Abstract
Hepatitis C Virus (HCV) is a leading cause of liver disease and represents a significant public health challenge. Treatments for this disease are inadequate and improved antiviral therapies are necessary. Several such antivirals are in development, most of which target the well-characterized NS3 protease or the NS5B polymerase. In contrast, the nonstructural 4B (NS4B) protein, though essential for HCV RNA replication, has been the subject of few pharmacological studies. One of the functions ascribed to this protein is the ability to form intracellular membrane-associated foci (MAF), which are believed to be related to the sites of viral replication. Here, we report the identification of a small molecule that inhibits HCV replication and disrupts the organization of these MAF. Genetic analysis links the compound’s mode of action to the NS4B gene product, and transient transfections of NS4B-GFP demonstrate that treatment with this compound can lead to the formation of novel elongated assemblies of NS4B. Furthermore, an in vitro dynamic light scattering assay provides evidence that the second amphipathic helix of NS4B may be the target of the drug. Our results demonstrate that this molecule represents a new potential class of HCV inhibitors and also provides us with a useful tool for studying the HCV life cycle.
Keywords: HCV, inhibitor, NS4B, amphipathic helix
1. Introduction
Worldwide, 170 million people are infected with Hepatitis C Virus (HCV), a (+) sense, single-stranded RNA virus (Pawlotsky, 2004). These individuals experience significant morbidity related to their infection, and mortality rates are high. Moreover, treatment options for this disease are inadequate for the majority of patients. The standard of care regimen, which relies on pegylated interferon-α plus ribavirin, is successful in only about 50% of patients; moreover, it is associated with significant side effects (Moradpour et al., 2007). Therefore, better antiviral therapies are needed to address this public health problem.
As a better picture of the molecular biology of HCV has emerged, a number of specific antivirals have been identified, and several pharmaceuticals that target the viral nonstructural proteins NS3 and NS5B have progressed into phase II clinical trials (Manns et al., 2007). Notably, resistant mutants have already been observed for both these types of compounds (Mo et al., 2005); therefore, it is likely that a combination of antivirals will provide the best sustained response in patients.
One potential target for a complementary antiviral is the HCV nonstructural 4B (NS4B) protein. Several functions have been ascribed to this protein, including the ability to rearrange cellular membranes into a so-called membranous web, visualizable through electron microscopy and believed to represent the viral replication platform (Egger et al., 2002), and to form membrane-associated foci (MAF), believed to reflect, in part, the light microscopic equivalent of the membranous web (Gretton et al., 2005). Furthermore, NS4B can bind and hydrolyze GTP (Einav et al., 2004), bind to HCV RNA (Einav et al., 2008), and interact physically with other nonstructural components of the HCV replication complex (Gao et al., 2004). Any one of these functions represents a potential target for pharmacological disruption.
Recently, Chunduru et al. have identified 23 small molecules that bind to NS4B with a dissociation constant (KD) to NS4B of 5 µM or less (Chunduru et al., 2005). Of those leads, 4 molecules were shown to inhibit HCV replication with an EC50 of 1 µM or less, as measured by an ELISA for NS5A protein levels. Here, we focus on one of these compounds, which we designate “anguizole.” We show that this compound indeed inhibits HCV RNA replication with little toxicity to host cells. Furthermore, we provide genetic and biochemical evidence that NS4B is the target of this drug, and we demonstrate that this compound can disrupt the subcellular distribution of MAF. Finally, our in vitro dynamic light scattering assay suggests that the second N-terminal amphipathic helix (AH2) of NS4B interacts with this compound, and we identify a mutation in the N-terminal region that confers resistance to the drug.
2. Materials and Methods
2.1 DNA constructs and peptides
Standard recombinant DNA technology was used to clone all constructs. The plasmid pEF-NS4B-GFP was cloned previously (Einav et al., 2004). To generate the H94R mutation in this construct, site-directed PCR mutagenesis was performed as described (Einav et al., 2004). The first round of amplification used the primer sets [pEF-6879F, 5’-GGCCAAGATCTGCACACTGGTATT-3’, to H94R_R, 5’-TAAACAGGAGGGTACGTTGGGTGGTGAGCGG-3’] and [H94R_F, 5’-CCGCTCACCACCCAACGTACCCTCCTGTTTA-3’, to pEF-1783R, 5’-AGGCTGATCAGCGGGTTTA-3’]. The second round of PCR amplification was achieved using the primer set pEF-6879F to pEF-1783R.
The Bart-Luciferase plasmid, Bart79ILuc*, was cloned from the Bart79I parent (Einav et al., 2004), an HCV genotype 1b Con1 sequence containing the adaptive mutation S2204I, and the pGL3-Basic parent (Promega) as follows. A NotI site was introduced after the 15th amino acid of Core in Bart79I using PCR mutagenesis. The luciferase gene was PCR amplified from the pGA3Basic plasmid using the primers NotLucS, GAATGCGGCCGCAATGGAAGACGCCAAAAACATAAAG, and LucDraAS, CGATTTAAATTACACGGCGATCTTTCCGCCC. The PCR product was ligated into the Bart79I plasmid following restriction digestions of both components with NotI and DraI. In addition, the ScaI restriction site in the Luciferase coding region was removed by introducing a silent mutation into the site by PCR mutagenesis.
A peptide comprising the second amphipathic helix region of the genotype 1b NS4B protein was synthesized (Anaspec). This peptide, Pep4BAH2, contained the sequence, H-WRTLEAFWAKHMWNFISGIQYLA-NH2.
2.2 Drugs and antibodies
7-[chloro(difluoro)methyl]-5-furan-2-yl-N-(thiophen-2-ylmethyl)pyrazolo[1,5-a]pyrimidine-2-carboxamide (Chemical Block A2828/0119446), which we refer to here as anguizole, was solubilized in DMSO, and aliquots of 10mM stocks were maintained at −20° C. Immediately prior to use, these stocks were diluted to 200-fold the desired final concentration, and this solution was added directly to the dishes.
Reagents used for fluorescence microscopy included the following antibodies: rabbit anti-calnexin (Stressgen SPA-860) rabbit anti-PDI (Stressgen SPA-890), mouse anti-α-tubulin (Sigma T9026), and Alexa Fluor® 594 conjugated secondary antibodies (Invitrogen A11012, A21044). Actin filaments and mitochondria were visualized with phalloidin-TRITC (Sigma P1951) and the Mitotracker dye (Invitrogen M7512), respectively.
