Abstract
The Helicobacter pylori virulence factor CagA targets a variety of host proteins to alter different cellular responses, including the induction of pro-inflammatory cytokines. We have previously shown that CagA-facilitated lysine 63-linked ubiquitination of TAK1 is essential for the H. pylori-induced NF-κB activation and the expression of proinflammatory cytokines. However, the molecular mechanism for TAK1 ubiquitination and activation in H. pylori-mediated NF-κB activation remains elusive. Here, we identify lysine 158 of TAK1 as the key residue undergoing lysine 63-linked ubiquitination in response to H. pylori infection. Mutation of lysine 158 to arginine prevents the ubiquitination of TAK1 and impairs H. pylori-induced TAK1 and NF-κB activation. Moreover, we demonstrate that E2 ubiquitin conjugating enzyme Ubc13 is involved in H. pylori-mediated TAK1 ubiquitination. Suppressing the activity of Ubc13 by a dominant-negative mutant or siRNA abolishes CagA-facilitated and H. pylori-induced TAK1 and NF-κB activation. These findings further underscore the importance of lysine 63-linked ubiquitination of TAK1 in H. pylori-induced NF-κB activation and NF-κB-mediated inflammatory response.
Keywords: CagA, NF-κB, TAK1, Ubc13, ubiquitination
Introduction
Infection with Helicobacter pylori and the resulting chronic inflammation is a major risk factor for the development of gastric cancer [Hatakeyama, 2008; Peek and Blaser, 2002]. H. pylori infection up-regulates the expression of pro-inflammatory cytokines, including IL-8, IL-6 and TNF-α, which contribute to an environment which promotes increased cell turnover, mutagenesis, and growth [McGee and Mobley, 2000]. The enhanced expression of pro-inflammatory cytokines is believed to be associated with the ability of H. pylori to activate transcription factor NF-κB, which plays a key role in modulating the expression of various cytokines [Ghosh and Karin, 2002].
Various virulence components of H. pylori, including flagella, lipopolysaccharide (LPS), vacuolating toxin VacA, and cytotoxin-associated gene pathogenicity island (cagPAI), have been shown to be able to affect the expression of cytokines [Yamaoka, 2010]. Of all these components, the most potent factor is cagPAI, which encodes a type 4 secretion system (T4SS) and the virulence factor CagA. CagA is injected into gastric epithelial cells via the T4SS [Peek, 2005]. By interacting with a variety of cellular signaling molecules, CagA induces a series of cellular events, including actin remodeling and IL-8 production [Peek, 2005]. Translocation of CagA into host cells induces higher levels of IL-8 production by activating transcription factors such as NF-κB and AP-1 [Backert and Naumann, 2010]. Ectopic expression of CagA is able to stimulate nuclear translocation of NF-κB and production of IL-8 in gastric epithelial cells [Backert and Naumann, 2010]. Furthermore, NF-κB activation and inflammation have been shown to be markedly reduced in the gastric antra of Mongolian gerbils infected with cagA-deficient H. pylori as compared to infection with wild-type H. pylori, emphasizing the essential role of CagA in NF-κB activation and H. pylori-mediated inflammation [Lamb and Chen, 2010; Shibata et al., 2006].
Transforming growth factor β–activated kinase 1 (TAK1) is a key regulator of signal transduction cascades leading to the activation of IκB kinases (IKKs) and NF-κB in response to various stimuli, including cytokines and bacterial and viral infections [Adhikari et al., 2007]. TAK1 phosphorylates and activates IKK2, which then phosphorylates IκBα, leading to its ubiquitination and degradation; subsequently, NF-κB is translocated into the nucleus and NF-κB target gene expression is activated [Chen and Greene, 2004; Wang et al., 2001]. Recent studies demonstrate that lysine (K) 63-linked ubiquitination actively participates in the activation of TAK1 and IKKs, and that many upstream signaling molecules are subject to K63-linked ubiquitination [Wang et al., 2001]. For example, IL-1β receptor-associated kinase 1 (IRAK1) is K63-linked ubiquitinated upon IL-1β stimulation, and the ubiquitination is important for IKK activation in the IL-1β signaling pathway [Conze et al., 2008; Windheim et al., 2008]. Ubiquitination of TAK1 by TNF-α receptor associated factor (TRAF) 6 is essential for TAK1 auto-phosphorylation and activation (Fan et al, 2010; Yamazaki et al, 2009; Sorrentino et al, 2008). Several lysines within TAK1, including K34, K158 and K209, have been identified to be K63-linked ubiquitinated, and ubiquitination of these different lysines appears to determine stimulus-specific and context-dependent TAK1 activation [Fan et al., 2010; Sorrentino et al., 2008; Yamazaki et al., 2009]. Ubiquitination of K34 of TAK1 has been reported to induce TGF-β-mediated p38 and JNK activation [Sorrentino et al., 2008]. Ubiquitination of TAK1 at a different lysine, K158, is required for TNF-α or IL-1β-induced TAK1 activation and IKK/NF-κB activation [Fan et al., 2010]. We have also reported that TRAF6-mediated ubiquitination of TAK1 is essential for H. pylori-induced NF-κB activation and the induction of pro-inflammatory cytokines [Lamb et al., 2009]. H. pylori infection stimulates virulence factor CagA-dependent K63-linked ubiquitination and activation of TAK1. CagA facilitates TAK1 ubiquitination and potentiates TAK1-mediated NF-κB activation in H. pylori-infected gastric epithelial cells [Lamb et al., 2009]. Nevertheless, the detailed molecular mechanism by which CagA enhances the ubiquitination and activation of TAK1 remains unclear.
