid data was based on a Gibbs sampling strategy, using Arlequin software (Excoffier). Comparison between CFH genotypes and serum factor H was performed in PRISM via the two-sample t-test with Welch’s correction at 95% significance level.
3. Results
3.1. CFH genotyping
One hundred and twenty one DNA samples from well-characterized dengue (DENV-3 genotype III, Srilankan–India strain) patients were used to sequence the CFH promoter, exon 2, 14 and 19. Only the polymorphism –257T, previously deposited in the SNP database (rs3753394) was found in the CFH promoter in our population (the known –196 position was not found). Dengue patient genotypes according to C-257T variant locus were defined as CC (54%), CT (39%) or TT (7%) TT homozygous (Table 2). The control group (healthy subjects) genotyping was determined using Applied Genotyping Assay and the frequencies observed were very similar to the dengue patient group (CC 56%, CT 39% and TT 5%) (Table 2). Allelic frequencies of both healthy control subjects and dengue patients are in Hardy–Weinberg equilibrium.
Table 2.
Four CFH variant loci genotypes and alleles frequencies in dengue patients and healthy control.
| rs3753394 (C-257T) | Dengue (n = 121/242*) | Healthy (n = 93/186*) |
|---|---|---|
| CC | 66 (54%) | 52 (56%) |
| CT | 47 (39%) | 36 (39%) |
| TT | 8 (7%) | 5 (5%) |
| C | 179 (74%) | 140 (75%) |
| T | 63 (26%) | 46 (25%) |
| rs800292 (G257A) | ||
| GG | 72 (59%) | 52 (56%) |
| GA | 35 (29%) | 31 (33%) |
| AA | 14 (12%) | 10 (11%) |
| G | 179 (74%) | 135 (72.5%) |
| A | 63 (26%) | 51 (27.5%) |
| rs3753396 (A2089G) | ||
| AA | 91 (75%) | 64 (69%) |
| AG | 28 (23%) | 27 (29%) |
| GG | 2 (2%) | 2 (2%) |
| A | 210 (87%) | 155 (83%) |
| G | 32 (13%) | 31 (17%) |
| rs1065489 (G2881T) | ||
| GG | 89 (73%) | 67 (72%) |
| GT | 29 (24%) | 25 (27%) |
| TT | 3 (3%) | 1 (1%) |
| G | 207 (86%) | 159 (85.5%) |
| T | 35 (14%) | 27 (14.5%) |
Number of chromosomes.
Three others variant loci in the CFH gene were observed showing the following genotype frequencies: G257A GG (0.59), GA (0.29) and AA (0.12); A2089G AA (0.75), AG (0.23) and GG (0.02); G2881T GG(0.73), GT(0.24) and TT(0.03). The G257A non-synonymous locus was in H.W. equilibrium while the others two loci were not. The frequencies observed in the control group were similar to the dengue group (Table 2).
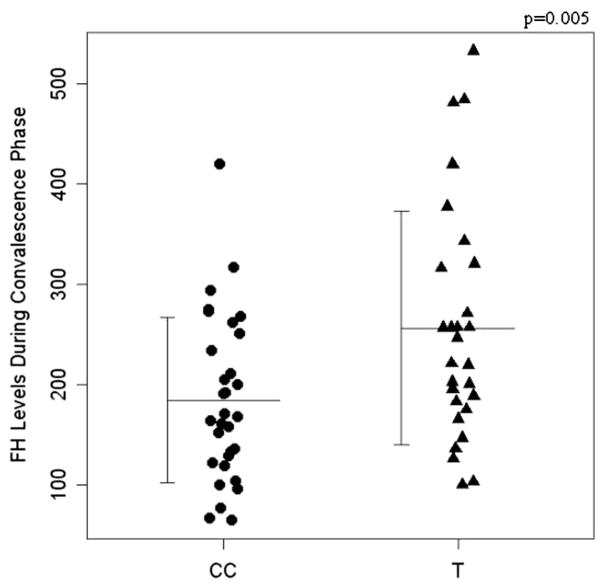
3.2. The CFH-257T allele is correlated with higher circulating levels of factor H during dengue convalescent phase
Factor H plasmatic levels were measured in healthy patients (206 ug/ml + 123) [11] and dengue samples during both acute and convalescent phases. Then, the protein level was analyzed according to their C-257T and G257A genotypes and respective haplotypes. Individuals bearing –257T variant allele (CT + TT genotypes) presented a plasmatic level of factor H that was on average about 20% higher than “CC” individuals (p = 0.005; Fig. 1) only during convalescent phase, suggesting that the –257T variant allele is correlated with the increased basal factor H production. To the 257A variant and 257T–257A haplotypes no association was found for factor H serum levels during convalescent or acute phases.
