Abstract
Background
Recent advances in genome wide studies have revealed the abundance of long non-coding RNAs (lncRNAs) in mammalian transcriptomes. The ENCODE Consortium has elucidated the prevalence of human lncRNA genes, which are as numerous as protein-coding genes. Surprisingly, many lncRNAs do not show the same pattern of high interspecies conservation as protein-coding genes. The absence of functional studies and the frequent lack of sequence conservation therefore make functional interpretation of these newly discovered transcripts challenging. Many investigators have suggested the presence and importance of secondary structural elements within lncRNAs, but mammalian lncRNA secondary structure remains poorly understood. It is intriguing to speculate that in this group of genes, RNA secondary structures might be preserved throughout evolution and that this might explain the lack of sequence conservation among many lncRNAs.
Scope of review
Here, we review the extent of interspecies conservation among different lncRNAs, with a focus on a subset of lncRNAs that have been functionally investigated. The function of lncRNAs is widespread and we investigate whether different forms of functionalities may be conserved.
Major conclusions
Lack of conservation does not imbue a lack of function. We highlight several examples of lncRNAs where RNA structure appears to be the main functional unit and evolutionary constraint. We survey existing genomewide studies of mammalian lncRNA conservation and summarize their limitations. We further review specific human lncRNAs which lack evolutionary conservation beyond primates but have proven to be both functional and therapeutically relevant.
General significance
Pioneering studies highlight a role in lncRNAs for secondary structures, and possibly the presence of functional “modules”, which are interspersed with longer and less conserved stretches of nucleotide sequences. Taken together, high-throughput analysis of conservation and functional composition of the still-mysterious lncRNA genes is only now becoming feasible.
Keywords: Long non-coding RNA, antisense RNA, conservation, secondary structure, polypurines, epigenetic
Background
Studies using the recent technical advances in genome-wide platforms have revealed the human genome to be vastly more complex than previously anticipated. While only ~1.2% of the human genome encodes for protein-coding genes1, it is becoming increasingly apparent that the large majority of the human genome is transcribed into non protein-coding RNAs (ncRNAs)2,3.Thousands of long ncRNAs (lncRNAs) have been identified, but very few have been assigned any function. The lack of functional studies and in many cases absence of evolutionary conservation have raised concerns about the importance of lncRNAs; some argue they are nothing more than transcriptional noise4. However, recent reports show thousands of lncRNAs being evolutionary conserved5, though not to the same extent as many protein-coding genes6. While the transcripts of lncRNAs appear less conserved than protein-encoding mRNAs, the promoter regions of lncRNAs are often just as conserved as the promoters of many protein-coding genes3,7. Furthermore as they are RNAs their conservation may be found in functional interactions with proteins and other RNAs, in contrast to the conservation of specific sequence stretches. Functional equivalency of lncRNAs that appear to lack conservation across species may be feasible thanks to the chemical properties of nucleotides and protein interaction affinities.
The function of RNA is indeed widespread; mRNAs encode proteins, rRNA and tRNA are in involved in translation, and microRNAs act by RNA:RNA interactions to modulate mRNA function. In contrast to microRNAs, almost all of which are post-transcriptional repressors, the diverse functions of lncRNAs include both positive and negative regulation of protein-coding genes, and range from lncRNA:RNA and lncRNA:protein to lncRNA:chromatin interactions8–11. Due to this functional diversity, it seems reasonable to presume that different evolutionary constraints might be operative for different RNAs, such as mRNAs, microRNAs, and lncRNAs.
The functional importance of lncRNAs is only now becoming revealed, and to date, of the tens of thousands of metazoan lncRNAs discovered from cDNA libraries and RNAseq data by high-throughput transcriptome projects, only a handful of lncRNAs have been functionally characterized. However, this number has been increasing, with more lncRNAs being found recently to be involved in disease8,10–13. Although the large majority of lncRNAs remain to be characterized there is no longer any doubt that at least some are of functional importance. Yet, the non-conservation conundrum remains: For many lncRNAs already proven functional, poor evolutionary conservation is paradoxical and in stark contrast to the conservation of protein-coding genes.
Lack of conservation does not imbue a lack of function
While conservation almost always indicates functionality, lack of sequence conservation does not directly imply the opposite10,14. The evidence that supports this statement arises from two vastly different classes of non-protein-coding genomic regions with completely opposite evolutionary properties; ultra conserved regions (UCR), which are highly conserved with near perfect sequence identity across all vertebrates, and human accelerated regions (HAR), which show unusually high sequence diversity between human and chimpanzee.
Ultra conserved regions
In a study by Bejerano et al, 481 segments longer than 200 nt were identified to have complete conservation among human, rat, and mouse genomes, and most also in chickens and dogs15. Some of these UCRs were found within protein-coding sequences (111 of 481), while others were found within introns and “gene deserts”. A subsequent study specifically addressed whether these UCRs were transcribed into RNA16. There, Calin et al found that the majority of the UCRs were indeed expressed as RNAs, so called transcribed UCRs (T-UCRs), and intriguingly, demonstrated differential expression in cancer16. While the function of the majority of these T-UCRs remains to be elucidated, it is clear that many of them give rise to non-protein-coding transcripts that do not host known small RNAs, and as such are categorized as lncRNAs. Initial reports suggest some T-UCRs are under microRNA mediated control and also dysregulated in several tumors such as chronic lymphocytic leukemia (CLL)16 and neuroblastoma17. However, further functional studies to elucidate and fully understand the role of T-UCRs remain necessary, in order to definitively determine the mechanistic role of T-UCRs. Additionally, it is imperative that transcriptome datasets from nonhuman species, including cDNA/EST libraries as well as RNAseq results from the modENCODE Consortium, be used to determine the presence, and the exact genomic structure, of any nonhuman orthologous T-UCR transcripts as a prerequisite for understanding their RNA secondary structure and hence their function.