Ubiquitination is an ATP-dependent, three-step enzymatic cascade involving an ubiquitin-activating enzyme (E1), an ubiquitin-conjugating enzyme (E2) and an ubiquitin-protein ligase (E3) [Adhikari et al., 2007]. TRAF family proteins, including TRAF2 and TRAF6, are known to be E3 ligases for K63-linked ubiquitination [Chen, 2012]. Biochemical studies also reveal that the heterodimeric Ubc13 and Uev1A are E2 conjugating enzymes for TRAF6-catalyzed polyubiquitination and activation of TAK1 [Wang et al., 2001]. Ubc13/Uev1A have been shown to be essential for the ubiquitination of TRAF6 in IL-1R/Toll-like receptor (TLR) signaling [Yamazaki et al., 2009]. In vivo studies also demonstrate that Ubc13 is a critical component of TRAF-mediated inflammatory responses since ubc13 heterozygous mice were resistant to LPS-induced lethality, and displayed reduced ubiquitination of TRAF6 [Fukushima et al., 2007]. Our previous studies have demonstrated that TRAF6 is critical for K63-linked ubiquitination of TAK1 and its subsequent kinase activation in response to H. pylori infection. However, the role of Ubc13 in the H. pylori-induced ubiquitination of TAK1 remains undetermined.
In this study, we further investigated the molecular mechanism by which TAK1 is ubiquitinated and activated by H. pylori. We identified lysine 158 of TAK1 as the pivotal residue for H. pylori CagA-facilitated TAK1 ubiquitination and activation. We also demonstrated that Ubc13 is responsible for H. pylori-induced ubiquitination and activation of TAK1, which further activates NF-κB and NF-κB-dependent inflammatory gene expression.
Results
Lysine 158 is critical for H. pylori-induced ubiquitination of TAK1
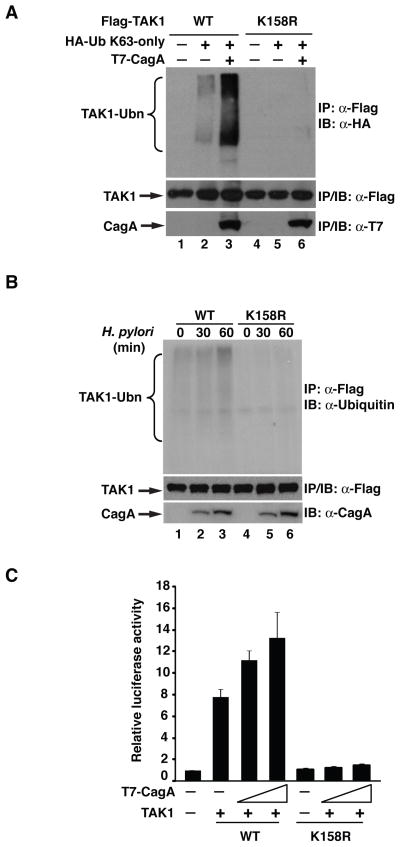
We have previously shown that H. pylori CagA facilitates K63-linked TAK1 ubiquitination and that this ubiquitination is essential for H. pylori-induced TAK1 and NF-κB activation [Lamb et al., 2009]. To further elucidate the role of TAK1 ubiquitination in NF-κB activation, we sought to identify the lysine residues that undergo K63-linked ubiquitination. Several lysine residues within TAK1, including lysines 34, 158 and 209, have been reported to be K63-linked ubiquitinated [Yamazaki et al., 2009]. Therefore, we generated TAK1 mutants with each of these individual lysines mutated to arginine and examined their ubiquitination with or without CagA. When co-transfected with K63-only ubiquitin, wild-type TAK1 was moderately ubiquitinated in the absence of CagA and co-expression of CagA significantly enhanced TAK1 ubiquitination (Figure 1A). In contrast, when lysine 158 was mutated to arginine, ubiquitination of this mutant TAK1 (designated as TAK-K158R) was barely detectable and co-expression of CagA failed to enhance any TAK1 ubiquitination (Figure 1A). Different from lysine 158, mutation of lysine 34 or 209 to arginine had no effect on the basal level or CagA-enhanced ubiquitination of TAK1 (data not shown). These data indicate that lysine 158 is critical for K63-linked TAK1 ubiquitination and that CagA facilitates TAK1 ubiquitination at lysine 158.
Figure 1. Lysine 158 is required for H. pylori-induced ubiquitination and activation of TAK1.
(A) Wild-type (WT) or K158R mutant TAK1 were expressed with K63-only ubiquitin and CagA in HEK293T cells. Flag immunoprecipitates from the lysates were immunoblotted for HA-ubiquitin. (B) WT or K158R mutant TAK1 were ectopically expressed in AGS cells, which were then infected with H. pylori as indicated. Flag immunoprecipitates from the lysates were immunoblotted for ubiquitin. (C) HEK293T cells were transfected with IL-8-luciferase reporter and either WT or K158R TAK1 and increasing amounts of CagA plasmids as indicated. Luciferase activity was measured 30 hrs post-transfection.
