Abstract
Functional magnetic resonance imaging (fMRI) technique with blood oxygenation level dependent (BOLD) contrast is a powerful tool for noninvasive mapping of brain function under task and resting states. The removal of cardiac- and respiration-induced physiological noise in fMRI data has been a significant challenge as fMRI studies seek to achieve higher spatial resolutions and characterize more subtle neuronal changes. The low temporal sampling rate of most multi-slice fMRI experiments often causes aliasing of physiological noise into the frequency range of BOLD activation signal. In addition, changes of heartbeat and respiration patterns also generate physiological fluctuations that have similar frequencies with BOLD activation. Most existing physiological noise-removal methods either place restrictive limitations on image acquisition or utilize filtering or regression based post-processing algorithms, which cannot distinguish the frequency-overlapping BOLD activation and the physiological noise. In this work, we address the challenge of physiological noise removal via the kernel machine technique, where a nonlinear kernel machine technique, kernel principal component analysis, is used with a specifically identified kernel function to differentiate BOLD signal from the physiological noise of the frequency. The proposed method was evaluated in human fMRI data acquired from multiple task-related and resting state fMRI experiments. A comparison study was also performed with an existing adaptive filtering method. The results indicate that the proposed method can effectively identify and reduce the physiological noise in fMRI data. The comparison study shows that the proposed method can provide comparable or better noise removal performance than the adaptive filtering approach.
Keywords: Physiological noise, aliasing, kernel, mutual information
1. Introduction
Advances in blood oxygenation level dependent (BOLD) functional magnetic resonance imaging (fMRI) are typically characterized by improved spatial resolution and detection of more subtle neuronal activity. However, BOLD contrast resulting from functional activation is usually small, so the advancements can depend heavily upon the signal-to-noise ratio (SNR). SNR is commonly improved by increasing the magnetic field strength [1–4], but cardiac- and respiration-induced physiological noise also increases with the field strength. Thus an increase in image SNR does not necessarily produce an equal improvement in contrast-to-noise ratio (CNR), a quantitative measure of imaging quality. Particularly, resting state studies of functional networks [5–9], which measure the baseline connectivity of functional networks, are vulnerable to reductions in CNR because they lack a clear stimulus paradigm to aid in detection and rely upon analysis of subtle, correlated signal fluctuations between brain regions.
The primary challenge to physiological noise removal is that the temporal sampling rates of most fMRI experiments are limited by the repetition time (TR), a major component governing signal intensity and image contrast, resulting in aliasing of physiological noise into frequencies of the BOLD signal. Additionally, changes of heartbeat and respiration patterns generate physiological fluctuations that have similar frequencies with the BOLD signal. Furthermore, physiological noise contaminates a wide range of frequencies whose power spectrum reflects not a purely sinusoidal variation but rather a distribution of frequencies about the peak, making them difficult to characterize in frequency domain [10].
Currently, physiological noise removal is approached either during acquisition, through gating and/or synchronization techniques [11,12] or during post-processing. Post-processing methods are desirable as they offer increased spatial and temporal specificity and place fewer limits upon the experimental design. Several post-processing approaches have been utilized previously, but each suffers from limitations. Navigator echo methods lack the specificity to localize the source of motion [13], which may lead to incomplete correction or introduce new artifacts. Retrospective correction methods [14,15], which fit a low-order Fourier series to fMRI data in either k-space or image domain based on the phase of respiration or cardiac cycle during each acquisition, have been shown to be effective [14,15]. However, these methods cannot remove the noise induced by changes in breathing pattern [7,8]. Other retrospective methods either require a short TR [16] or are limited to global fluctuations [17]. More importantly, none of these methods, including digital filtering and wavelet-based methods [18–20], can distinguish frequency components of the physiological noise that overlap with the BOLD activation. If the signal and noise occupy same frequency bands, then the removal of noise will also result in the attenuation of signal [21,22]. Efforts to increase the resolution and scope of BOLD-contrast fMRI, particularly in the area of characterizing resting-state functional networks, would therefore benefit greatly from new techniques better suited to separating the aliased physiological noise from the BOLD signal.