Human accelerated regions
In contrast to T-UCRs, which were found and defined by their high sequence conservation, Pollard et al used an opposite approach18. Instead of looking for highly conserved regions, they identified genomic regions with accelerated rate of nucleotide substitution between human and chimpanzee, with an emphasis on sequences whose substitution rates in evolution prior to the emergence of the human terminal lineage had been lower. Because of the latter property, these sequences were termed “human-accelerated” regions (HAR). A total of 4918 and 20219 HAR regions were initially identified, of which 96% were localized within non-coding segments18. The most divergent of these regions, which had multiple substitutions distinguishing humans and chimpanzees but surprisingly tight sequence conservation between chimpanzees and nonprimate species, was named HAR1. HAR1 was identified to be bidirectionally transcribed as part of two longer lncRNAs in a sense-antisense pair: the lncRNA HAR1A (HAR1 forward) on the forward genomic strand, and the lncRNA HAR1B (HAR1 reverse) on the opposite strand. The HAR1 region was found to be 118 nt long, to reside precisely in the exon-to-exon sense-antisense overlap of these two lncRNA genes (whose reference transcripts range from 900 to nearly 3,000 nt in length, including the HAR1 118 nt sequence), and to fold into an organized secondary RNA structure whose differences between human and chimpanzee have been biochemically confirmed by independent studies 18,20. Interestingly, it was suggested that the mutations in the human HAR1 compared to the chimpanzee sequence, stabilized this RNA structure further and were therefore evolutionary produced through positive selection20. Alternatively, this varied secondary structure may be involved in sense antisense pairing of HAR1B and HAR1A, which are reverse complement and overlapping one another, thus allowing for RNA:RNA pairing and higher ordered secondary structures to form. The HAR1 ncRNA was found to be expressed in developing neocortex early in human embryonic development and to co-localize with Reelin, an important brain protein with functions in schizophrenia and aging. Therefore, the authors speculated whether the increased rate of nucleotide substitutions within this region is of importance for human brain evolution. This example illustrates that poorly-conserved ncRNAs can have specific spatiotemporal gene expression patterns that strongly suggest function, and that major aspects of lncRNA secondary structure can undergo drastic changes during evolutionary events, such as during the emergence of modern humans. Neanderthal and Denisovan genomes, which recently became publicly available, collectively provide an invaluable resource that will allow more precise timing of sequence substitutions concomitant with RNA secondary structure changes within the last 50,000 years of human evolution.
Many more HARs, as well as T-UCRs, remain to be investigated, as improved bioinformatics and high-throughput RNA sequencing approaches make it possible to discover additional rapidly evolving regions and additional evidence of transcriptional activity, respectively. It will be of great interest to gauge the extent to which these regions are transcribed as ncRNAs and the role that these regions may have in cellular function and evolution19.
LncRNAs and secondary structures
The vast majority of post-genomic lncRNA experimental biology has been an observational science, a modern equivalent to Darwin’s voyage on The Beagle: high-throughput cDNA library construction and next-generation RNA sequencing have provided deep and comprehensive catalogs of lncRNA genes and transcripts, while the inherent bottleneck between the large size of these datasets and the low throughput of experimental validation methods has ensured that functional validation lags far behind. For this reason, only a relatively few lncRNAs have been functionally characterized to date, and even fewer have been investigated for their secondary structure and the interplay between structure and function.
Primary sequence conservation of lncRNA genes, across species, has already been studied genomewide in mammals21–23. Jointly, these three studies establish that genomic sequence conservation and gene structure conservation are rare at orthologous and positionally-equivalent lncRNA loci, and that intergenic lncRNAs are subjected to rapid turnover during evolution. The presence and absence of apparently species-specific lncRNAs at orthologous loci in related species, and the gene structure differences that affect even conserved lncRNAs in these studies, are suggestive of lncRNA functional differences between species as well. These three genomewide studies collectively provide thousands of lncRNA loci affected by such differences. There is a need for additional global studies of lncRNA evolution. In order to motivate the field to carry out such studies and in-depth analyses of specific functional lncRNAs, we have canvassed the existing literature in order to show the potential for these types of studies to enhance our understanding of RNA structure and human disease. Accordingly, here, we highlight some of the lncRNAs for which these questions have been addressed.
Steroid receptor RNA activator
The steroid receptor RNA activator (SRA) is an lncRNA which has, partially, undertaken increased rate of mutation in the human lineage24. The SRA locus expresses several different RNA isoforms, including the protein coding mRNA (SRAP) as well as several lncRNAs, which exhibit a wide array of alternatively spliced variants. Here, we focus on the structural analysis of the primarily expressed lncRNA isoform (ncSRA).
The ncSRA has been shown to be a co-activator for several nuclear receptors and to interact with several proteins such as the nuclear co-activator SRC-125, the nuclear repressors SHARP26 and SLIRP27. Moreover, increased expression of ncSRA has been linked to breast cancer28–31, concordant with the original discovery of SRA as a co-activator of the estrogen receptor alpha, a nuclear hormone receptor whose signaling is central to estrogen-dependent breast cancer pathogenesis25. Novikova et al performed extensive analysis of the secondary structure of the 0.87kb long ncSRA. Using chemical probing as well as enzymatic treatment with RNase V1, which cleaves base paired regions, some remarkable observations were generated24. A four-domain structure (domain I-IV) with 25 helices was identified and different segments of this structure appeared to have evolved separately with clear differences on the level of sequence conservation. Specific helices are highly conserved, while one junction with branching helices has 57% of its bases 100% conserved in all mammals, down to marsupials and monotremes. Hence overall the SRA lncRNA structure is deeply conserved across 45 species at a variety of secondary-structure elements throughout the SRA sequence. While terminal loops, bulges and looping regions were in general well conserved, base paired regions appeared less conserved. Moreover, it was also observed that the majority of single stranded regions were rich in purines (adenine and guanine), so-called polypurine regions. Covariance analysis among 45 eukaryotic species showed 14 of 25 helices to have at least one covariant base pair, thus indicating selection for preserving the secondary structure. A detailed conservation analysis of ncSRA between mouse and human showed 99 positions which had mutated throughout the sequence, of which 58 were predicted to stabilize the secondary structure.
The SRA locus encodes several ncRNAs and protein-coding mRNAs. In the protein-coding SRA RNAs, frame-preserving indels are widespread and out-of-frame indels are also surprisingly frequent in unrelated mammalian lineages. There are significant disruptions of protein-coding potential across lineages in the interspecies ORF comparison of SRA. This suggests that selective constraint is preserving the RNA secondary structure more than the protein sequence. Therefore, Novikova et al found that SRA protein function appears to be dispensable24. These observations represent one of the few studies which thoroughly investigate the structural aspects of a lncRNA. It would be of great interest to further map the interactions between the SRA lncRNA and the proteins that are already known to associate with it, such as SRC-1, SHARP and SLIRP25–27, and to study whether any of the more conserved (or non-conserved) bulges, stems, loops, or other domains in SRA’s secondary structure specifically interact with certain proteins, thus acting as scaffolds for forming protein complexes.