Since H. pylori infection induces the ubiquitination of TAK1 [Lamb et al., 2009], we next explored the importance of lysine 158 in H. pylori-induced TAK1 ubiquitination. When expression vectors for wild-type TAK1 or TAK1-K158R were transfected into gastric AGS epithelial cells and H. pylori-induced ubiquitination of TAK1 was examined, we found that infection with H. pylori stimulated the ubiquitination of wild-type TAK1 (Figure 1B), consistent with our previous findings [Lamb et al., 2009]. However, mutation of lysine 158 to arginine completely abolished H. pylori-induced TAK1 ubiquitination (Figure 1B), suggesting that lysine 158 is critically involved in H. pylori-induced TAK1 ubiquitination.
Ubiquitination of TAK1 is important for its ability to activate NF-κB, and CagA facilitates TAK1-mediated NF-κB activation by enhancing TAK1 ubiquitination [Lamb et al., 2009]. We next determined whether ubiquitination of lysine 158 was indeed responsible for TAK1-mediated NF-κB activation. We compared the abilities of wild-type TAK1 and TAK1-K158R to activate NF-κB in a κB-luciferase reporter assay with or without CagA. Expression of wild-type TAK1 activated the κB-luciferase reporter, and the NF-κB activity was further enhanced by the co-expression of CagA in a dose-dependent manner (Figure 1C). Conversely, TAK1-K158R displayed a significantly reduced NF-κB activation, and co-expression of CagA barely enhanced its ability to activate NF-κB (Figure 1C). These data suggest that ubiquitination of lysine 158 of TAK1 is essential for CagA-facilitated TAK1-mediated NF-κB activation.
Lysine 158 is critical for H. pylori CagA-facilitated TAK1 activation
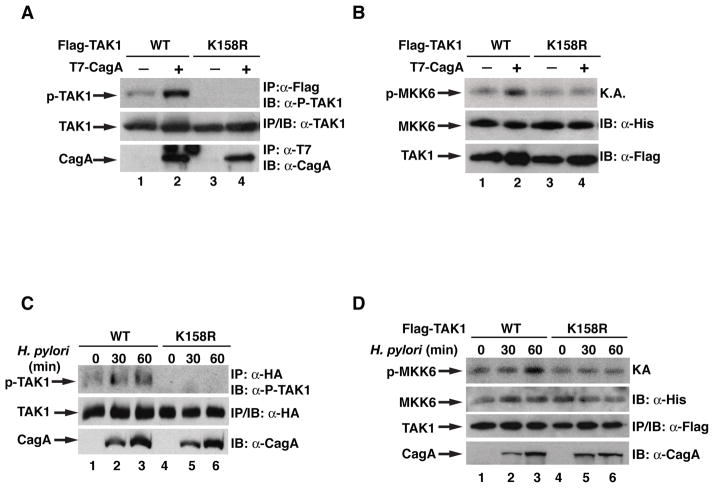
K63-linked ubiquitination of TAK1 has been shown to be important for its kinase activation in response to various stimuli including H. pylori infection [Fan et al., 2009; Lamb et al., 2009]. We next assessed whether ubiquitination of lysine 158 was involved in H. pylori-induced TAK1 activation. Over-expressed TAK1 displayed a basal level of kinase activity which is demonstrated by its auto-phosphorylation of threonine 187 (Figure 2A). Co-expression of CagA further enhanced the kinase activity of wild-type TAK1. However, co-expression of CagA did not enhance the kinase activity of TAK1-K158R, which displayed almost undetectable basal kinase activity (Figure 2A). The importance of ubiquitination of K158 in TAK1 kinase activation was further demonstrated by an in vitro kinase assay using MKK6 as the substrate (Figure 2B). Wild-type TAK1 phosphorylated MKK6, and this phosphorylation was further enhanced by co-expression of CagA. However, mutation of lysine 158 to arginine demolished TAK1’s ability to phosphorylate MKK6, and co-expression of CagA failed to enhance the levels of phosphorylated MKK6 (Figure 2B). More importantly, when we examined the activation of TAK1 in response to H. pylori infection, we found that H. pylori infection stimulated the auto-phosphorylation of wild-type TAK1 but not the TAK1-K158R mutant (Figure 2C), indicating that K158 is critical for H. pylori infection-induced TAK1 activation. Consistently, when TAK1 activity was examined by the in vitro kinase assay, we observed that H. pylori infection activated wild-type TAK1 but not the TAK-K158R mutant (Figure 2D). All together, these data suggest that ubiquitination of lysine 158 is essential for the activation of TAK1 in vivo in response to H. pylori infection.
Figure 2. Ubiquitination at lysine 158 of TAK1 is important for its kinase activity.
(A) WT or K158R TAK1 were expressed in HEK293T cells with or without CagA. Flag-TAK1 immunoprecipitates were immunoblotted for phosphorylated TAK1. (B) WT or K158R TAK1 were expressed in HEK293T cells with or without CagA. Flag-TAK1 immunoprecipitates were subjected to an in vitro kinase assay using His-MKK6(K82A) as a substrate. Levels of phosphorylated MKK6, TAK1, and MKK6 are shown. (C) AGS cells expressing WT or K158R TAK1 were infected with H. pylori for indicated times. Flag-TAK1 immunoprecipitates were immunoblotted for phosphorylated TAK1. (D) AGS cells expressing WT or K158R TAK1 were infected with H. pylori for indicated times. Flag-TAK1 immunoprecipitates were subjected to an in vitro kinase assay as in (B).