Another type of techniques, such as principal component analysis (PCA) and independent component analysis (ICA), represent a fundamentally different approach to the noise removal problem. These techniques decompose fMRI data into multiple components, and feature projections on these components share same frequency bands but could be originated from different signal and noise sources [10, 23–26]. Since these feature projections are not implemented in the frequency domain, they are potentially capable of distinguishing frequency-overlapping signal and noise. In this work, we report a new fMRI physiological noise removal procedure based on the kernel principle component analysis (KPCA) [27]. KPCA is a nonlinear extension of principal component analysis (PCA) and has been used in fMRI data analysis [28–31]. Nonlinear PCA can characterize high order dependence among multiple voxels, and can provide more complete characterization of fMRI data structure than linear PCA [32,33]. KPCA provides a controllable signal-noise differentiation analysis via a kernel and its parameter. While KPCA has been successfully used to remove Gaussian noise by excluding the least significant components from reconstruction [34], the same approach is not directly applicable to the removal of physiological noise that is usually characterized by both the most and least significant components. Therefore, we further develop a KPCA-based method for physiological noise removal. This method aims to differentiate and attenuate the aliased physiological noise that could overlap with the BOLD signal. The method was compared to an adaptive filtering method [35], which is an improved version of the RETROICOR method [15], for task-related and resting state fMRI data. The results indicate that the proposed method can provide comparable or better noise removal performance than the adaptive filtering method, implying promising applications in fMRI studies.
2. Material and Methods
2.1. Data Acquisition
Both task and resting state fMRI data were used in this study. Task-related data were obtained from three different experiments. The first experiment was performed using a 3 Tesla GE system with an 8-channel coil at Duke University Medical Center. Four data sets were acquired from a healthy adult on a same day using T2*-weighted parallel echo planar imaging (EPI) with an acceleration factor of 2, while the subjects was performing a right finger-tapping motor task with a blocked-design paradigm, which consisted of four 25 sec task blocks and five 25 sec off blocks. There was a 15 sec dummy scan at the beginning of each run, which was removed before the analysis. EPI parameters included a TR of 2 sec, an echo time (TE) of 30 msec, and a flip angle of 90°. 30 axial-slices were collected for each volume with 4 mm slice thickness and 1 mm gap, FOV was 24 cm × 24 cm, and image matrix size is 120 × 120 after the sensitivity encoding reconstruction, corresponding to an in-plane resolution of 2 × 2 mm2. The second experiment was performed using the same scanner used in the first experiment. Two fMRI data sets were collected from two subjects using a T2*-weighted EPI sequence with SENSE acceleration factor of 2 while the subjects were performing the right finger-tapping motor task with a blocked-design paradigm, which consisted of five 30 sec task blocks and five 30 sec off blocks. The scan time for each run was 5 min. TR=2 sec, TE= 25 msec. 35 axial-slices were collected in each volume with 3 mm slice thickness. FOV was 24 cm × 24 cm, and image matrix size is 64 × 64. Another three sets of task fMRI data are from the website of New York University Center for Brain Imaging (cbi.nyu.edu). The data were acquired from a subject using a 3T Siemens Allegra scanner. The first two sets were collected using a single channel head coil, and another set was collected using a surface coil. Imaging parameters include a TR of 1.5 sec, a TE of 30 msec, and a flip angle of 75°. A visual stimulation was applied to the subject by alternatively showing a left and right circular hemifield stimulus of alternating checks at full contrast. In each set 150 volumes were acquired with 25 axial-slices in each volume. Each slice is represented by 64 × 80 3 mm isotropic voxels.