Fig. 1.
The promoter C-257T variant locus and CFH gene expression. Complement factor H plasma levels in CC and CT/TT genotype patients, during dengue convalescence phase (p = 0.005).
3.3. Dengue patients bearing CFH-257T and 257A alleles and variant haplotypes are less likely to develop SD
We tested the hypothesis that individuals encoding one of four CFH variant alleles (the promoter C-257T, exon2 G257A, exon 14 A2089G or exon 19 G2881T) could be less likely to develop severe dengue symptoms, especially if individuals with these variant genotypes could have rapidly increase of local concentration of factor H at the inflammatory sites. In order to perform such analysis, the dengue group was stratified taking into account the dengue clinical outcomes. Out of the 121 dengue patients genotyped, 34 (28%) were diagnosed with DF and 87 (72%) with severe dengue (Table 1), and the possibility of correlation between SNPs and clinical outcome was then explored.
For the C-257T locus, the results indicate association between “T” allele and mild dengue disease, suggesting that this allele (CT + TT genotypes) and related higher levels of factor H may be protective factors against the SD development by a margin larger than 2-fold. In order to test this hypothesis, we performed a two-tailed two-sample Fisher’s test (DF vs. SD; CC vs. CT + TT) that results in estimated odds ratio OR = 2.53 (using the conditional maximum-likelihood method) for protection of the T allele against severe phenotype (p = 0.001, CI = 1.38–4.69) (Table 3). In a previous study [10] we perform the DENV-3 genotype III cohort stratification using RLM method considering some biological features like sex, type of infection, age and genotypes. In that study, only patient’s age was significantly related with DHF risk development beyond genetic background.
Table 3.
Four CFH variant loci effect in a Brazilian severe dengue cohort.
| Locus | OR | p | CI | x2 | p |
|---|---|---|---|---|---|
| rs3753394 C-257T (TT + CT × CC) | 2.53 | 0.001 | 1.38–4.69 | 9.42 | 0.002148 |
| rs800292 G257A (AA + AG × GG) | 1.99 | 0.02 | 1.09–3.67 | 5.45 | 0.01957 |
| rs3753396 A2089G (GG + AG × AA) | 0.94 | 1 | 0.47–1.89 | 0.0173 | 0.8952 |
| rs1065489 G2881T (TT + GT × GG) | 1 | 1 | 0.50–1.97 | 0.0208 | 0.8853 |
The same analysis was performed to G257A (exon 2), A2089G (exon 14) and G2881T (exon 19) CFH gene polymorphisms. The results showed a weak, but significant, correlation between the 257A non-synonymous SNP (OR = 1.99, p = 0.02, CI = 1.09–3.67) and protection to SD (Table 3). The other two SNPs studied showed no relevant effect on dengue development.
Haplotypes analysis using relevant –257T and 257A CFH variations showed the presence of four haplotypes in dengue cohort: CG, CA, TG and TA (Table 4). The haplotype genotype frequencies are shown on in Tables 5 and 6. The ancestral CG/CG genotype showed a significant risk effect to SD either in a general comparison (ancestral × all variant genotype forms: OR = 2.90 (p = 0.002, CI = 1.40–6.24) (Table 5), as well in individual genotypes comparison (ancestral × each variant genotypes), where the most prevalent effect was observed in the CG/CG × CA/TG comparison with OR = 11.79 (p = 1.29−13, CI = 5.53–26.91) (Table 6).
Table 4.
C-257T/G257A CFH haplotypes frequencies in dengue patients and healthy control.
| Haplotypes | Dengue (n = 242) | Healthy (n = 186) |
|---|---|---|
| CG | 122 (50.5%) | 89 (47.85%) |
| CA | 57 (23.5%) | 51 (27.42%) |
| TG | 57 (23.5%) | 46 (24.73%) |
| TA | 6 (2.5%) | 0 |
Table 5.