Growth arrest-specific 5 RNA
The spliced and poly-adenylated Growth Arrest-Specific 5 (GAS5) RNA was initially identified as a putative tumor suppressor gene due to its accumulation during growth arrest32. Sequence comparison between lncRNA-GAS5 exons in humans and mice indicated poor conservation. In contrast, some parts of the introns contained highly conserved regions, which were revealed to be the locus for several small nucleolar RNAs (snoRNAs)33. However, as intriguing as these observations were they did not address the function of the spliced lncRNA-GAS5. Notably, differentially spliced lncRNAs may interact with different protein complexes and affect gene functions including splicing34. The function of the lncRNA-GAS5 remained unknown until a study by Kino et al revealed that lncRNA-GAS5 acts as a decoy for the glucocorticoid receptor (GR)35. Upon binding to a glucocorticoid agonist, GR translocates from the cytoplasm to the nucleus where it binds to glucocorticoid response elements (GREs) via its DNA binding domain (DBD) and influences many cell functions including metabolism, cell survival and the response to apoptotic stimuli. Intriguingly, lncRNA-GAS5 is predicted to fold into a secondary RNA structure36, which exposes an RNA sequence that mimics the genomic DNA GRE. The GRE mimic sequences of lncRNA-GAS5 reside at nt 539–559. They are located in the stem part of the 5th (of 6 total) stem-loop structure of the RNA, toward the 3’end of the RNA35. This part of the lncRNA-GAS5 sequence then physically binds to the DNA-binding domain (DBD) of the GR, titrating out bioavailable GR molecules by preventing them from binding genomic-DNA GREs. This RNA:protein interaction blocks the binding between GR and GRE and the lncRNA-GAS5 thus ultimately acts as a decoy and transcriptional repressor for the GR. Although the mouse and human GAS5 exonic sequences share ~70% nucleotide homology, the GRE-mimic sequences in human GAS5 are conserved in mouse GAS5, which is the only other species in which the GAS5 sequence that has been reported to date35. Only experimental work in mammals outside of human and mouse would show whether any species in which GRE-mimic sequences are <100% conserved still have GAS5 interactions with GR.
Both GAS5 and SRA belong to the emerging class of lncRNAs that function as endogenous riboregulators by directly interacting with dual RNA- and DNA-binding proteins that serve as transcription factors: in this case, the nuclear hormone receptors GR and, ER respectively. GAS5 has been shown to possess a wealth of functions related to cellular growth arrest and apoptosis37,38. GR-mediated function of the unprocessed lncRNA-GAS5 is not related to the short snoRNAs which can be processed out of GAS5. There are two other snoRNA hosts whose unprocessed lncRNAs are known to possess distinct functions39,40. Rigorous and deep concurrent short-RNA and long-RNA sequencing, such as that being performed by the ENCODE Consortium and the FANTOM Consortium, should enable future computational analysis of RNAseq data from all snoRNA host loci, required to establish the extent to which these loci give rise to stable long transcripts, in addition to processed short molecules.
The X inactive specific transcript
One of few lncRNAs which has been extensively characterized on both the functional and structural level, is the X inactive specific transcript (Xist). Xist is a ~17kb lncRNA essential for mammalian X chromosome inactivation41–45. Xist RNA spreads along the inactive X chromosome, and this is followed by induction of a series of PRC2-mediated repressive chromatin marks. Interestingly, the most conserved regions of Xist correspond to low-copy repetitive elements, where the repetitive element A (repA) is the most highly conserved46,47. RepA localizes at the 5’ end of Xist, a region which has been found to be essential for X chromosome inactivation48. RepA binds, and recruits the chromatin remodeling polycomb repressor complex 2 (PRC2), consisting of EZH2, SUZ12 and EED, which initiate the X inactivation by chromatin remodeling49. The RNA structure within the repA element has been investigated and shown to contain two loops, linked together by a uracil rich linker sequence, which is divergent between humans and mice. While the entire repA region appears essential for SUZ12 interactions and subsequent efficient silencing, deletion constructs demonstrated that both EZH2 and EED bind sub regions of the repA region. Taken together, this indicates that the repA region may act as a scaffold, whereby different parts of the secondary RNA structure recruit certain proteins and bring them together into one complex. Supporting this notion, Wutz et al generated transcripts with different modifications, both on the sequence level as well as the length of the linker between the two loop structures and found that these modifications had no effect on the repA capacity to induce chromatin remodeling and X chromosome inactivation50. Such observations suggest that the repA linker is preferentially involved in bridging the two protein binding modules. Xist and other lncRNAs at the X-Inactivation Center have arisen from a mosaic combination of pseudogenized protein-coding genes and repetitive element insertions, and while parts of the Xist locus arise from ancestral sequences that are autosomal in birds and marsupials, the Xist lncRNA is specific to eutherian mammals51,52. Although Xist is processed to small RNAs, this processing is likely non-essential, because X-inactivation is Dicer-independent53,54.