2.3 Cell culture, NS4B-GFP transfections, and fluorescence microscopy
Cells of the human hepatoma cell line, Huh7.5, were cultured in monolayers as described (Sklan et al., 2007), with media consisting of DMEM (Mediatech) supplemented with 1% L-glutamine (Mediatech), 1% penicillin, 1% streptomycin (Mediatech), and 10% fetal bovine serum (Omega Scientific). Transfections of pEFNS4B-GFP constructs were performed with Lipofectamine-2000 (Invitrogen), according to the manufacturer’s instructions. Following transfection, the cells were cultured in standard media containing 5 µM anguizole and 0.5% (70 mM) DMSO. Controls were cultured in media containing 0.5% DMSO. Unless noted otherwise, the cells were treated for 48 hours, fixed in 4% formaldehyde, and mounted with Prolong® Gold antifade reagent with DAPI (Invitrogen). Slides were visualized under a Nikon E600 fluorescence microscope and images were captured with a SPOT digital camera and Openlab image acquisition software (Improvision).
Immunofluorescence was performed as above and as described (Matto et al., 2004), with the following variations: following fixation, cells were permeabilized with 0.2% saponin, and subcellular structures were visualized using the primary antibodies, rabbit anti-calnexin (1:1000), rabbit anti-PDI (1:2000), and mouse anti-α-tubulin (1:1000), followed by the appropriate secondary antibody (1:600). Actin was visualized with TRITC-conjugated phalloidin (50 µg/ml), and mitochondria were visualized with the Mitotracker mitochondrial dye (100 nM), according to the manufacturer’s instructions.
2.4 Stable luciferase replication assays
The ET cell line was established by stably transfecting Huh7 cells with RNA transcripts harboring a firefly luciferase-ubiquitin-neomycin phosphotransferase fusion protein and EMCV IRES-driven genotype 1b Con1 NS3-5B polyprotein containing the cell culture adaptive mutations E1202G, T1280I, and K1846T (Lohmann et al., 2003). Cells were plated at 0.5-1.0 x104 cells/well in 96-well plates and incubated for 24 hrs. Then, anguizole was added to the cells to achieve 10 final concentrations ranging from 0.097 to 50 μM. Luciferase activity was measured 48 hours later by adding a lysis buffer and the substrate (Promega E2661 and E2620). Under the same conditions, cytotoxicity of the compounds was determined using the cell proliferation reagent, WST-1 (Roche), according to the manufacturer’s instructions. Percent inhibition of replication was determined relative to a no compound control. EC50 values were calculated using the equation: % inhibition = 100%/[1 + 10(log EC50 – log (I)) * b)], where b is Hill’s coefficient.
2.5 Transient luciferase replication assays
Bart-Luciferase RNA was generated by linearizing the Bart79ILuc* vector with the ScaI restriction enzyme (NEB). Subsequently, in vitro transcription was performed with the Megascript T7 kit according to the manufacturer’s instructions (Ambion).
In vitro-transcribed Bart-Luciferase RNA was transfected into Huh7.5 cells by electroporation, as previously described (Lindenbach et al., 2005). Briefly, 5 µg of RNA was transfected into 6×106 cells with five 99 μs pulses at 0.82kV over 1.1 seconds, using a BTX-830 electroporator. Cells were seeded into 6-well plates at 3×105 cells/well. For EC50 measurements, serial dilutions of anguizole were added to each well to achieve six final concentration of anguizole ranging from 0.001 to 3.125 µM (WT) or 0.01 to 10.24 µM (H94R), while the percentage of DMSO in each well remained constant, at 0.5%. Luciferase activity was measured after 5 days of treatment.
For comparing replication kinetics between the wild-type and H94R replicons, Bart-Luciferase RNA from the wild-type or mutant constructs was electroporated into Huh7.5 cells as above. Bart-Luciferase RNA of a construct containing an NS5B polymerase lethal mutation (Einav et al., 2004) served as a negative control. Cells were seeded into 6-well plates at either 5-, 3-, or 1.5×105 cells/well. Cells were lysed at 48 hour timepoints throughout a 6-day experiment.
Prior to lysis of each sample, cell viability was measured with the alamarBlue® reagent (TREK Diagnostic Systems), according to the manufacturer’s recommendations.
Relative levels of replication were measured using the Luciferase Assay System (Promega). Briefly, cells were lysed in 150 µl Cell Culture Lysis Buffer, scraped from the well, transferred into microcentrifuge tubes, and centrifuged at 16,000×g for 1 minute. 20 µl of the resulting supernatant was mixed with 100 μl of the Firefly luciferase assay buffer containing the assay substrate and luminescence was measured using a Berthold LB 96 V luminometer.
In all cases, replication levels were normalized to viability measurements. EC50 values were determined by fitting normalized replication data points to the equation Y=Bottom + (Top-Bottom) / [1+10^(X-Log[EC50])], using the Graphpad Prism software (GraphPad Software).
2.6 Analysis of resistance mutants
Mutations conferring resistance to anguizole treatment were identified as previously described (Trozzi et al., 2003). Briefly, ET cells stably harboring a subgenomic genotype 1b HCV replicon were cultured in T150 flasks with standard Huh7.5 media containing either 1.5 µM or 3 µM of anguizole and 1 mg/mL G418 for 3 – 4 weeks. An initial round of cell death was followed by proliferation of replicon cells in the presence of the compound. RNA was extracted from the cells with Trizol (Invitrogen), according to the manufacturer’s protocol. RNA was then electroporated into 4×106 cells of the human hepatoma cell line, Huh-Lunet, with a BioRad Gene Pulser at 270V, 960 uF and infinite resistance. Transfected cells were plated in 25 cm2 dishes and selected with 1 mg/mL G418 and 1.5 or 3 µM of anguizole until colonies proliferated (3 – 5 weeks). Colonies were picked and propogated until growth had exceeded a T25 flask, approximately 1 – 2 weeks. Cells were then trypsinized and lysed in Trizol. RNA was isolated according to the manufacturer’s instructions. HCV replicon RNA was reverse-transcribed and amplified with Superscript II (Invitrogen), using a primer specific to the HCV 3’UTR, and NS4B was amplified using gene-specific primers. The PCR product was cloned, sequenced, and compared to wild type NS4B sequence.