General comparison of C-257T/G257A CFH haplotype effect on severe dengue phenotype.
| Genotypes (C-257T e G257A) | Severe dengue (n = 87) | Dengue fever (n = 34) |
|---|---|---|
| CG/CG | 30 (34.5%) | 5 (14.7%) |
| CG/CA, CG/TG, CG/TA, CA/CA, CA/TG, CA/TA, TG/TG, TG/TA, TA/TA) | 57 (65.5%) | 29 (85.3%) |
| OR = 2.90 | CI = 1.40–6.24 | p = 0.002 |
Table 6.
Individual comparison of C-257T/G257A CFH haplotype effect on severe dengue phenotype.
| Haplotypes genotypes | Severe dengue | Dengue fever | OR | CI | p |
|---|---|---|---|---|---|
| CG/CG | 30 (66.7%) | 5 (45.4%) | 2.46 | 1.34–4.58 | 0.002 |
| CG/CA | 15 (33.3%) | 6 (54.6%) | |||
| CG/CG | 30 (60%) | 5 (31.2%) | 3.31 | 1.79–6.24 | 6.28E–05 |
| CG/TG | 20 (40%) | 11 (68.8%) | |||
| CG/CG | 30 (78.9%) | 5 (71.4%) | 1.53 | 0.76–3.10 | 0.25 |
| CA/CA | 8 (21.1%) | 2 (28.6%) | |||
| CG/CG | 30 (88.2%) | 5 (38.5%) | 11.79 | 5.53–26.91 | 1.29E–13 |
| CA/TG | 4 (11.8%) | 8 (61.5%) | |||
| CG/CG | 30 (83.3%) | 5 (100%) | 0 | 0.00–0.21 | 7.26E–06 |
| TG/TG | 6 (16.7%) | 0 (0) | |||
| CG/CG | 30 (93.7%) | 5 (71.4%) | 6.34 | 2.41–19.71 | 2.46E–05 |
| CA/TA | 2 (6.3%) | 2 (28.6%) | |||
| CG/CG | 30 (100%) | 5 (100%) | 0 | 0–inf | 1 |
| CG/TA | 0 (0) | 0 (0) | |||
| CG/CG | 30 (93.7%) | 5 (100%) | 0 | 0.00–0.82 | 0.02 |
| TG/TA | 2 (6.3%) | 0 (0) | |||
| CG/CG | 30 (100%) | 5 (100%) | 0 | 0–inf | 1 |
| TA/TA | 0 (0) | 0 (0) |
4. Discussion
Factor H is a major regulator of the alternative pathway of the complement system, and abnormal complement responses seem to be involved in dengue pathogenesis [7,11,36]. In our study, we have analyzed the genetic polymorphisms in the promoter and encoding region of CFH gene encoding the human factor H, in a dengue patient cohort. The CFH-257T variant (CT + TT genotypes) was associated with a 2.5-fold lower risk to develop severe dengue disease and seems to have a significant impact on levels of factor H circulating in the blood. The results also showed a weak correlation between exon 257A allele and protection to SD (Table 3). When CFH haplotypes were evaluated, we observed that the homozygous ancestral genotype (CG/CG) promotes higher risk to develop severe symptoms both in general comparison (ancestral × all variant genotype forms) and individual genotypes comparison (ancestral × each variant genotypes) (Tables 5 and 6). These results are consistent with the hypothesis that regulation of the alternative pathway activity and production of factor H by immune cells at the site of inflammation may be important to prevent the development of vascular leakage and other SD symptoms [37].
Our experiments demonstrate a strong association between the –257T variant (alone) and higher complement factor H serum levels during the convalescent phase (20% more). No association was found to the other analyzed polymorphisms/haplotypes with complement factor serum levels. This suggests the existence of other functional polymorphisms forming haplotypes with the –257T variation. The chromosome region of CFH gene has a high linkage disequilibrium level and several known polymorphisms. Besides, non-synonymous variations such as G257A (Val/Ile) can promote functional protein modification without changes in gene expression (i.e. protein synthesis). In this case, a functional factor H analysis is needed to perform a real association between SNPs/haplotypes and active factor H levels.