Three resting state fMRI experiments were implemented at Duke University Medical Center. In the first experiment, four data sets were collected from a subject using the same T2*-weighted parallel EPI sequence as that used in the first task related experiment while the subjects were instructed to look at a crosshair. The scan time for each run was 4 min. Inversion-recovery (IR) prepared spin-echo EPI was also acquired to provide an anatomic reference with identical voxel geometry and geometric distortions as in fMRI. IR-EPI scan parameters included TR = 5 sec, TE = 24 msec, IR time = 1 sec, flip angle = 90°, slice thickness = 4 mm (with 1 mm gap), FOV = 24 cm × 24 cm, in-plane matrix size = 120 × 120 (with 2 segments), and 30 axial slices. In the second experiments, six sets of resting state fMRI data were collected from six healthy adults on different days. The imaging parameters are TR=4 sec, TE=35 msec. flip angle =90°, FOV=24 cm × 24 cm, the image matrix size is 140 × 140. 56-axial slices were acquired in each volume with a 3 mm slice thickness = 3 mm, and 74 volumes were collected in each data set (about 5 minutes time duration). In the third experiment, two data sets were collected from two subjects using a T2*-weighted EPI sequence with SENSE acceleration factor of 2 while the subjects were instructed to look at a crosshair. The scan time for each run was 5 min. TR=2 sec, TE= 25 msec. 35 axial-slices were collected in each volume with 3 mm slice thickness. FOV was 24 cm × 24 cm, and image matrix size is 64 × 64.
The cardiac and respiration cycles were simultaneously recorded using Biopac MRI-compatible transducers at a sampling rate of 100 Hz during the fMRI data acquisition. The cardiac cycles were measured by a fiber-optic finger pulse-oximeter cuff. The respiration data were collected by a stretch transducer on an elastic belly belt placed around the abdomen. All electrical connections are grounded and pass through MRI filters in the magnet room shield. Cardiac signals were amplified outside the magnet room using Biopac amplifiers. All acquired physiological signals are connected to an analog/digital data acquisition device (Measurement Computing Inc.) connected to the computer via a USB interface. The acquired physiological data were down-sampled and synchronized to the slice-acquisition timing of fMRI. The physiological data from New York University Center for Brain Imaging were obtained from their website (cbi.nyu.edu). The experiments on human subjects were compliant with the standards established by the Institutional Review Boards of Duke University.
2.2. Data Analysis
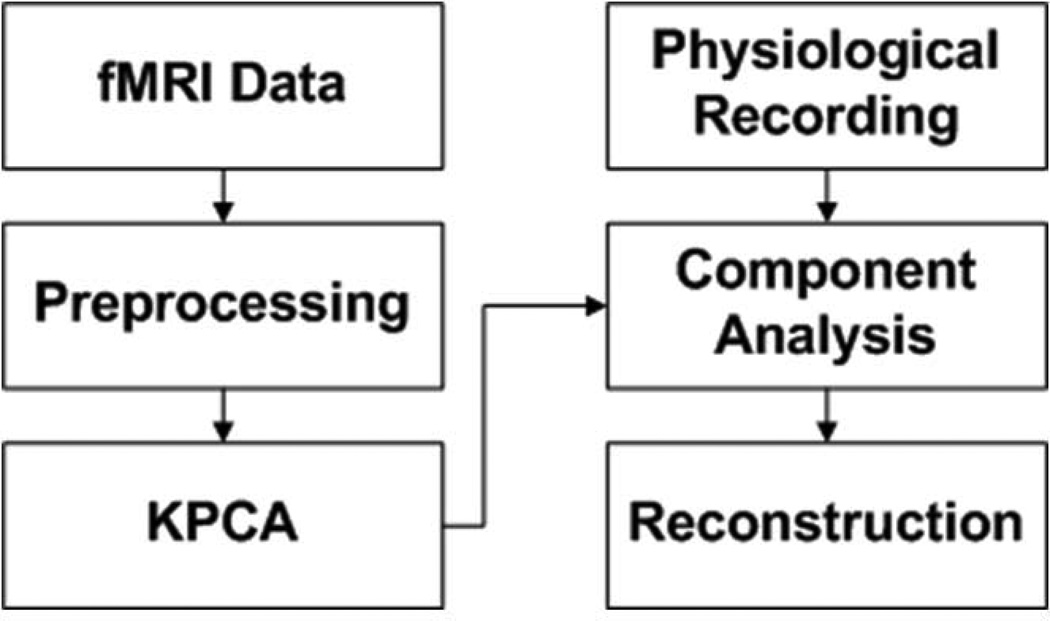
A block diagram of the proposed method is shown in Figure 1. fMRI data is first motion corrected and spatially smoothed. The preprocessed fMRI data is decomposed into multiple principal components (PC) using KPCA. The feature projections on PCs characterizing significant physiological variations are identified to locate frequencies of aliased physiological cycles, as indicated by simultaneously recorded physiological data. The identified feature projections are filtered by digital filters designed to attenuate the power at these frequencies. Finally KPCA reconstruction is implemented to obtain the denoised data for the subsequent analysis.