2.7 Dynamic Light Scattering (DLS)
Unilamellar light vesicles were prepared using the extrusion method, as previously described (Cho et al., 2007). Vesicle size distribution was determined by quasi-elastic dynamic light scattering (DLS) as previously described (Kolchens et al., 1993). Briefly, DLS measurements were performed by a 90Plus particle size analyzer from Brookhaven Instrument Corp. (Kolchens et al., 1993) and results were analyzed by digital autocorrelator software (Brookhaven Instruments Corp.). All measurements were taken at a scattering angle of 90°, where the reflection effect is minimized. The hydrodynamic diameter of the vesicles was determined via nonnegatively constrained least squares fitting (Kolchens et al., 1993). The lipid and peptide concentrations for DLS measurements were 300 µM and 13 µM, respectively. For inhibitor experiments, either anguizole or clemizole was first incubated with the peptide at a 2:1 molar ratio. Then, the mixture was added to the lipid vesicles. All of the experiments were thermostatically controlled at 25°C.
3. Results
3.1 The small molecule, anguizole, inhibits HCV RNA replication
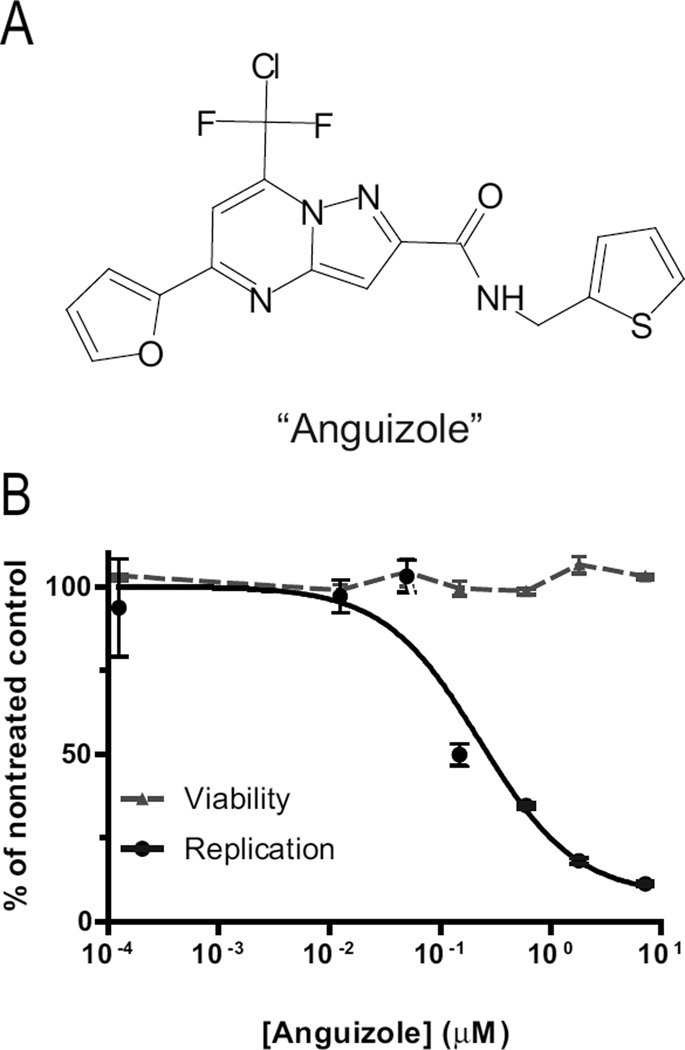
Chunduru et al. (2005) have recently performed a large-scale screen for small molecules that could bind to recombinant NS4B. This assay was based on monitoring changes in intrinsic protein fluorescence of recombinant NS4B as an indirect measure of candidate ligand binding. One of the compounds identified in the screen, 7- [chloro(difluoro)methyl]-5-furan-2-yl-N-(thiophen-2-ylmethyl)pyrazolo[1,5- a]pyrimidine-2-carboxamide, is shown in Figure 1A, and is hereon referred to as “anguizole”. In the screen, this compound was shown to inhibit HCV protein expression, as detemined by an ELISA-based HCV replication assay. Initially, we sought to confirm that anguizole does indeed possess specific anti-HCV activity in the HCV replicon system.
Figure 1.
A small molecule inhibits HCV replication. (A) Structure of the compound, referred to as “Anguizole,” which was originally reported as a ligand of NS4B (Chunduru et al., 2005). (B) Mean 50% effective concentrations (EC50) of the compound were determined by treating luciferase-linked HCV replicons with various concentrations of anguizole and assaying luciferase activity as a measure of HCV replication. Pictured above is a representative experiment for genotype 1b, in which anguizole concentrations ranging from 0.0001 to 7.2 µM were tested. Replication and cell viability levels are reported as a percentage of the nontreated control. The mean EC50 was 310 nM, while the CC50 was greater than 50 µM. Each data point is the mean of three replicates, and error bars indicate SEM.
Luciferase-linked HCV replicons were treated with various concentrations of anguizole, and luciferase activity was assayed as a measure of HCV replication. A representative experiment testing genotype 1b is shown in Figure 1b. Treatment with anguizole significantly inhibited viral RNA replication in both genotype 1b and 1a replicons. The mean 50% effective concentration (EC50) of the compound in genotype 1b was 310 nM and in genotype 1a was 560 nM. Anguizole appeared to have no significant cytotoxic effect at the concentrations studied, with a mean 50% cytotoxicity concentration (CC50) of > 50 µM, as measured by the cell proliferation reagent, WST- 1(Roche). Notably, we found that the EC50 for genotype 2a replicons was > 50 µM, the highest concentration tested, suggesting that this compound’s effect on viral replication may be genotype-specific. As a control for the sensitivity of our replicon cell line, we tested these replicons with a control compound, 1-{[6-carboxy-2-(4-chlorophenyl)-3- cyclohexyl-1H-indol-1-yl]acetyl}-N,N-diethylpiperidin-4-aminium chloride, previously reported as compound 55 in (Harper et al., 2005). We measured the EC50 of this compound to be 0.5 µM in our cell line, as compared to the 0.13 µM value previously reported.