Infected cells can activate the alternative complement pathway and the expression of factor H at inflammation sites may be critical to restrict the complement activation to the surfaces of the infected cell. The human factor H is expressed by several cell types, including monocytes, dendritic cell, endothelial cells, B-cells and keratinocytes [38]. Factor H expression in these cells can be differentially induced by interleukin-1 and 1, interferon-(IFN), IFN-, IFN-, TNF- and other cytokines [38–41]. Studies of the CFH promoter region have indicated that the C-257T variant site is located within a putative NF-B responsive element containing the sequence: GGATAT[ T/C]ACC [29]. Since signal transduction of these cytokine receptors are mediated by the NF-B, it is likely that this NF-B responsive element is involved in the transcription regulation of factor H.
During DENV infection, dengue NS1 (non structural protein 1) protein is secreted in enormous amounts in the patient’s plasma and soluble dengue NS1 has been shown to directly activate the complement in absence of antibodies, suggesting activation of complement by the alternative pathway [8,9]. In addition, secreted dengue NS1 adheres to the surfaces of several non-infected cells types such as hepatocytes, endothelial cells, monocytes, and fibroblasts and preferentially binds to cultured human microvascular [37]. Recent study has shown that the West Nile (flavivirus) NS1 protein can bind to soluble factor H, bringing it to the infected host cells as a complement-mediated lyses evading mechanism [24,42]. Although this mechanism does not seem to be present in other flavivirus such as in dengue and JEV [42], we postulate that the increased expression of factor H at systemic and local level may enable better complement regulation and protection of non-infected bystander cells from complement attack.
CFH polymorphisms have been associated with hemolytic uraemic syndrome [43], including the promoter, exon 14 and exon 19 SNPs evaluated by us [35]. HUS is a non-immune haemolytic anemia that promotes microcirculation disorders including vascular leakage and edema, ranging from thrombocytopenia to kidney failure. Although the HUS and dengue are quite different diseases, they are also characterized by some common clinical manifestations involving thrombocytopenia and vascular abnormalities. It is noticeable that in a previous report the same –257T variant allele was strongly related to HUS development [35], but in that case the T allele means increased risk. However, in the HUS study the factor H levels were similar in different CFH C-257T allele bearing patients. However, since the exon 14 and exon 19 CFH SNPs were not found associated with dengue outcome, like in HUS, it suggests that the HUS development may be defined by a different haplotype combination then the one we found in dengue.
Based on this evidence we suggest that higher basal levels of factor H associated with CFH-257T variant can lead to better complement regulation in the plasma during dengue infection and protection of non-infected immune cells, restricting the complement activation to the virus infected cells, which results in better control of infection control and reduced pathology.
Acknowledgments
Funding
The author also acknowledge the support of the National Institute of Allergy and Infectious Diseases (NIAID/NIH), under Grants U19 AI56541 and N01 AI 40085, the Brazilian National Research Council (CNPq), under Grant DCR 35.0382/2004.2 (to Dr. Braga-Neto), and the FACEPE (Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco, Brazil) for A. Pastor‘s fellowship.
The authors thank all staff of the Instituto Materno Infantil de Permanbuco, Hospital Esperança, and Hospital Santa Joana for their valuable cooperation with this project.
Abbreviations
- DENV
dengue virus
- DF
dengue fever
- SD
severe dengue
- CFH
complement factor H
- OR
odds ratio
References
- 1.Gubler DKG. Dengue and dengue hemorrhagic fever. London: 1997. [Google Scholar]
- 2.Paradoa Perez ML, Trujillo Y, Basanta P. Association of dengue hemorrhagic fever with the HLA system. Haematologia Budap. 1987;20:83–7. [PubMed] [Google Scholar]