Ubc13 mediates the ubiquitination and activation of TAK1
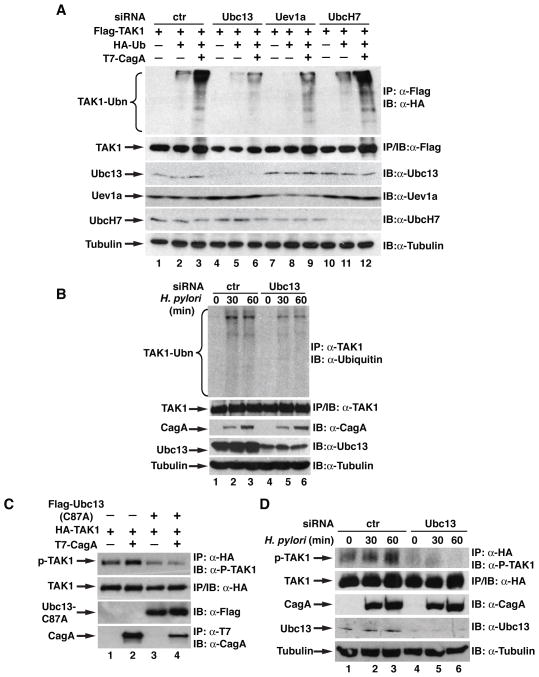
We have previously shown that E3 ligase TRAF6 is involved in H. pylori-induced TAK1 ubiquitination [Lamb et al., 2009]. Recent studies indicate that TRAF6 couples with Ubc13/Uev1a E2 enzymes to catalyze K63-linked ubiquitination [Wang et al., 2001], so we next examined whether Ubc13/Uev1a was also involved in the CagA-facilitated ubiquitination of TAK1. First, we employed small interference RNA (siRNA) to knock down the expression of Ubc13, Uev1a or UbcH7 and examined the ubiquitination of TAK1. Compared to control siRNA, depletion of Ubc13 or Uev1a impaired the basal as well as the CagA-enhanced ubiquitination of TAK1 (Figure 3A). Depletion of UbcH7, an E2 enzyme which is not involved in TRAF6-mediated ubiquitination [Wang et al., 2001], had no effect on the ubiquitination of TAK1 (Figure 3A). These data suggest a specific involvement of Ubc13/Uev1a in the CagA-facilitated ubiquitination of TAK1. To further dissect the role of Ubc13 in the H. pylori-induced TAK1 ubiquitination, we examined the ubiquitination of TAK1 in AGS cells transfected with control or Ubc13 siRNA. H. pylori infection induced ubiquitination of TAK1 in cells transfected with control siRNA; however, ubiquitination was attenuated when expression of Ubc13 was inhibited by siRNA (Figure 3B). These data reveal that Ubc13 plays an important role in H. pylori-induced ubiquitination of TAK1.
Figure 3. Ubc13 is required for TAK1 ubiquitination and activation by H. pylori.
(A) siRNA against different E2 enzymes were transfected into HEK293T cells, then TAK1 was overexpressed along with HA-ubiquitin and CagA. Anti-Flag immunoprecipitates were immunoblotted for HA for ubiquitination of TAK1. (B) AGS cells transfected with control or Ubc13 siRNA were infected as indicated with H. pylori. Anti-TAK1 immunoprecipitates were immunoblotted with anti-ubiquitin antibodies for the ubiquitination of TAK1. (C) TAK1 and CagA were expressed in HEK293T cells with or without Ubc13-C87A. HA-TAK1 immunoprecipitates were immunoblotted for phosphorylated TAK1. (D) AGS cells expressing TAK1 and either control or Ubc13 siRNA were infected with H. pylori as indicated. HA-TAK1 immunoprecipitates were immunoblotted for phosphorylated TAK1.
Since ubiquitination of TAK1 is critical for its activation and Ubc13 is involved in H. pylori-induced TAK1 ubiquitination, we next determined whether Ubc13 is important for H. pylori-mediated TAK1 activation. We first examined the effect of a catalytically inactive form of Ubc13 (Ubc13-C87A) on the phosphorylation of TAK1. Ubc13-C87A with cysteine 87 mutated to alanine lacks E2 activity and functions as a dominant negative form to suppress activity of Ubc13 [Deng et al., 2000]. Ubc13-C87A not only blocked the basal levels of TAK1 phosphorylation but also inhibited CagA-enhanced TAK1 phosphorylation (Figure 3C), indicating that Ubc13 is involved in the activation of TAK1. To further investigate the role of Ubc13 in H. pylori-induced TAK1 activation, we knocked down the expression of Ubc13 by siRNA in AGS cells and examined the H. pylori-induced TAK1 phosphorylation. H. pylori infection stimulated the phosphorylation of TAK1 in cells transfected with control siRNA (Figure 3D). However, depletion of Ubc13 dramatically reduced TAK1 phosphorylation (Figure 3D). Together, these data demonstrate that Ubc13 is indeed involved in the ubiquitination and activation of TAK1 in response to H. pylori infection.