3.2 Mutations conferring resistance to anguizole map to NS4B
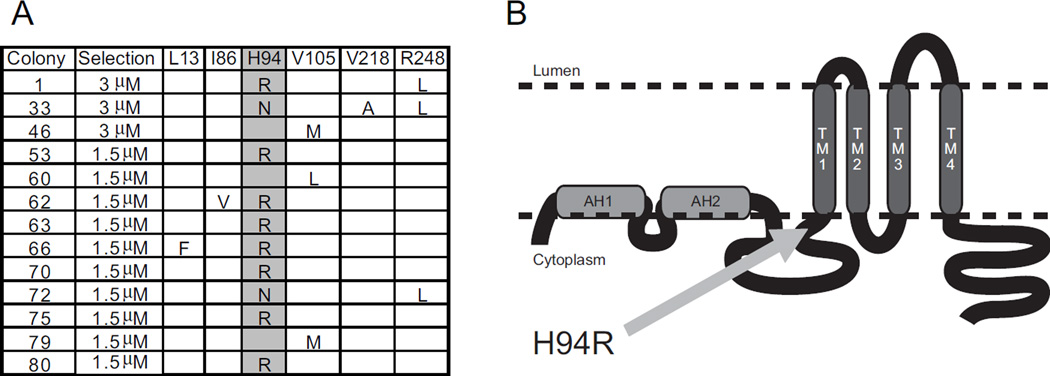
In order to determine anguizole’s mechanism of action, we first sought to obtain genetic evidence that anguizole interacts with NS4B. Therefore, we selected for anguizole-resistant mutants. Genotype 1b replicon cells were grown in the presence of the drug at either 5- or 10-fold the EC50, and resistant colonies were isolated and propagated. RNA was extracted from 18 colonies, and the NS4B coding region was reverse-transcribed, amplified, and sequenced. Several mutations within NS4B were identified in the resistant colonies (Fig. 2A), while no NS4B mutations were observed in untreated replicon cells grown in parallel.
Figure 2.
Resistant mutations map to NS4B. Colony formation assays were performed at either 5- or 10 times the EC50 for this compound. Replicating colonies were isolated and cDNA from these colonies was sequenced. (A) Several point mutations in the genotype 1b NS4B coding region were identified in resistant colonies. Colonies containing such mutations and the stringency of selection are indicated. At the top are listed the wild-type amino acid found at the indicated location in the NS4B polypeptide sequence. Observed mutations are listed below. Highlighted is the most common mutation observed, a mutation of NS4B’s 94th amino acid—corresponding to the HCV polyprotein’s 1805th amino acid—from a histidine to either an arginine or an asparagine. (B) A predicted topology of the NS4B protein, as modified from (Lundin et al., 2006). The location of the H94 mutation is indicated. Two predicted N-terminal amphipathic helices are shown (AH1 and AH2), as are the four predicted transmembrane domains (TM1-4).
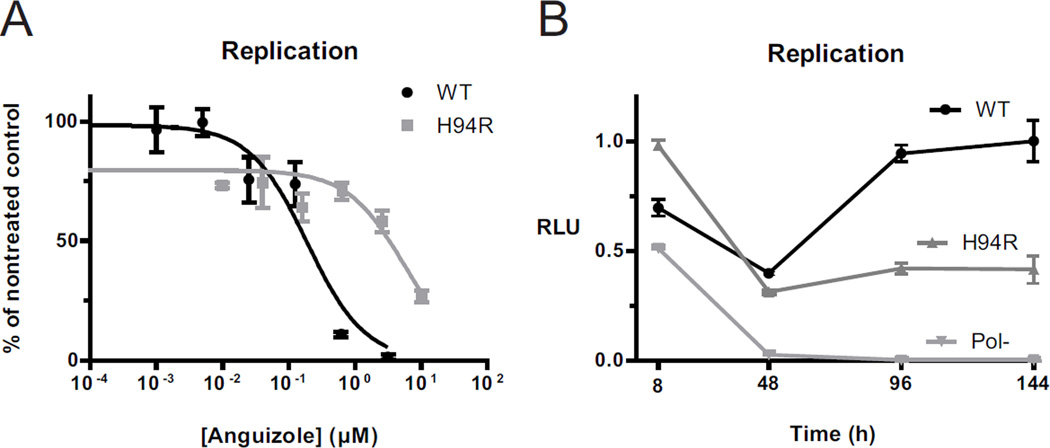
Present in 14 of the 18 isolated clones, the most commonly selected mutation conferring anguizole resistance was a substitution of histidine to either arginine (H94R) or asparagine in position 94 of the NS4B amino acid sequence, corresponding to position 1805 of the HCV polyprotein. To confirm that it was responsible for the phenotypic resistance to anguizole, the H94R mutation was cloned into a firefly luciferase-linked HCV reporter construct and electroporated into Huh7.5 cells. The cells were then dosed with various concentrations of anguizole solubilized in DMSO, and cells were treated with DMSO alone as a negative control. Luciferase expression levels were measured after 5 days of treatment. As shown in Figure 3A, the replicon harboring the H94R mutation was indeed much less sensitive to anguizole treatment (EC50 = 7.5 µM, 95% CI = 3.2-17.4 µM), relative to the wild-type replicon (EC50 = 0.20 µM, 95% CI = 0.08-0.52 µM).
Figure 3.
Characterization of the H94R resistance mutation. (A and B) Transient luciferase replication assays were performed with genotype 1b HCV replicon constructs containing either a histidine (WT, black) or an arginine (H94R, dark gray) at amino acid 94 in the NS4B sequence. (A) Luciferase assays were performed following 5 days of treatment with various concentrations of anguizole. Replication levels (RLU) are shown relative to the maximal luminescence observed for each electroporation, and they are normalized to cell viability measurements for each sample. EC50 values were calculated to be 0.20 µM for WT and 7.5 µM for the H94R mutant. (B) Replication kinetics were tested for these constructs, along with a polymerase defective control (Pol-, light gray), over a 6-day period in the absence of anguizole. Replication levels are shown relative to the maximal luminescence observed for all electroporations. Compared to wild-type, the H94R mutant is impaired in its replication ability. Each data point is the mean of three replicates and error bars represent SEM.
To evaluate the replication capacity of the H94R NS4B mutant replicon, nontreated WT and H94R replicons were monitored as a function of time over the course of 6 days post-electroporation (Fig. 3B). Replication kinetics of the luciferase-linked H94R replicon were similar to those of the wild-type replicon; however, the efficiency of replication was roughly two-fold lower for the mutant. Therefore, the H94R mutation appears to confer a fitness cost on the replication of the virus.