- 3.Soundravally R, Hoti SL. Immunopathogenesis of dengue hemorrhagic fever and shock syndrome: role of TAP and HPA gene polymorphism. Hum Immunol. 2007;68:973–9. doi: 10.1016/j.humimm.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Sakuntabhai A, Turbpaiboon C, Casademont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–13. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang WK, Chao DY, Kao CL, Wu HC, Liu YC, Li CM, et al. High levels of plasma dengue viral load during defervescence in patients with dengue hemorrhagic fever: implications for pathogenesis. Virology. 2003;305:330–8. doi: 10.1006/viro.2002.1704. [DOI] [PubMed] [Google Scholar]
- 6.Libraty DH, Endy TP, Houng HS, Green S, Kalayanarooj S, Suntayakorn S, et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185:1213–21. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 7.Bokisch VA, Muller-Eberhard HJ, Dixon FJ. The role of complement in hemorrhagic shock syndrome (dengue) Trans Assoc Am Physicians. 1973;86:102–10. [PubMed] [Google Scholar]
- 8.Avirutnan P, Malasit P, Seliger B, Bhakdi S, Husmann M. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J Immunol. 1998;161:6338–46. [PubMed] [Google Scholar]
- 9.Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, Auethavornanan K, et al. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J Infect Dis. 2006;193:1078–88. doi: 10.1086/500949. [DOI] [PubMed] [Google Scholar]
- 10.Acioli-Santos B, Segat L, Dhalia R, Brito CA, Braga-Neto UM, Marques ET, et al. MBL2 Gene polymorphisms protect against development of thrombocytopenia associated with severe dengue phenotype. Hum Immunol. 2008;69:122–8. doi: 10.1016/j.humimm.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Nascimento EJ, Silva AM, Cordeiro MT, Brito CAA, Gil LHVG, Braga-Neto U, et al. Alternative complement pathway deregulation is correlated with dengue severity. PLoS ONE. 2009;4(8):e6782. doi: 10.1371/journal.pone.0006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sierra B de la C, Kouri G, Guzman MG. Race: a risk factor for dengue hemorrhagic fever. Arch Virol. 2007;152:533–42. doi: 10.1007/s00705-006-0869-x. [DOI] [PubMed] [Google Scholar]
- 13.Loke H, Bethell DB, Phuong CX, Dung M, Schneider J, White NJ, et al. Strong HLA class I–restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. 2001;184:1369–73. doi: 10.1086/324320. [DOI] [PubMed] [Google Scholar]
- 14.Burke DS, Nisalak A, Johnson DE. Scott RM A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–80. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 15.Stephens HA, Klaythong R, Sirikong M, Vaughn DW, Green S, Kalayanarooj S, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–18. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 16.Chiewsilp P, Scott RM, Bhamarapravati N. Histocompatibility antigens and dengue hemorrhagic fever. Am J Trop Med Hyg. 1981;30:1100–5. doi: 10.4269/ajtmh.1981.30.1100. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Mestre MT, Gendzekhadze K, Rivas-Vetencourt P, Layrisse Z. TNF-alpha- 308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64:469–72. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 18.Loke H, Bethell D, Phuong CX, Day N, White N, Farrar J, et al. Susceptibility to dengue hemorrhagic fever in vietnam: evidence of an association with variation in the vitamin d receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67:102–6. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- 19.Fang X, Hu Z, Shang W, Zhu J, Xu C, Rao X. Genetic polymorphisms of molecules involved in host immune response to dengue virus infection. FEMS Immunol Med Microbiol. 2012:1–13. doi: 10.1111/j.1574-695X.2012.00995.x. [DOI] [PubMed] [Google Scholar]
- 20.Tan GK, Alonso S. Pathogenesis and prevention of dengue virus infection: state-of-the-art. Curr Opin Infect Dis. 2009;22:302–8. doi: 10.1097/QCO.0b013e328329ae32. [DOI] [PubMed] [Google Scholar]
- 21.Nishioka K. Serum complement level in dengue hemorrhagic fever. Allerg Immunol (Leipz) 1974;20–21:385–92. [PubMed] [Google Scholar]
- 22.Wang WK, Chen HL, Yang CF, Hsieh SC, Juan CC, Chang SM, et al. Slower rates of clearance of viral load and virus-containing immune complexes in patients with dengue hemorrhagic fever. Clin Infect Dis. 2006;43:1023–30. doi: 10.1086/507635. [DOI] [PubMed] [Google Scholar]