Ubc13 is required for H. pylori-induced NF-κB activation
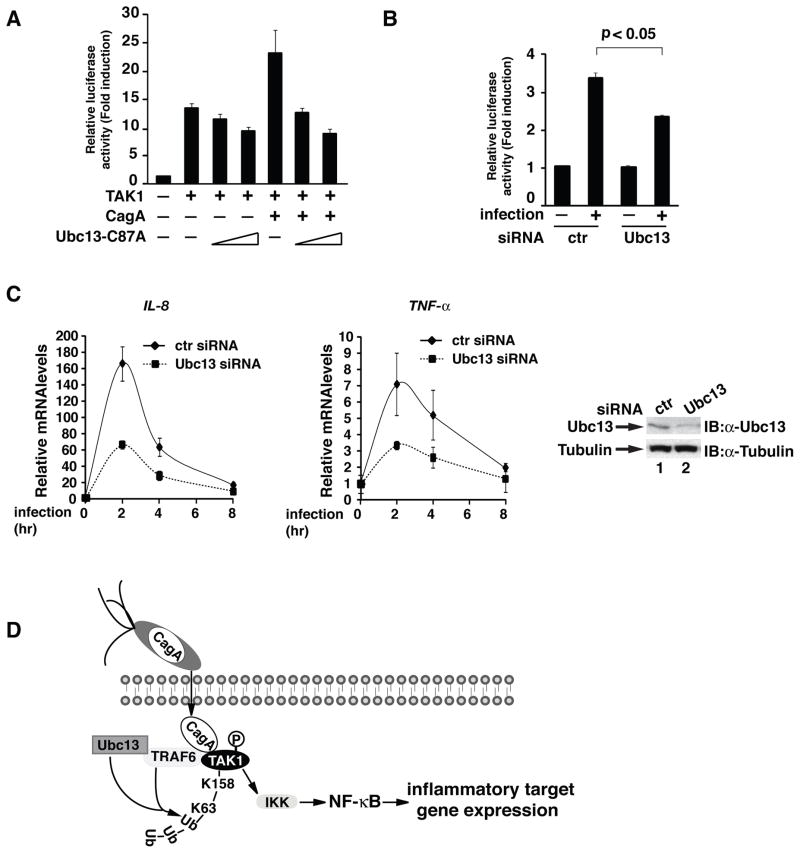
Since our previous studies demonstrate that TAK1 ubiquitination and activation are essential for NF-κB activation in response to H. pylori infection [Lamb et al., 2009], we therefore next evaluated the role of Ubc13 in H. pylori-induced NF-κB activation. We first examined the effect of Ubc13-C87A on the TAK1-mediated and CagA-enhanced NF-κB activation. Consistent with its ability to suppress TAK1 activation (Figure 3C), which is essential for NF-κB activation, Ubc13-C87A inhibited TAK1-mediated and CagA-facilitated NF-κB activation in the κB-luciferase reporter assay in a dose-dependent fashion (Figure 4A). Knocking-down the expression of Ubc13 in AGS cells also inhibited the H. pylori-induced κB-luciferase activity (Figure 4B). To further demonstrate the role of Ubc13 in H. pylori-induced NF-κB activation, we knocked down the expression of Ubc13 in AGS cells and examined the expression of NF-κB target genes after H. pylori infection. Compared to control siRNA, depletion of Ubc13 by siRNA against Ubc13 impaired H. pylori-induced mRNA expression of two NF-κB target genes, TNF-α and IL-8 (Figure 4C). Collectively, these data demonstrate that Ubc13-mediated ubiquitination of TAK1 plays an essential role in H. pylori-induced NF-κB activation and NF-κB-dependent inflammatory target gene expression.
Figure 4. Ubc13 is required for H. pylori-induced activation of NF-κB.
(A) IL-8-luciferase, Renilla, and TAK1 were expressed in HEK293T cells with and without CagA along with increasing amounts of Ubc13-C87A. Luciferase activity was measured 30 hrs post-transfection. (B) Ubc13 or control siRNA was transfected into AGS cells, along with IL-8-luciferase and Renilla. Cells were infected with H. pylori for 6 hrs, then lysates were assayed for luciferase activity. Statistical analysis was performed using a student’s t-test for 2 variables. (C) AGS cells were transfected with control or Ubc13 siRNA, then infected with H. pylori as indicated. mRNA was collected from cells and RT-PCR was performed to determine NF-κB gene transcription. (D) CagA is injected by H. pylori into host epithelial cells, where it interacts with TAK1 and TRAF6. This interaction induces the TRAF6- and Ubc13-dependent ubiquitination of TAK1 at lysine 158. This ubiquitination is required for the activation of the downstream NF-κB pathway by TAK1.
Dicussion
Virulence factor CagA plays an essential role in the H. pylori-mediated NF-κB activation and inflammatory response. CagA associates with TAK1 and enhances its TRAF6-mediated K63-linked ubiquitination [Lamb et al., 2009]. TAK1 ubiquitination is critical for its activation and the subsequent activation of downstream IKKs and NF-κB [Lamb et al., 2009]. In this study, we have identified lysine 158 as the key residue for the ubiquitination and activation of TAK1 in response to H. pylori infection. We also demonstrate that Ubc13 is involved in H. pylori-induced TAK1 ubiquitination and the subsequent NF-κB activation. The identification of the specific lysine and the E2 enzyme for the K63-linked ubiquitination of TAK1 underscores the importance of TAK1 ubiquitination in H. pylori-induced NF-κB activation and NF-κB-dependent expression of pro-inflammatory cytokines.