HOX antisense intergenic RNA
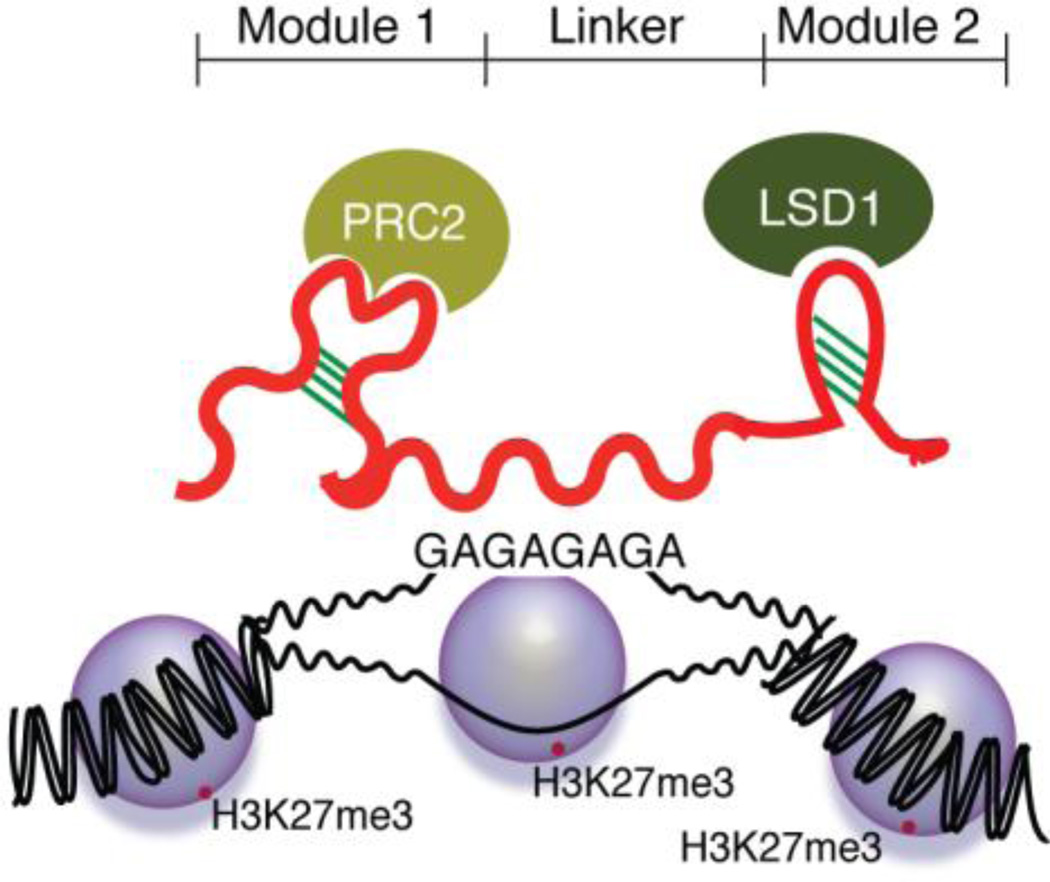
HOX antisense intergenic RNA (HOTAIR) is an lncRNA encoded within the HOXC locus and has been shown to mediate chromatin remodeling of the HOXD locus55. Increased expression of HOTAIR has also been observed in primary breast tumors and metastases8. To date, it has been shown that HOTAIR consists of two different modules, which are connected by a linker sequence. No particular RNA folding has been reported for HOTAIR, but it has been shown that one module on the 5’ end binds the chromatin remodeling complex PRC2, while the other module binds the lysine-specific demethylase 1 (LSD1)8,56, which specifically demethylates Histone 3 Lysine 4 (H3K4me2)57. Such a transcript might be active as a localized chromatin regulator, a smaller scale enhancer RNA for instance58, involved in chromatin and/or domain looping based interactions. It is currently unclear whether the linker functions to connect the two modules, possibly at a predetermined spacial distance, or whether other yet-to-be determined functions are maintained within this region (Figure 1). It would thus be interesting to generate deletion constructs of HOTAIR where the linker sequence is modified and the length is altered in order to investigate the functional characteristics of this region.
Figure 1. HOTAIR mediated chromatin remodeling.
LncRNA HOTAIR functions as a scaffold and brings the chromatin remodeling factors PRC2 and LSD1 in close proximity to each other. PRC2 and LSD1 interact with two separate RNA modules in HOTAIR, which are connected with a linker. The HOTAIR:protein complex is recruited to polypurines by a so far unknown mechanism, whereby suppressive epigenetic marks, such as H3K27me3 is induced.
Adding to the importance of this lncRNA-regulated locus are recent observations from studies on HOTAIR in mice (mHOTAIR) that call into question the concept and over-importance placed on nucleotide conservation among lncRNAs. Human HOTAIR is intergenic and localized between HOXC12 and HOXC11. By looking for mouse orthologues for HOXC12 and HOXC11, a corresponding mouse intergenic region encoding the mHOTAIR was found59. The authors specifically addressed whether the sequence and function of HOTAIR/mHOTAIR is conserved among human and mouse. While human HOTAIR consists of six exons, mHOTAIR only contains two exons. Although peaks of higher conservation were observed, the overall sequence conservation was low. Moreover, the 5’ end of the transcript, which has been described to contain the PRC2 interacting module, did not appear in mouse55. In addition, the LSD1 binding domain also showed poor sequence conservation. Indeed, absence of mHOTAIR only showed minor effects, if any, in mice and in addition, poorly overlapped with changes on the chromatin level59. This is consistent with the absence, in mice, of the human exons that contain the PRC2-interacting domain. It is interesting to speculate whether the function of HOTAIR has emerged specifically in the human lineage, and whether mHOTAIR maintains other functions in mice, still not characterized. It would for example be of great interest to investigate if the mHOTAIR maintains binding capacity to PRC2 and/or LSD1, inspite of the lack of sequence conservation. Such investigations would clearly illustrate the interplay between structural and sequence conservation. Taken together, despite the presence of orthologs, these observations collectively suggest that careful considerations should be taken when making the assumption of ortholog functions.
MALAT1
The lncRNAs MALAT-1 (metastasis-associated lung adenocarcinoma transcript 1, also known as NEAT2) has been identified to be evolutionary conserved within multiple mammalian species, while no homologues was present in non-mammalian species60. MALAT-1 is involved in the formation of nuclear speckles, which are thought to be involved in the processing of pre-mRNAs (reviewed in61) and has further been reported to be dysregulated in numerous different cancers62,63. Interestingly, MALAT-1 exhibit a particular so-called cloverleaf structure at the 3’ ends of its transcript. This cloverleaf structure is evolutionary conserved and appears important for 3’ end processing and generation of two mature transcripts64. The 3’ end of MALAT-1, including the downstream region that is cleaved by RNAse P and processed into the tRNA-like small RNA known as mascRNA, is conserved from humans to fish65,66. Future studies of transcriptomes – not genomes – of nonmammalian model organisms are essential for resolving the questions that are still outstanding, such as whether the conserved MALAT-1 3’ end gives rise to mascRNA-like RNAs in nonmammalian species. Although nuclear speckles, which contain MALAT-1, appear to be unique to mammals, the deep evolutionary conservation is consistent with the findings that specific RNAs – though not homologous to MALAT-1 – are involved in subnuclear structure formation in nonmammalian vertebrates and in other metazoa67. The cloverleaf structure is a four-way-junction structure, which mimics the structure of a pre-tRNA. In a similar fashion as tRNAs, the MALAT1 cloverleaf is recognized, and cleaved on its 5’end by RNase P, followed by cleavage on the 3’ by RNase Z68,69. The RNase P/RNase Z processing thus generates a 7kb long nuclear lncRNA and also a 61 nt long ncRNA transcript, which localizes to the cytoplasm64,70. The cloverleaf structure of the MALAT1-associated small cytoplasmic RNA (mascRNA) has been conserved between human and mouse and the four mutations which are present still maintain the same structure64. The function of both MALAT-1 and mascRNA still remains poorly understood. MALAT-1 is not essential for the formation of nuclear speckles. Although the roles of MALAT-1 in human cancer and in an increasing number of human neurological diseases have been thoroughly investigated71, mouse Malat-1 knockouts are phenotypically normal, without any differences in either behavior or cancer predisposition relative to wildtype72. Functional characterization of mascRNA in colorectal malignancies suggests a role during cell proliferation, migration and invasion73.