- 23.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat immunol. 2010;11(9):785–97. doi: 10.1038/ni.1923. New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, et al. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci USA. 2006;103:19111–6. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Cordoba SR, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin Exp Immunol. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams SA, Vik DP. Characterization of the 5′ flanking region of the human complement factor H gene. Scand J Immunol. 1997;45:7–15. doi: 10.1046/j.1365-3083.1997.d01-364.x. [DOI] [PubMed] [Google Scholar]
- 27.Cordeiro MT, Schatzmayr HG, Nogueira RM, Oliveira VF, Melo WT, Carvalho EF. Dengue and dengue hemorrhagic fever in the State of Pernambuco, 1995–2006. Rev Soc Bras Med Trop. 2007;40:605–11. doi: 10.1590/s0037-86822007000600001. [DOI] [PubMed] [Google Scholar]
- 28.Cordeiro MT, Silva AM, Brito CA, Nascimento EJ, Magalhaes MC, Guimarães GF, et al. Characterization of a dengue patient cohort in Recife. Brazil Am J Trop Med Hyg. 2007;77:1128–34. [PubMed] [Google Scholar]
- 29.Warwicker P, Goodship TH, Goodship JA. Three new polymorphisms in the human complement factor H gene and promoter region. Immunogenetics. 1997;46:437–8. doi: 10.1007/s002510050300. [DOI] [PubMed] [Google Scholar]
- 30.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–51. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gubler DJ, Kuno G, Sather GE, Velez M, Oliver A. Use of mosquito cell cultures and specific monoclonal antibodies in surveillance for dengue virus. Am J Trop Med Hyg. 1984;33:158–65. doi: 10.4269/ajtmh.1984.33.158. [DOI] [PubMed] [Google Scholar]
- 32.Marques ET, Jr, Maciel Filho R, August PN. Overcoming health inequity: potential benefits of a patient-centered open-source public health infostructure. Cad Saude Publica. 2008;24:547–57. doi: 10.1590/s0102-311x2008000300008. [DOI] [PubMed] [Google Scholar]
- 33.Organization WH. The World Health Report. 2004. Dengue: guidelines for diagnosis treatment, prevention, and control. New ed. [PubMed] [Google Scholar]
- 34.Alencar LXE, Braga-Neto UL, Nascimento EJM, Cordeiro MT, Silva AM, et al. HLA-B/44 is Associated with Dengue Severity Caused by DENV-3 in a Brazilian Population. Journal of Tropical Medicine. 2013;2013:11. doi: 10.1155/2013/648475. Article ID 648475. http://dx.doi.org/10.1155/2013/648475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caprioli J, Castelletti F, Bucchioni S, Bettinaglio P, Bresin E, Pianetti G, et al. Complement factor H mutations and gene polymorphisms in haemolytic uraemic syndrome: the C-257T, the A2089G and the G2881T polymorphisms are strongly associated with the disease. Hum Mol Genet. 2003;12:3385–95. doi: 10.1093/hmg/ddg363. [DOI] [PubMed] [Google Scholar]
- 36.Shresta S. Role of Complement in Dengue Virus Infection: Protection or Pathogenesis? mBio. 2012;3(1):e00003–12. doi: 10.1128/mBio.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, et al. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007;3:e183. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friese MA, Hellwage J, Jokiranta TS, Meri S, Peter HH, Eibel H, et al. FHL-1/reconectin and factor H: two human complement regulators which are encoded by the same gene are differently expressed and regulated. Mol Immunol. 1999;36:809–18. doi: 10.1016/s0161-5890(99)00101-7. [DOI] [PubMed] [Google Scholar]
- 39.Timar KK, Pasch MC, van den Bosch NH, Jarva H, Junnikkala S, Meri S, et al. Human keratinocytes produce the complement inhibitor factor H: synthesis is regulated by interferon-gamma. Mol Immunol. 2006;43:317–25. doi: 10.1016/j.molimm.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Luo W, Vik DP. Regulation of complement factor H in a human liver cell line by interferon-gamma. Scand J Immunol. 1999;49:487–94. doi: 10.1046/j.1365-3083.1999.00528.x. [DOI] [PubMed] [Google Scholar]
- 41.Halme J, Sachse M, Vogel H, Giese T, Klar E, Kirschfink M. Primary human hepatocytes are protected against complement by multiple regulators. Mol Immunol. 2009;46:2284–9. doi: 10.1016/j.molimm.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Krishna VD, Rangappa M, Satchidanandam V. Virus-specific cytolytic antibodies to nonstructural protein 1 of Japanese encephalitis virus effect reduction of virus output from infected cells. J Virol. 2009;83:4766–77. doi: 10.1128/JVI.01850-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furlan M, Lammle B. Haemolytic-uraemic syndrome and thrombotic thrombocytopenic purpura–new insights into underlying biochemical mechanisms. Nephrol Dial Transplant. 2000;15:1112–4. doi: 10.1093/ndt/15.8.1112. [DOI] [PubMed] [Google Scholar]