Several lysine residues within TAK1, including K34, K158 and K209, have been reported to be K63-linked ubiquitinated in response to various stimuli, and ubiquitination at these lysines is essential for TAK1 activation [Fan et al., 2009; Sorrentino et al., 2008; Yamazaki et al., 2009]. Sorrentino et al first reported that K34 was ubiquitinated in response to TGF-β stimulation and that the ubiquitination of this lysine correlated with the TAK1 activation and the downstream activation of JNK [Sorrentino et al., 2008]. K209 was shown to be ubiquitinated by TRAF6 in the IL-1β signaling pathway and was required for IL-1β-induced TAK1 and NF-κB activation [Yamazaki et al., 2009]. Intriguingly, K158, but not K34 or K209, was demonstrated by another group to be important for TGF-β, TNF-α or IL-1β-mediated TAK1 activation [Fan et al., 2011a; Fan et al., 2010; Fan et al., 2009]. Of all these lysines, K158 of TAK1 appears to play a very important role in the ubiquitination and activation of TAK1 in response to H. pylori infection. Mutation of K158, but not K34 or K209, to arginine completely ablated K63-linked ubiquitination of TAK1 and CagA-enhanced TAK ubiquitination (Figure 1 and data not shown). The kinase activity and autophosphorylation of TAK1-K158R was also much less than the wild-type TAK1 (Figure 2), suggesting a possible conformation change of TAK1-K158R to completely inactivate its kinase activity. However, crystal structure of the TAK1 kinase domain, where K158 resides, reveals that lysine 158 is on the surface of the protein, arguing against this possibility [Brown et al., 2005].
While it is clear that ubiquitination of K158 of TAK1 is critical for TAK1 activation, it is still not clear how the ubiquitination of TAK1 activates the kinase. It is expected that, like other K63-linked ubiquitinated signaling molecules, the polyubiquitin chain functions as a scaffold to recruit other signaling molecules which are essential for the activation of TAK1 [Chen, 2012]. For example, MEKK3 has been shown to be recruited to the ubiquitinated TAK1, and this recruitment is required for the activation of TAK1 [Yamazaki et al., 2009]. TAK1 associated proteins, such as TAB1 and TAB2/3, activate TAK1 and specifically bind to K63-linked ubiquitin chains through their zinc fingers [Kanayama et al., 2004]. Ubiquitination of TAK1 may also facilitate the formation of TAK1-TAB2/3 complexes, and the interaction of multiple complexes allows for cross-activation of TAK1, although a recent study shows that free K63-linked ubiquitin chains, not attached to any other proteins, can bind the ubiquitin-binding domain of TAB2 and activate the TAK1 complex in vitro [Xia et al., 2009]. On the other hand, TAK1 ubiquitination may aid in its activity by recruiting substrates, such as the IKK complex. The ubiquitin-binding domain of the regulatory subunit NEMO has been shown to be important in the activation of IKK [Adhikari et al., 2007; Windheim et al., 2008], and it may be that it binds to the ubiquitin chains on TAK1. Supporting this possibility, Fan et al showed that ubiquitinated TAK1 recruited NEMO and the IKK complex, leading to the activation of IKKs [Fan et al., 2009]. Lysine 158 resides in the kinase domain of TAK1, raising another possibility that TAK1 ubiquitination might simply change the conformation of the kinase, allowing for the exposure of its catalytic domain. But the exact molecular mechanism for how ubiquitinated TAK1 activates it kinase activity needs to be further determined.
We have previously shown that E3 ligase TRAF6 is involved in H. pylori-induced TAK1 ubiquitination and activation [Lamb et al., 2009]. TRAF6 functions together with the E2 enzymes Ubc13/Uev1a to catalyze the synthesis of K63-linked ubiquitination [Deng et al., 2000]. For example, Ubc13 mediates the K63-linked ubiquitination of TAK1 in IL-1β signaling [Yamazaki et al., 2009]. Ubc13 is also involved in the K63-linked ubiquitination of ELKS in response to genotoxic stress [Wu et al., 2010]. Consistent with its function as an E2 enzyme, we found that Ubc13 was involved in H. pylori-induced TAK1 ubiquitination, since depletion of Ubc13 impaired H. pylori-induced and CagA-facilitated TAK1 ubiquitination (Figure 3). Furthermore, Ubc13 was required for the activation of TAK1, activation of NF-κB, and NF-κB-dependent target gene expression (Figure 3&4). The identification of Ubc13 as the E2 enzyme for H. pylori-induced TAK1 ubiquitination and activation further supports the notion that K63-linked ubiquitination of TAK1 is essential for the H. pylori-mediated inflammatory response.
Although CagA enhances Ubc13/TRAF6-mediated K63-linked ubiquitination of TAK1 (Figure 4D), how this occurs is still not clear. CagA fails to enhance the interaction between TRAF6 and TAK1 (data not shown), excluding the possibility that CagA might function as a bridging factor to facilitate the TAK1-TRAF6 interaction. It is possible that CagA could target TRAF6 and enhance its E3 ligase activity. Supporting this argument, we also found that CagA interacts with TRAF6 in vitro and enhances its auto-ubiquitination [Lamb et al., 2009]. CagA is associated with the membrane when it enters the host cell, and has been shown to oligomerize [Higashi et al., 2005; Ren et al., 2006]. It is possible that the membrane-bound and oligomerized CagA recruits TRAF6, enhancing its E3 ligase activity and thus the ubiquitination of TAK1. Alternatively, CagA might facilitate the formation of the Ubc13/TRAF6 complex to determine linkage-specific ubiquitination of TAK1. Studies suggest that linkage specificity may be determined either by an E2 alone or by E2–E3 combinations in vivo [Ye and Rape, 2009]. Another possibility is that CagA may inhibit deubiquitination by de-ubiquitinases (DUBs) such as A20 or CYLD [Lamb and Chen, 2013]. These DUBs are capable of removing ubiquitin chains from TRAF6 and TAK1 [Boone et al., 2004; Fan et al., 2011b; Kovalenko et al., 2003; Reiley et al., 2007], thereby enhancing the overall ubiquitination of TAK1.