Taken together, these findings suggest that anguizole does not merely target NS4B in vitro, but also likely in the context of viral replication in cells; furthermore, its antiviral activity appears to depend on the sequence of NS4B.
3.3 Anguizole treatment leads to an altered subcellular distribution pattern of the NS4B protein
Having obtained genetic evidence that NS4B is the target of this drug, we sought to examine functions of NS4B that might be affected by the compound. One of these potential functions is NS4B’s ability to assemble into membrane-associated foci (MAF) that are believed to correlate to the sites of viral replication (Gretton et al., 2005). We hypothesized that anguizole might alter the integrity of the MAF.
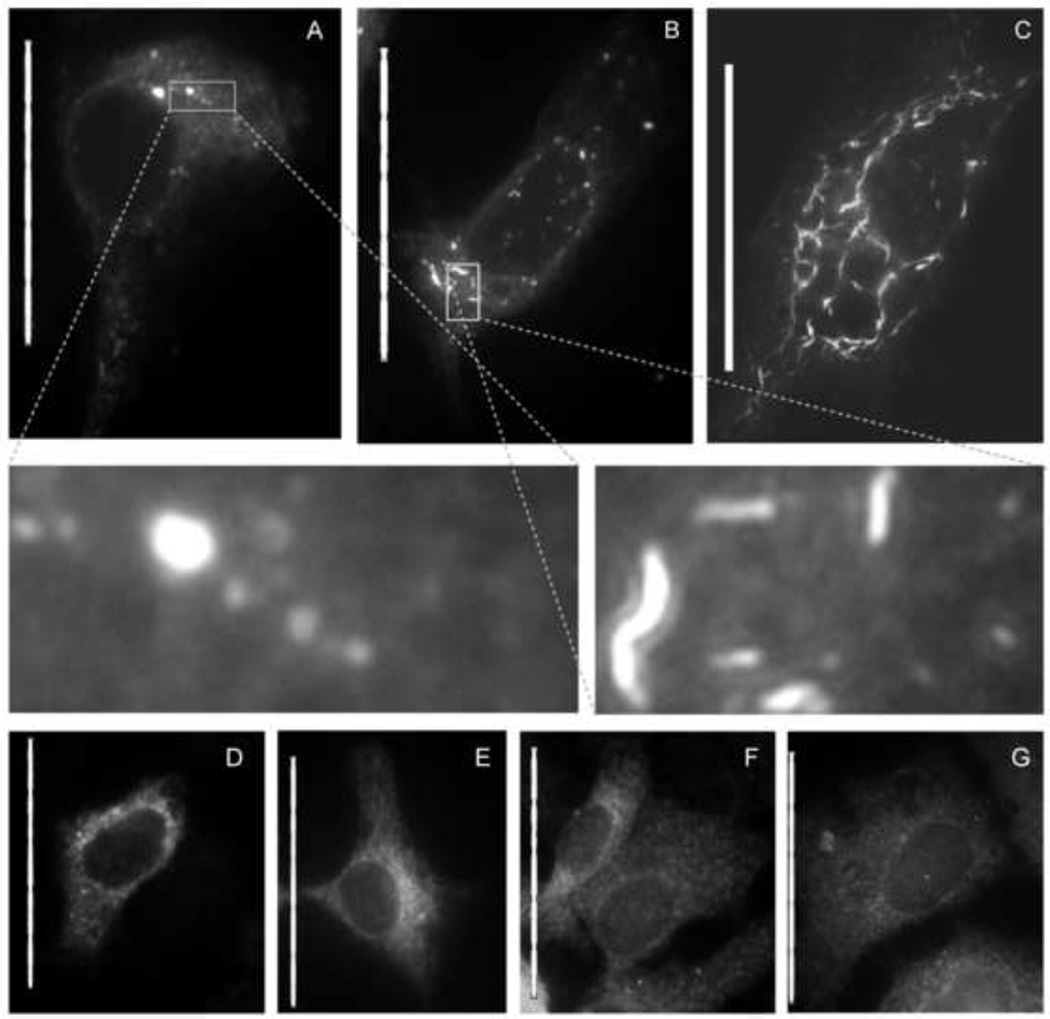
To test this hypothesis, a plasmid expressing NS4B fused in frame with a C-terminal green fluorescent protein (GFP) was transfected into Huh7.5 cells. As shown in Figure 4, cells treated with 5 µM anguizole for 48 hours displayed a NS4B distribution pattern distinct from the MAF visible in non-treated cells. Rather than appearing as small foci (Fig. 4A), a significant amount of the NS4B proteins in these treated cells appeared to form elongated snake-shaped structures (Fig. 4B & 4C). As shown in the insets, these “snakes” were both longer and shaped differently than the MAF observed in nontreated cells, suggesting that anguizole treatment can alter the distribution pattern of NS4B MAF.
Figure 4.
Anguizole alters the subcellular distribution of NS4B-GFP in transiently transfected cells. Huh7.5 cells were cultured in the absence (A, D, F) or presence (B, C, E, G) of 5 µM anguizole, either alone (F, G) or following transfection with NS4B-GFP (A, B, C) or GFP-NS5A (D, E). (A) In nontreated cells, NS4B-GFP displays an ER-associated pattern of localization, along with several membrane-associated foci (MAF) of varying size throughout the cytoplasm. (B and C) In the presence of anguizole, NS4B-GFP appears to form elongated, curved structures (“snakes”), most commonly observed to be small and dispersed (B), but occasionally observed to be dramatically long (C). (Insets of A and B) Zoomed-in images highlight the differences in shape and length between MAF and snakes. (D and E) The distribution pattern of a GFP-NS5A fusion protein is equivalent in untreated (D) and treated (E) samples. (F and G) The pattern of calnexin staining is equivalent in untreated (F) and treated cells (G). Scale bars represent 50 µm.
Notably, there was variation in the extent of the snake phenotype. A pattern resembling a network of extremely elongated snakes (Fig. 4C) was visible in rare cells, while moderately elongated snakes (Fig. 4B) were visible in roughly one fifth of the cells expressing NS4B. In the remaining cells in which NS4B-GFP was expressed, the distribution pattern of NS4B-GFP either adopted a diffuse cytoplasmic pattern (Fig. S1A) or displayed a pattern of small, bright foci (Fig. S1B) unlike the MAF observed in untreated cells.