Polypurine elements: (one) missing link in lncRNA function
Recent observations have begun to highlight the importance of polypurine elements (repeats of guanine and adenine) for lncRNA mediated regulation9,74,75. A newly developed method by Chang and colleges made it possible to study physical interactions of lncRNAs with other RNAs, chromatin and proteins on the genome wide level. By tiling the lncRNA of interest with a number of biotinylated antisense oligos, efficient pulldown of the lncRNA HOTAIR and its interactome with chromatin (Chromatin isolation by RNA Purification = ChIRP) was successfully performed74. ChIRP on the lncRNA HOTAIR revealed binding to more than 800 loci. These loci significantly overlapped with the presence of the PRC2 subunits EZH2 and SUZ12 and enrichment of the suppressive chromatin mark H3K27me3, strongly supporting the involvement of HOTAIR in chromatin remodeling. Interestingly, the authors further investigated the HOTAIR binding regions and revealed the presence of polypurine elements, suggesting guanine and adenine repetitive elements being involved in the recruitment process.
In another study, Kretz el al studied the terminal differentiation-induced ncRNA (TINCR), which is a 3.7 kb lncRNA, expressed during human epidermal differentiation9. A similar tiling approach as described for HOTAIR above was applied and modified for TINCR and putative RNA:RNA interactions. TINCR was found to interact with ALU elements of mRNAs, causing a destabilizing effect of the targeted mRNAs. This destabilization was shown to be mediated by the RNA binding protein Staufen 1 (STAU1)76–78. Interestingly, binding motif analysis of the TINCR interacting RNAs also revealed the presence of a polypurine-binding motif, thus very similar to the binding motif of HOTAIR.
Taking advantage of the observation that polypurine motifs appear important for the function of lncRNAs, an algorithm was generated to detect and target such sequences using small antisense RNAs (sasRNAs)75. This algorithm was found to be useful in the design sasRNAs capable of modulating RNA directed epigenetic silencing. The mechanism of the polypurines remains to be investigated in detail, but it is tempting to speculate whether such elements are frequently occurring throughout numerous lncRNAs. Although the functional importance, if any, still has to be investigated, such polypurines may either undergo secondary folding, or alternatively link functional RNA domains together. Moreover, it would be of great interest to study whether such elements are conserved among different species. Revealing functional elements of lncRNAs, such as polypurines, will be of great importance in order to mimic, or possible disrupt, the action of lncRNAs, which could be of therapeutically interest in order to modulate gene expression.
AntisenseRNAs
Cis acting asRNAs
It has been estimated that approximately 20–40% of all protein-coding genes have antisense RNA (asRNA) transcription21,79,80. AsRNAs share complementarity to a sense-expressed transcript, which is usually a protein-coding gene. Promoters, untranslated regions, protein-coding regions, and introns all can be overlapped by antisense RNAs transcribed from the same locus as the protein-coding gene. With the exception of intronic overlaps, all these scenarios confer the possibility of post-transcriptional, including cytoplasmic, regulation of sense mRNAs by antisense lncRNAs. The genomic structure of mRNA-lncRNA sense-antisense overlaps has been surveyed in numerous genomewide studies21,81.
Different modes of regulation have been suggested for asRNA transcripts, mainly acting as concordant or discordant regulators of its sense counterpart80,82. Antisense RNAs can also act as suppressive regulators by recruiting repressive chromatin remodeling proteins10–12, as well as positive regulators, by stabilizing the corresponding sense transcript through RNA:RNA interactions10,83. Since in cis expressed sense:asRNA (SA) pairs share the same locus, they are ultimately tightly linked with each other through evolution. This raises several interesting questions regarding their function and sequence conservation.
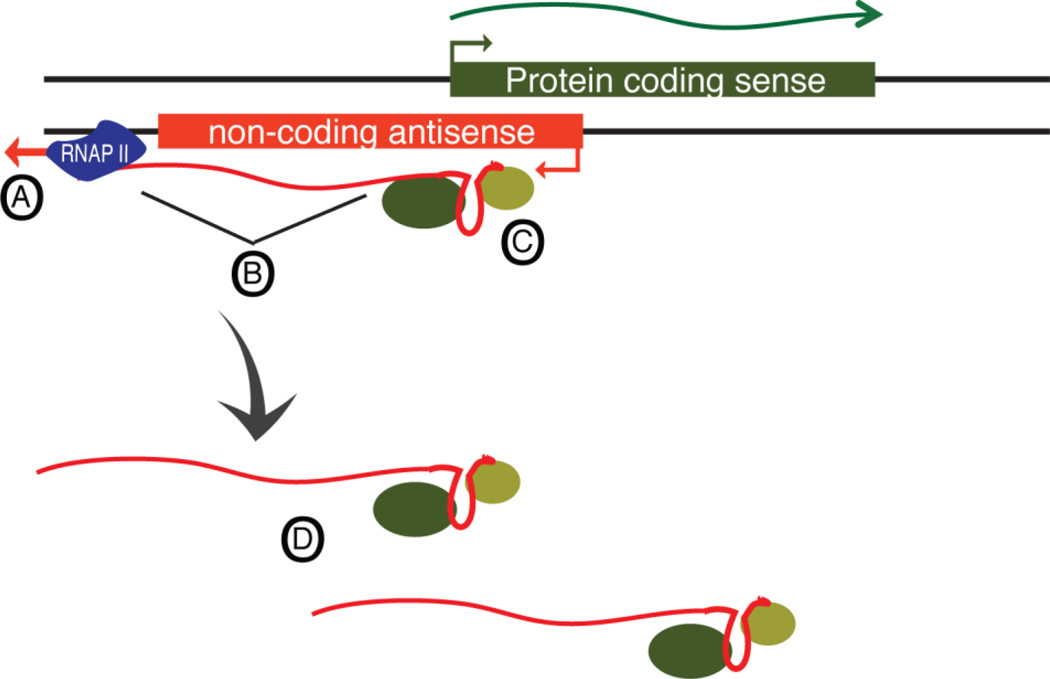
Being expressed from the same locus ultimately generates sequence overlap and genomic proximity. First, SA pairs evolve together, making the sequence overlap continuous over time, even though sequence mutations may arise. Evidence for functional importance of active transcription of lncRNAs, regardless of their sequence, is surveyed across multiple model organisms84. Second, the transcription per se may generate proximity to the genomic locus where it is being transcribed by tethering the asRNA transcript to the DNA through ongoing transcription (Figure 2). Tethering refers to a stable RNA-DNA hybrid that remains in place after the RNA is transcribed and that causes epigenetic remodeling of the DNA allele that gave rise to the transcript. The asRNA of interest may have a protein binding domain where conservation at the structural and/or sequence level is of importance (Figure 2C), while the majority of the sequence rather maintain tethering. The ongoing transcription will mediate a RNA:RNPII:DNA hybrid through transcription, where the proximity is maintained as long as transcription is active (Figure 2A–D).This promotes the idea that asRNAs may, at least in part, act independently of their actual sequence (Figure 2B). Ongoing transcription of the asRNA at the locus in question may be important for tethering the asRNA to the region which it is transcribed from, while SA pair regulation occurs due to their shared genomic location and thus overlapping sequences.