TAK1 is activated by a wide range of stimuli, including TGF-β, TNF-α, IL-1β, T cell receptor (TCR), and TLR signaling, to modulate immune and inflammatory responses, and K63-linked ubiquitination plays a pivotal role in its activation [Chen, 2012]. It has to be noted that TAK1 and its ubiquitination can be targeted by a variety of pathogens to either enhance or suppress the cellular immune response. In this study, we demonstrate that ubiquitination of TAK1 at K158 is targeted by CagA for H. pylori-induced TAK1 activation and NF-κB-dependent inflammatory target gene expression. TAK1 ubiquitination and activation has also been shown to be necessary for NF-κB activation by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor [Bottero et al., 2011]. Therefore, understanding the molecular mechanism for H. pylori-induced TAK1 ubiquitination and activation not only provides new insights into activation of the NF-κB signaling cascade, but also provides new information for how pathogens might hijack host signaling molecules to activate or suppress NF-κB activation. Inhibiting the ubiquitination of TAK1 by small molecules could be a potential therapeutic approach to ameliorate H. pylori-induced inflammation and the resulting gastric cancer.
Materials and Methods
Cell lines, recombinant proteins, plasmids and antibodies
Human AGS gastric adenocarcinoma cells and human HEK293T cells were purchased from ATCC and cultured in DMEM supplemented with 10% fetal bovine serum (FBS). Recombinant proteins of His-MKK6 (K82A) and expression vectors of CagA, TAK1, and ubiquitin have been previously described [Lamb et al., 2009]. TAK1 mutants were generated using QuikChange site-directed mutagenesis kit from Stratagene. Flag-Ubc13-C87A was received as a gift from Dr. Zhijian Chen (UT Southwestern). Antibodies against CagA, TAK1, Flag, HA, ubiquitin, and His were from Santa Cruz Biotechnology Inc.. Anti-T7 antibody was from Covance and anti-phospho-T187 TAK1 was from Cell Signaling.
H. pylori culture and infection
H. pylori G27 strain was cultured in bisulfite-free Brucella broth on agar media containing Ham’s F-12 medium supplemented with 10% FBS and 5μg/mL vancomycin at 37°C in the presence of 10% CO2. H. pylori was added to AGS cells for infection at an MOI (Multiplicity of Infection) of 50–100.
Transient transfection and luciferase reporter assay
Transient transfection and luciferase reporter assays were performed as previously described [Lamb et al., 2009]. In each luciferase experiment, cells were also co-transfected with EF1-Renilla luciferase reporter plasmid which was used as an internal control. Firefly and renilla luciferase activities were measured with the dual luciferase assay system from Promega.
Immunoprecipitation and immunoblotting analysis
Immunoprecipitation and immunoblotting analysis were performed as previously described [Chen et al., 2002].
siRNA knockdown
Ubc13, Uev1a, and UbcH7 siRNA from Ambion (Ubc13 sense: CCAGAUGAUCCAUUAGCAAtt; Uev1a sense: ACUUACAAGAUGGACAGGGtt; UbcH7 sense: AUGUGGGAUGAAAAACUUCtt) was transfected into AGS cells using Lipofectamine 2000 according to the manufacturer’s protocol. 24 hrs post-transfection, cells were transfected with expression vectors for Flag-TAK1, HA-TAK1, HA-Ub, and/or T7-CagA as described, and 24 hrs later cells were infected or harvested for further experiments.
Quantitative real-time PCR analysis
AGS cells were transfected with control or Ubc13 siRNA using Lipofectamine 2000 (Invitrogen) 48 hrs before being infected with H. pylori for various time-points. Total RNA was extracted using RNeasy Mini kit (Qiagen). Complementary DNA was synthesized with Omniscript RT kit (Qiagen). Quantitative real-time PCR was performed using Qiagen SYBR green PCR kit by 7300 Real-time PCR system (ABI). PCR primers for human β-actin, IL-8 and TNF-α were purchased from Qiagen.
In vitro kinase assay
TAK1 immunoprecipitated from transfected HEK293T cells or H. pylori infected AGS cells, was washed 3 times with IP buffer [Chen et al., 2002] and once with 1X kinase assay buffer (10mM HEPES pH7.4, 1mM MnCl2, 5mM MgCl2, 12.5mM glycerol-2-phosphate, 50μM Na3VO4, 2mM NaF, 0.5mM DTT), and incubated with MKK6(K82A) (1μg) in kinase assay buffer with 70μCi [γ-32P] ATP (3,000 Ci/mmol) for 10 min at 30°C. Samples were separated by 10% SDS-PAGE and visualized by autoradiography.