To test the specificity of anguizole’s interaction with NS4B, we examined the effect of anguizole treatment on another viral protein and on subcellular structures in Huh7.5 cells. First, when we treated cells expressing an NS5A-GFP fusion protein, no snake-like structures were observed (Fig. 4E). Also, when markers of the ER, actin filaments, microtubules, or mitochondria were visualized, no differences between the cellular structure of treated and nontreated cells were observed, both in cells transfected with NS4B-GFP and in those not transfected (Fig. 4G, S2). Therefore, we hypothesized that the observed effect is likely the result of a specific interaction between anguizole and the NS4B protein. In total, these data show that anguizole specially impairs a function of NS4B—its ability to assemble into the typical MAF distribution pattern.
3.4 A resistance mutation also alters the subcellular distribution pattern of the NS4B protein
Having identified a mutation in NS4B that confers resistance to anguizole treatment, we hypothesized that this mutation may also affect NS4B’s ability to assemble into a MAF distribution pattern. To test this hypothesis, we characterized the subcellular distribution of H94R NS4B-GFP, both in the presence and absence of anguizole.
Transient transfections were performed with a construct of NS4B-GFP carrying the H94R mutation, as described for Figure 4. In the absence of anguizole, cells expressing the H94R NS4B-GFP mutant displayed a distribution pattern quite different from the wild-type MAF pattern (Fig. 5A vs. Fig. 4A). Notably, this pattern was similar to the snake-like pattern seen in wild-type cells treated with anguizole, with the exception that the H94R snakes tended to be longer and more pronounced than those observed in the wild-type samples. These results suggest that the H94R mutation itself is sufficient to alter the distribution pattern of NS4B MAF.
Figure 5.
The H94R mutation alters NS4B-GFP’s subcellular distribution. The H94R mutation was cloned into the NS4B-GFP construct, and transient transfections were performed as in Figure 4. (A) In the absence of anguizole, the subcellular distribution of H94R NS4B-GFP adopts a snake-like pattern. (B) In cells treated with 5 µM anguizole, the snake-like pattern is not observed; instead, NS4B appears to assemble into a novel pattern of many bright cytoplasmic foci. Scale bars are 50 µm.
In contrast, cells treated with 5 µM anguizole displayed a different distribution of H94R NS4B-GFP (Fig. 5B). In these cells, no snake patterns were detected, and many large foci were present throughout the cytoplasm. These foci are reminiscent of MAF, but their expression pattern is distinct from that of wild-type NS4B-GFP in that H94R foci tend to be somewhat larger and present in higher numbers than wild-type NS4B foci.
3.5 Anguizole interacts with the second amphipathic helix of NS4B
Previously, our laboratory identified an N-terminal amphipathic helix (AH1) in NS4B that is essential for wild-type distribution of NS4B throughout the cell (Elazar et al., 2004). We have also identified a second, smaller amphipathic helix (AH2), consisting of amino acids 43 – 65 of the NS4B protein. Recently, we have found that upon addition to a monodisperse population of lipid vesicles, a synthetic peptide comprising this AH2 dramatically alters the size distribution of the vesicles by inducing massive vesicle aggregation (N.J. Cho, H. Dvory-Sobol, C.H. Lee, S.J. Cho, P.D. Bryson, C. Frank, and J.S. Glenn, Science Translational Medicine, in press). Furthermore, this activity appears to be important for both the formation of the membranous web replication platform and for HCV RNA replication. We hypothesized that the AH2 represents a target for anguizole and that addition of anguizole might disrupt AH2’s lipid vesicle aggregating activity.
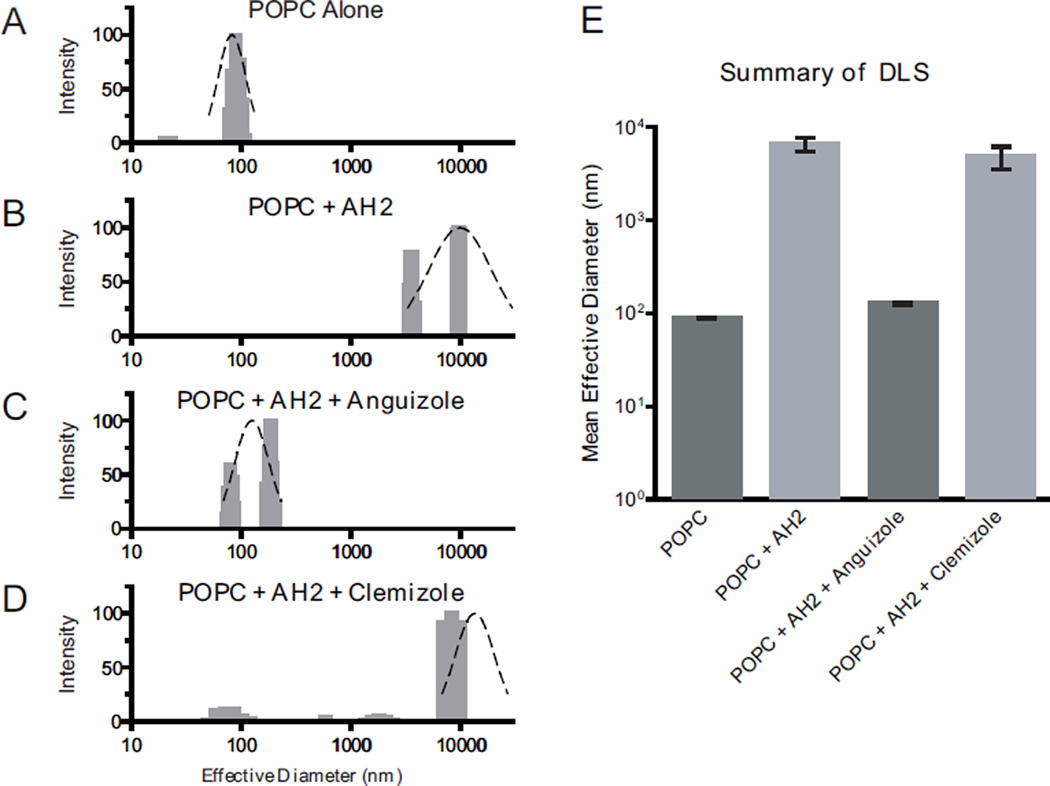
To test this hypothesis, dynamic light scattering (DLS) measurements were performed with POPC vesicles and AH2 peptide in the presence or absence of anguizole. Clemizole hydrochloride—which has been recently shown to inhibit HCV RNA:NS4B binding, for which a C-terminal domain mediating RNA binding is essential (Einav et al., 2008)—was used as a negative control. As shown in Figure 6, the addition of AH2 peptide to the POPC vesicles radically affected the effective diameter of vesicles in solution, changing the 88 nm diameter untreated vesicles to aggregates with a mean effective diameter of 5377 nm (Fig 6B vs. 6A). While clemizole exerted no significant effect on AH2’s vesicle aggregating activity (Fig. 6D), anguizole markedly inhibited AH2’s ability to aggregate vesicles (Fig. 6C). Specifically, vesicles incubated with AH2 and anguizole adopted a mean effective diameter of 126 nm, comparable to what is observed for vesicles without AH2 (88 nm, Fig 6E). These results suggest that anguizole specifically interacts with the AH2 segment of NS4B.