Figure 2. In-cis mediated regulation may be controlled by tethering.
(A) Ongoing transcription tethers the asRNA transcript to the genomic locus. A DNA-RNPII-RNA complex maintains the tethering and its cis location. (B) The genomic sequence is not of importance, as long as the transcription, and tethering, is ongoing. (C) The asRNA contain a structure with the capacity to bind and recruit RNA biding proteins and ultimately regulates the expression of the protein coding sense gene. (D) Once transcription of the asRNA stops, the asRNA loses location and capacity to regulate the sense gene.
The FANTOM3 Consortium, which generated a set of 600,000 mouse full-length cDNA and EST sequences that remains the most comprehensive experimentally derived full-length transcript catalog in any mammalian system to date, investigated the conservation of cis SA pairs among human and mouse21. Surprisingly, only 17% of the pairs were found conserved between these species. Even though not addressed within this study, it would be of interest to investigate the degree of sequence conservation among these 17% SA pairs and also determine if there are any conserved motifs. In a similar fashion as HOTAIR, it may be speculated that asRNAs may also consist of different modules, coupled together by linker sequences that may show less sequence, but possible size dependent, constraints.
Many of these above suppositions have not yet been addressed to date. Without a doubt, though, it will be of great interest to reveal the role of structure, conservation and composition among SA pairs, and also to understand the reason for the lack of conservation among most human and mouse SA pairs.
Trans acting asRNAs
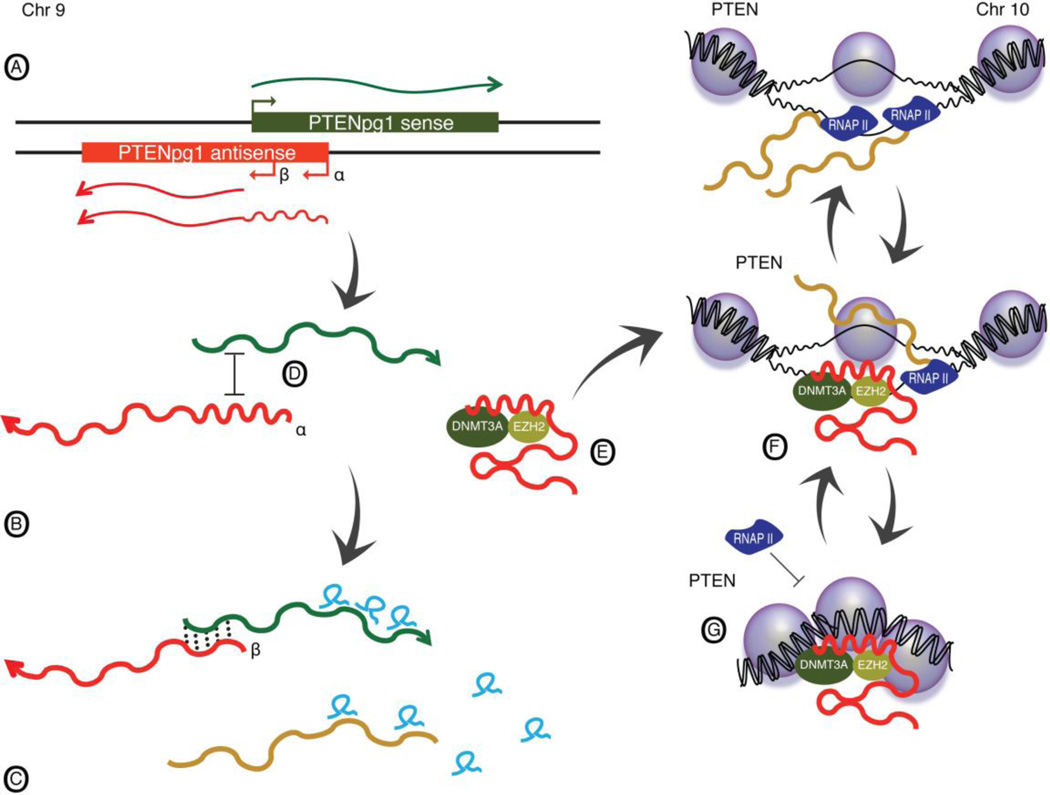
A recent example of trans acting asRNA-mediated regulation was presented by Johnsson et al10. In this body of work an asRNA to the tumor suppressor gene PTEN was found transcribed from a pseudogene to PTEN (PTENpg1, also called PTENp1)(Figure 3). Although expressed in trans, this asRNA exhibited high sequence homology with PTEN (>95%). Interestingly, PTENpg1 was found to express two different asRNAs, alpha and beta (Figure 3A). Through its shared sequence homology, the alpha transcript recruits chromatin-remodeling complexes to the PTEN promoter (Figure 3 E–G). In contrast, the beta transcript was observed to interact with the PTENpg1 sense through RNA:RNA based interactions. This RNA:RNA interaction increased the stability of PTENpg1, thus affecting the sponging of PTEN related miRNAs and consequently translation of PTEN (Figure 3B–C). Even though the alpha transcript exhibited greater overlap on the 5’ end with PTENpg1 sense, the alpha isoform was not found to stabilize the PTENpg1 sense transcript. Albeit not addressed within the study, it is intriguing to speculate that the longer alpha transcript folds into a secondary structure, which covers part of the sequence and makes it unavailable for interactions with the PTENpg1 sense (Figure 3D). Such variation in RNA folding allows the PTENpg1 alpha variant an alternative function, independent of the PTENpg1 sense, thus increasing the inherent complexity of PTEN regulation.