Acknowledgments
Grant sponsor: NIH/NIDDK; Grant number: DK-085158.
We would like to thank Z.J. Chen (HHMI) for the gift of reagents, and members of the Chen lab for discussion. This work is supported in part by NIH grant DK-085158 (to L.F.C.).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26:3214–26. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- Backert S, Naumann M. What a disorder: proinflammatory signaling pathways induced by Helicobacter pylori. Trends in Microbiology. 2010;18:479–86. doi: 10.1016/j.tim.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Bottero V, Kerur N, Sadagopan S, Patel K, Sharma-Walia N, Chandran B. Phosphorylation and polyubiquitination of transforming growth factor beta-activated kinase 1 are necessary for activation of NF-kappaB by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. J Virol. 2011;85:1980–93. doi: 10.1128/JVI.01911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Vial SC, Dedi N, Long JM, Dunster NJ, Cheetham GM. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. Journal of Molecular Biology. 2005;354:1013–20. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 2002;21:6539–48. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunological Reviews. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28:3538–47. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Fan Y, Yu Y, Mao R, Zhang H, Yang J. TAK1 Lys-158 but not Lys-209 is required for IL-1beta-induced Lys63-linked TAK1 polyubiquitination and IKK/NF-kappaB activation. Cellular Signalling. 2011a;23:660–5. doi: 10.1016/j.cellsig.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Yu Y, Shi Y, Sun W, Xie M, Ge N, Mao R, Chang A, Xu G, Schneider MD, Zhang H, Fu S, Qin J, Yang J. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J Biol Chem. 2010;285:5347–60. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Yu Y, Shi Y, Sun W, Xie M, Ge N, Mao R, Zhang A, Xu G, Schneider MD, Zhang H, Fu S, Qin J, Yang J. Lysine 63-linked polyubiquitination of transforming growth factor-[beta]-activated kinase 1 at lysine 158 is required for tumor necrosis factor [alpha] and interleukin-1 [beta]-induced I[kappa]B kinase/nuclear factor-[kappa]B and c-JUN N-terminal kinase/activator protein 1 activation. J Biol Chem. 2009;285:5347–60. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YH, Yu Y, Mao RF, Tan XJ, Xu GF, Zhang H, Lu XB, Fu SB, Yang J. USP4 targets TAK1 to downregulate TNFalpha-induced NF-kappaB activation. Cell Death and Differentiation. 2011b;18:1547–60. doi: 10.1038/cdd.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Matsuzawa S, Kress CL, Bruey JM, Krajewska M, Lefebvre S, Zapata JM, Ronai Z, Reed JC. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc Natl Acad Sci U S A. 2007;104:6371–6. doi: 10.1073/pnas.0700548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. Curr Opin Microbiol. 2008;11:30–7. doi: 10.1016/j.mib.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Higashi H, Yokoyama K, Fujii Y, Ren S, Yuasa H, Saadat I, Murata-Kamiya N, Azuma T, Hatakeyama M. EPIYA motif is a membrane-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J Biol Chem. 2005;280:23130–7. doi: 10.1074/jbc.M503583200. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Lamb A, Chen LF. The many roads traveled by Helicobacter pylori to NFkappaB activation. Gut Microbes. 2010;1:109–113. doi: 10.4161/gmic.1.2.11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb A, Chen LF. Role of the Helicobacter pylori-Induced inflammatory response in the development of gastric cancer. J Cell Biochem. 2013;114:491–7. doi: 10.1002/jcb.24389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb A, Yang XD, Tsang YH, Li JD, Higashi H, Hatakeyama M, Peek RM, Blanke SR, Chen LF. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10:1242–9. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee DJ, Mobley HL. Pathogenesis of Helicobacter pylori infection. Curr Opin Gastroenterol. 2000;16:24–31. doi: 10.1097/00001574-200001000-00005. [DOI] [PubMed] [Google Scholar]
- Peek RM., Jr Orchestration of aberrant epithelial signaling by Helicobacter pylori CagA. Sci STKE. 2005;2005:pe14. doi: 10.1126/stke.2772005pe14. [DOI] [PubMed] [Google Scholar]
- Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, Leonard TO, Norbury CC, Fitzpatrick L, Zhang M, Sun SC. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–85. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Higashi H, Lu H, Azuma T, Hatakeyama M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J Biol Chem. 2006;281:32344–52. doi: 10.1074/jbc.M606172200. [DOI] [PubMed] [Google Scholar]
- Shibata W, Hirata Y, Maeda S, Ogura K, Ohmae T, Yanai A, Mitsuno Y, Yamaji Y, Okamoto M, Yoshida H, Kawabe T, Omata M. CagA protein secreted by the intact type IV secretion system leads to gastric epithelial inflammation in the Mongolian gerbil model. J Pathol. 2006;210:306–14. doi: 10.1002/path.2040. [DOI] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH, Landstrom M. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol Cell Biol. 2008;28:1783–91. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, Tergaonkar V. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell. 2010;40:75–86. doi: 10.1016/j.molcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629–41. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, Gohda J, Kanayama A, Miyamoto Y, Sakurai H, Yamamoto M, Akira S, Hayashi H, Su B, Inoue J. Two mechanistically and temporally distinct NF-kappaB activation pathways in IL-1 signaling. Sci Signal. 2009;2:ra66. doi: 10.1126/scisignal.2000387. [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]