Figure 6.
Anguizole interacts with the second amphipathic helix (AH2) of NS4B. Extruded synthetic vesicles composed of 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine (POPC) lipids were incubated with synthetic peptide comprising the AH2 region of NS4B and with either anguizole or clemizole hydrochloride. Dynamic light scattering was employed to measure the effective diameter of vesicles in a sample. (A – D) The size distribution of vesicle diameters is shown (gray bars), as well as a gaussian fit of the data (black dashed line). Samples contained (A) 300 µM POPC lipid vesicles only, (B) POPC vesicles + 13 µM AH2 peptide, (C) POPC vesicles + AH2 peptide + 26 µM anguizole, and (D) POPC vesicles + AH2 peptide + 26 µM clemizole hydrochloride, as a negative control compound. The x-axis is presented on a logarithmic scale. (E) The mean values of the effective vesicle diameters in at least 3 independent experiments are shown for all four samples. Error bars indicate SEM.
4. Discussion
In this study, we present evidence concerning the mechanism of a novel means of inhibiting HCV replication. Anguizole, the small molecule studied here, is a member of a previously uncharacterized class of inhibitors for this pathogen, and we demonstrate that mutations in the viral NS4B coding region are sufficient to confer resistance to this compound. Furthermore, we show that anguizole can disrupt the intracellular distribution of NS4B membrane-associated foci and that it also disrupts the interaction of a specific peptide segment of NS4B with lipid vesicles, suggesting that anguizole may directly bind this region of the protein. Thus, this compound likely employs a novel mechanism of action for inhibiting HCV replication.
In addition to the data from the original screen that identified anguizole (Chunduru et al., 2005), three pieces of evidence presented in this study suggest that anguizole’s inhibition of viral replication is a result of directly targeting the viral NS4B protein. First, the mutation of a single amino acid in the NS4B coding region is sufficient to reduce the virus’ susceptibility to drug treatment (Fig. 3). Second, treatment with anguizole significantly alters the subcellular distribution pattern of NS4B (Fig. 4), but not that of another viral protein or cellular proteins. Third, anguizole specifically blocks the function of a portion of the NS4B polypeptide, the amphipathic helix comprising amino acids 43 – 65. Taken together, these data point to NS4B as the target with which anguizole interacts. Because NS4B induces the formation of membranous viral replication structures (the “membranous web”) (Egger et al., 2002) and is required for the proper localization of other nonstructural viral components (Elazar et al., 2004; Gao et al., 2004), it is logical that the disruption of functional NS4B would lead to a reduction in viral replication. We propose that this is what happens upon either the addition of anguizole or the mutation of NS4B histidine 94 (Fig. 4C and Fig. 5A).
Our genetic analysis identified histidine 94 as an important determinant of anguizole’s effect on HCV replication. In patient isolates, this position is not strictly conserved, as glutamine (1a), asparagine (1b), serine (1b), and threonine (2a) have each been observed in reference sequences of the indicated genotype (Kuiken et al., 2005). Of the replicons we tested in the luciferase replication assay, we found that those that contained histidine (1b) or glutamine (1a) at position 94 were highly sensitive to anguizole treatment (EC50 <0.6 µM), one that contained arginine (1b mutant) was moderately resistant (EC50 = 7.5 µM), and one that contained threonine (2a) was highly resistant to the drug (EC50 > 50 µM). Although our analysis has identified position 94 as an important determinant to anguizole’s mode of action, it is likely that other regions of the protein may be involved as well.
For example, our in vitro dynamic light scattering experiments (Fig. 6) suggest that anguizole interacts directly with the second amphipathic helix of NS4B (amino acids 43-65). Interestingly, to date our resistant mutant analysis (Fig. 2) has not identified any mutations that map within this region of NS4B, while it has shown that a mutation at amino acid 94 in the NS4B sequence is sufficient to confer resistance to anguizole. We interpret these potentially conflicting results as follows: (1) mutations in the AH2 region are very poorly tolerated by the virus; (2) mutations at position 94, in contrast, are well tolerated; and (3) the H94R mutation, in particular, might cause a conformational change in the NS4B protein, one which counteracts anguizole’s ability to inhibit HCV replication. In support of point (2), we note that position 94 in NS4B is not strictly conserved in patient isolates, as described above. In support of point (3), we found that the H94R mutation alone is sufficient to drastically alter NS4B’s subcellular distribution (Fig. 5A). The nature of such a conformational change is unclear, but one possibility is that it partially blocks anguizole’s binding site on the AH2. Structural studies may help to further clarify the molecular interactions occurring in this situation.
In addition to identifying a novel mechanism of HCV inhibition, our results have enhanced our understanding of the biological processes that occur during HCV replication. For example, we have shown that anguizole can disrupt both NS4B AH2:lipid interactions in vitro (Fig. 6) and the distribution pattern of MAF in cell culture (Fig. 4). These results are consistent with the hypothesis that NS4B’s AH2 is a key determinant for the formation of MAF, a correlate of HCV replication complexes. In addition, we have shown that the assembly of NS4B into MAF can be altered both pharmacologically and genetically (Fig. 4 and 5), resulting in the cytoplasmic distribution of elongated snake-like structures. Previous work has shown that NS4B can physically interact with other nonstructural components of the virus, including itself (Gao et al., 2004; Yu et al., 2006), and we hypothesize that these snakes represent assemblies of NS4B proteins that have lost their ability to form the proper protein:protein or protein:lipid interactions. Future studies may reveal more insight into these interactions and, more generally, the processes involved in replication complex formation.