Figure 3. In-trans mediated asRNA regulation of PTEN.
(A) The PTENpg1 locus encodes for three different lncRNAs; PTENpg1 sense, PTENpg1 asRNA alpha and beta. (B) The PTENpg1 asRNA beta interacts with and stabilizes the PTENpg1 sense through RNA:RNA interactions, (C) whereby microRNA sponging and consequently PTEN translation is affected. (D) The PTENpg1 asRNA alpha does not interact with PTENpg1 sense, presumably due to RNA secondary structures. (E) The PTENpg1 asRNA alpha binds the chromatin remodeling factors DNMT3a and EZH2 and (F) is recruited to the PTEN promoter where (G) transcriptional repression is induced by the formation of H3K27me3.
While observed to be functional in human cells, the formation of the PTENpg1 is a recent evolutionary event with this locus lacking in mice. The generation of pseudogenes, in particular so called processed pseudogenes, is thought to be caused by a recent burst of retrotranspositional activity in the ancestral primates about 40 million years ago85,86. Although only a few pseudogenes have been functionally investigated10,14,87, thousands have been shown to be transcribed, many of which lack orthologs in mice88, suggesting some level of uniqueness to primates89. Notably, those pseudogenes that have to date been found to be functionally active, are active in modulating the therapeutically and disease relevant OCT4 and PTEN protein-coding genes10,90.
Concluding remarks
Thousands of lncRNAs have been identified during the last couple of years. Functional studies for most of these lncRNAs are however still lacking with only a handful having been characterized in detail8,10,11,90. From these few studies it is apparent that some lncRNAs are important cellular effectors ranging from splice complex formation34 to chromatin and chromosomal complex formation43,46 to epigenetic regulators of key cellular genes11,12,90,91. Some lncRNAs have been found to act in cis, such as many antisense RNAs92, while others, such as lincRNAs and pseudogenes, often act in trans. In addition, some lncRNAs are positive regulators, while others are negative regulators of gene expression. Due to a lack of understanding, the functional characterization of lncRNAs is today challenging, with the main approach for investigation dependent more on functional experiments involving depletion and overexpression studies. This is most likely due to lncRNA function, and selective pressures thereon, residing predominantly in its structure and protein interaction repertoire, rather than primary sequence context.
It is becoming increasingly apparent that lncRNAs do not show the same pattern of evolutionary conservation as protein-coding genes. Many lncRNAs have been shown to be evolutionary conserved5; but they do not appear to exhibit the same evolutionary constraints as mRNAs of protein-coding genes3. This maybe is the result of expression patterns of many lncRNAs being conserved among different species due to similarities in their regulatory promoter elements.
It has been observed that several lncRNAs act as multi-modular regulatory units45,50,93. The lncRNAs HOTAIR and XIST (in its repA region) both have two different modules, while the ncSRA has four different modules. While certain regions of the lncRNAs appear to maintain the regulatory function, such as bulges and loops, the exact sequence in other regions of lncRNAs appear less important and possibly act as spacers in order to link functional units or modules. Depending on the function, e.g. whether the RNA sequence is a linker or a functional module, different patterns of conservation might be expected.
In order to address these questions, it will be of great importance to understand the RNA structure and the interplay between structure and sequence. Some of the examples highlighted within this review suggest that evolutionarily observed mutations could represent positive selection for instance by favoring stabilizing RNA structures within lncRNAs. Furthermore, a recent study using PARS (Parallel Analysis of RNA Structures) investigated RNA structures on the genome wide level in yeast, showing that physiological stimuli largely changed RNA structures94. It was observed that stable RNA structures were more prevalent in ncRNAs, such as rRNA, tRNA, snoRNA and snRNA, compared to protein-coding mRNAs. This observation was persistent for both the coding region as well as the 3’ and 5’ UTRs, again indicating the importance of specific RNA secondary structures in the function of ncRNAs. In addition to this group’s PARS method for high-throughput, RNAseq-assisted determination of RNA secondary structure, competing approaches such as FragSeq95 and 3S96 have been developed that will soon afford important complementary insights into the structure of the mammalian lncRNAome. Taken together, understanding the structure aspects of lncRNAs will be of great importance to fully understand the evolution, form and function of these emerging regulatory elements.
Table 1.
Conservation of five known functional lncRNAs.
| lncRNA | most distant species from human where conservation was detected | conservation type (genomic, transcription, gene structure, RNA secondary structure) | reference |
| SRA | marsupials and monotremes | genomic sequence; and RNA secondary structure | 24 |
| GAS5 | mouse | genomic sequence; transcription; and RNA secondary structure | 35 |
| XIST | eutherian mammals | genomic sequence; transcription; and gene structure | 52 |
| HOTAIR | mouse | genomic sequence; transcription (but gene structure is different) | 59 |
| MALAT-1 | fish | genomic sequence at 3’end | 65 |
Highlights.
Recent genome wide studies have revealed the presence of thousands of lncRNAs.
Many lncRNAs do not show the same pattern of conservation as protein-coding genes.
Due to the lack of sequence conservation, functional interpretation is challenging.
The presence, and conservation, of secondary structural elements have been suggested.
This phenomenon remains poorly studied, and we explore what is currently known.
Acknowledgements
The project was supported by the National Institute of Allergy and Infectious Disease (NIAID) P01 AI099783-01 to KVM, the Swedish Childhood Cancer Foundation, The Swedish Cancer Society, Radiumhemmets Forskningsfonder, the Karolinska Institutet PhD support programme, and Vetenskapsrådet to DG. The Erik and Edith Fernstrom foundation for medical research to P.J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Collins FS, Lander ES, Rogers J, Waterston RH, Conso IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature Structural & Molecular Biology. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Research. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends in Genetics. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–U245. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnsson P, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipovich L, et al. Activity-Dependent Human Brain Coding/Noncoding Gene Regulatory Networks. Genetics. 2012;192 doi: 10.1534/genetics.112.145128. 1133-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–U1090. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, et al. Ultraconserved regions encoding ncRNAs are, altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 17.Mestdagh P, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29:3583–3592. doi: 10.1038/onc.2010.106. [DOI] [PubMed] [Google Scholar]
- 18.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 19.Pollard KS, et al. Forces shaping the fastest evolving regions in the human genome. Plos Genetics. 2006;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beniaminov A, Westhof E, Krol A. Distinctive structures between chimpanzee and human in a brain noncoding RNA. RNA. 2008;14:1270–1275. doi: 10.1261/rna.1054608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engstrom PG, et al. Complex loci in human and mouse genomes. Plos Genetics. 2006;2:564–577. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutter C, et al. Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression. Plos Genetics. 2012;8 doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood JE, Chin-Inmanu K, Jia H, Lipovich L. Sense-antisense gene pairs: sequence, transcription, and structure are not conserved between human and mouse. Front Genet. 2013 Sep; doi: 10.3389/fgene.2013.00183. In press (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Research. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 26.Shi YH, et al. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes & Development. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatchell EC, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Molecular Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 28.Hussein-Fikret S, Fuller PJ. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Molecular and Cellular Endocrinology. 2005;229:149–160. doi: 10.1016/j.mce.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Lanz RB, et al. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Molecular and Cellular Biology. 2003;23:7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Research. 1999;59:4190–4193. [PubMed] [Google Scholar]
- 31.Murphy LC, et al. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Research. 2000;60:6266–6271. [PubMed] [Google Scholar]
- 32.Schneider C, King RM, Philipson L. Genes Specifically Expressed at Growth Arrest of Mammalian-Cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 33.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5 '-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Molecular and Cellular Biology. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry G, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 35.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Science Signaling. 2010;3 doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dimitrov RA, Zuker M. Prediction of hybridization and melting for double-stranded nucleic acids. Biophysical Journal. 2004;87:215–226. doi: 10.1529/biophysj.103.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochemical Society Transactions. 2011;39:482–486. doi: 10.1042/BST0390482. [DOI] [PubMed] [Google Scholar]
- 38.Pickard MR, Mourtada-Maarabouni M, Williams GT. Long noncoding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochimica Et Biophysica Acta-Molecular Basis of Disease. 2013;1832:1613–1623. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Powell WT, et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Askarian-Amiri ME, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown CJ, et al. A Gene from the Region of the Human X-Inactivation Center Is Expressed Exclusively from the Inactive X-Chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 42.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 43.Lucchesi JC, Kelly WG, Parming B. Chromatin remodeling in dosage compensation. Annual Review of Genetics. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 44.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 45.Jeon Y, Lee JT. YY1 Tethers Xist RNA to the Inactive X Nucleation Center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown CJ, et al. The Human Xist Gene - Analysis of a 17 Kb Inactive X-Specific Rna That Contains Conserved Repeats and Is Highly Localized within the Nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 47.Brockdorff N. X-chromosome inactivation: closing in on proteins that bind XistRNA. Trends in Genetics. 2002;18:352–358. doi: 10.1016/s0168-9525(02)02717-8. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maenner S, et al. 2-D Structure of the A Region of Xist RNA and Its Implication for PRC2 Association. Plos Biology. 2010;8 doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature Genetics. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 51.Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011;93:1935–1942. doi: 10.1016/j.biochi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Shevchenko AI, Zakharova IS, Zakian SM. The evolutionary pathway of x chromosome inactivation in mammals. Acta Naturae. 2013;5:40–53. [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanellopoulou C, et al. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci U S A. 2009;106:1122–1127. doi: 10.1073/pnas.0812210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai MC, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi YJ, et al. Histone demethylation mediated by the nuclear arnine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013 doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schorderet P, Duboule D. Structural and Functional Differences in the Long Non-Coding RNA Hotair in Mouse and Human. Plos Genetics. 2011;7 doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. Bmc Genomics. 2007;8 doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nature Reviews Molecular Cell Biology. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 62.Ji P, et al. MALAT-1, a novel noncoding RNA, thymosin beta 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 63.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 64.Wilusz JE, Freier SM, Spector DL. 3 ' End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-like Cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilusz JE, et al. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stadler PF. Evolution of the Long Non-coding RNAs MALAT1 and MEN beta/epsilon. Advances in Bioinformatics and Computational Biology. 2010;6268:1–12. [Google Scholar]
- 67.Sasaki YTF, Ideue T, Sano M, Mituyama T, Hirose T. MEN epsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobson MR, et al. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110(Pt 7):829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 69.Vogel A, Schilling O, Spath B, Marchfelder A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- 70.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Research. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutschner T, Hammerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 73.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 74.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Molecular Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ackley A, et al. An Algorithm for Generating Small RNAs Capable of Epigenetically Modulating Transcriptional Gene Silencing and Activation in Human Cells. Mol Ther Nucleic Acids. 2013;2:104. doi: 10.1038/mtna.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dugre-Brisson S, et al. Interaction of Staufen1 with the 5 ' end of mRNA facilitates translation of these RNAs. Nucleic Acids Research. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gong CG, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3 ' UTRs via Alu elements. Nature. 2011;470 doi: 10.1038/nature09701. 284-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kiebler MA, et al. The mammalian Staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. Journal of Neuroscience. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen JJ, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Research. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 81.Grinchuk OV, Jenjaroenpun P, Orlov YL, Zhou J, Kuznetsov VA. Integrative analysis of the human cis-antisense gene pairs, miRNAs and their transcription regulation patterns. Nucleic Acids Res. 2010;38:534–547. doi: 10.1093/nar/gkp954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morris KV. The emerging role of RNA in the regulation of gene transcription in human cells. Semin Cell Dev Biol. 2011 doi: 10.1016/j.semcdb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Faghihi MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biology. 2010;11 doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohshima K, et al. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biology. 2003;4 doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang ZL, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends in Genetics. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 87.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pei BK, et al. The GENCODE pseudogene resource. Genome Biology. 2012;13 doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tay SK, Blythe J, Lipovich L. Global discovery of primate-specific genes in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12019–12024. doi: 10.1073/pnas.0904569106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcr. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature biotechnology. 2012 doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4 doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wan Y, et al. Genome-wide Measurement of RNA Folding Energies. Molecular Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novikova IV, Dharap A, Hennelly SP, Sanbonmatsu KY. 3S: Shotgun secondary structure determination of long non-coding RNAs. Methods. 2013 doi: 10.1016/j.ymeth.2013.07.030. [DOI] [PubMed] [Google Scholar]