To the best of our knowledge, anguizole is the first pharmacological compound shown to affect NS4B subcellular distribution. Because targeting multiple stages of the viral life cycle is likely to be essential for effective HCV therapy (De Francesco & Migliaccio, 2005), anguizole is thus representative of an important new potential class of HCV inhibitors. Not only is such a class likely to be complementary to inhibitors that target other viral proteins (Lin et al., 2006; Malcolm et al., 2006), but it may also complement inhibitors that target other functions of the same NS4B protein. In support of such an outcome, our results show that clemizole hydrochloride does not inhibit AH2’s vesicle-altering properties while anguizole does (Fig. 6). Similarly, anguizole does not inhibit NS4B RNA binding, while clemizole does (S. Einav, unpublished data). Development of such compounds may lead to an efficacious antiviral cocktail and help manage this disease for the millions of people it affects.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (to J.S.G.) and National Institutes of Health (NIH) RO1 DK066793 and RO1 AI087917.
P.B. is supported by the Cell and Molecular Biology Training Program, NIH T32-GM0072726, N.C by an American Liver Foundation postdoctoral award and a Roche global postdoctoral fellowship, and S.E. by an NIH K08-AI079406-01 Award, a Stanford Digestive Disease Center (DDC) Pilot/Feasibility Award, NIH P30 DK56339 (“Molecular Pathogenesis of Digestive Diseases”) and a Stanford Institute for Immunity, Transplantation, and Infection (ITI) Young Investigator Innovation Award.
We thank Ella Sklan for providing reagents, Lene Nejsum and James Nelson for their assistance with the NS4B-GFP assays, and Robert Hamatake for reviewing the manuscript.
J.S.G. is a consultant to, and has an equity interest in, Eiger BioPharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cho NJ, Kanazawa KK, Glenn JS, Frank CW. Employing two different quartz crystal microbalance models to study changes in viscoelastic behavior upon transformation of lipid vesicles to a bilayer on a gold surface. Anal Chem. 2007;79:7027–7035. doi: 10.1021/ac0709504. [DOI] [PubMed] [Google Scholar]
- Chunduru SK, Benetatos CA, Nitz TJ, Bailey TR, Chunduru S, Benetatos C, Nitz T, Bailey T, Benatatos CA. Composition useful in the prophylaxis or treatment of viral infection e.g. hepatitis C infection in living host comprises amides or N and/or S containing heterocyclic compounds and carrier patent. WO 2005051318-A2; EP1686949-A2; US2007269420-A1. 2005 [Google Scholar]
- De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S, Elazar M, Danieli T, Glenn JS. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol. 2004;78:11288–11295. doi: 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotech. 2008;26:1019–1027. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78:11393–11400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretton SN, Taylor AI, McLauchlan J. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membrane-associated foci. J Gen Virol. 2005;86:1415–1421. doi: 10.1099/vir.0.80768-0. [DOI] [PubMed] [Google Scholar]
- Harper S, Avolio S, Pacini B, Di Filippo M, Altamura S, Tomei L, Paonessa G, Di Marco S, Carfi A, Giuliano C, Padron J, Bonelli F, Migliaccio G, De Francesco R, Laufer R, Rowley M, Narjes F. Potent inhibitors of subgenomic hepatitis C virus RNA replication through optimization of indole-N-acetamide allosteric inhibitors of the viral NS5B polymerase. J Med Chem. 2005;48:4547–4557. doi: 10.1021/jm050056+. [DOI] [PubMed] [Google Scholar]
- Kolchens S, Ramaswami V, Birgenheier J, Nett L, O'Brien DF. Quasi-elastic light scattering determination of the size distribution of extruded vesicles. Chem Phys Lipids. 1993;65:1–10. doi: 10.1016/0009-3084(93)90076-f. [DOI] [PubMed] [Google Scholar]
- Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos hepatitis C sequence database. Bioinformatics. 2005;21:379–384. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- Lin C, Kwong AD, Perni RB. Discovery and development of VX-950, a novel, covalent, and reversible inhibitor of hepatitis C virus NS3.4A serine protease. Infect Disord Drug Targets. 2006;6:3–16. doi: 10.2174/187152606776056706. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M, Lindstrom H, Gronwall C, Persson MA. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J Gen Virol. 2006;87:3263–3272. doi: 10.1099/vir.0.82211-0. [DOI] [PubMed] [Google Scholar]
- Malcolm BA, Liu R, Lahser F, Agrawal S, Belanger B, Butkiewicz N, Chase R, Gheyas F, Hart A, Hesk D, Ingravallo P, Jiang C, Kong R, Lu J, Pichardo J, Prongay A, Skelton A, Tong X, Venkatraman S, Xia E, Girijavallabhan V, Njoroge FG. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother. 2006;50:1013–1020. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, Foster GR, Rockstroh JK, Zeuzem S, Zoulim F, Houghton M. The way forward in HCV treatment--finding the right path. Nat Rev Drug Discov. 2007;6:991–1000. doi: 10.1038/nrd2411. [DOI] [PubMed] [Google Scholar]
- Matto M, Rice CM, Aroeti B, Glenn JS. Hepatitis C virus core protein associates with detergent-resistant membranes distinct from classical plasma membrane rafts. J Virol. 2004;78:12047–12053. doi: 10.1128/JVI.78.21.12047-12053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H, Lu L, Pilot-Matias T, Pithawalla R, Mondal R, Masse S, Dekhtyar T, Ng T, Koev G, Stoll V, Stewart KD, Pratt J, Donner P, Rockway T, Maring C, Molla A. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob Agents Chemother. 2005;49:4305–4314. doi: 10.1128/AAC.49.10.4305-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282:36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- Trozzi C, Bartholomew L, Ceccacci A, Biasiol G, Pacini L, Altamura S, Narjes F, Muraglia E, Paonessa G, Koch U, De Francesco R, Steinkuhler C, Migliaccio G. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J Virol. 2003;77:3669–3679. doi: 10.1128/JVI.77.6.3669-3679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GY, Lee KJ, Gao L, Lai MM. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J Virol. 2006;80:6013–6023. doi: 10.1128/JVI.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.