Figure 1.
The block diagram of the proposed method.
2.2.1. Preprocessing
All fMRI data were first registered using a 2-D rigid body registration method [36] as part of FSL tools [37] to remove small head motion artifacts. Then the data were detrended to remove low frequency drift and spatially smoothed with a Gaussian kernel (FWHM=2 mm) using in-house Matlab programs. The resting state data were low-pass filtered at the cut-off frequency of 0.1 Hz to remove high-frequency signal variations from the slowly-varying signal fluctuations of interest in resting state. All data were normalized to zero mean and unit variance.
2.2.2. KPCA decomposition
KPCA introduces nonlinearity into PCA through a kernel that implicitly projects the input to a higher dimensional feature space [27]. A linear PCA performed in the feature space is equivalent to a nonlinear PCA in the input space. Given M voxels sampled at T time points, we may form M T-dimensional vectors for spatial analysis, or T M-dimensional vectors for temporal analysis [24]. In this work, the temporal analysis is used and each vector is represented as xi ∈ RM,i = 1,2,…,T, the PCs are obtained by computing eigenvalues λ > 0 and eigenvectors V satisfying:
| (1) |
where Φ is a nonlinear mapping from RM to a higher dimensional feature space F and 〈•〉 is the inner product operator. Because V = span(Φ(xi):i = 1,…,T) , we have . If we define a T×T matrix K with each entry as Ki, j= 〈Φ(xi)•Φ(xj)〉 and replace the inner product in each entry with a kernel k(xi,xj) , the eigen problem becomes λα = Kα, where α = (αl,α2,…, αT)’. After computing the single value decomposition on K, the feature projection on the kth PC in F can be obtained by:
| (2) |
where Vk is the kth eigenvector in F.
2.2.3. Kernel Evaluation
It is not clear how much separation between the BOLD signal and physiological noise could be achieved by KPCA. The KPCA’s capability relies on the kernel it uses. Different kernels and parameters will provide distinct performances. The selection of kernels and parameters has been discussed for support vector machine-based applications [38], but rarely addressed for KPCA-based applications. In this work, two widely used kernels, the polynomial and radial basis function (RBF) kernels, as defined in equation(3), were examined with different parameters regarding their performance in separating the physiological noise and BOLD signal.
| (3) |
where σ is the RBF kernel width, 〈x,xi〉 is the inner product of vectors x and xi, and p is the polynomial kernel order. Given a kernel and its parameter, fMRI data is decomposed into multiple PCs using KPCA. The mutual information (MI) between the feature projection on each PC and the expected haemodynamic response, the MI between the feature projection on the PC and the synchronized cardiac and respiration cycles were calculated. Since projections showing high MI with the haemodynamic response are expected to have low MI with the cardiac and respiratory recording, an index S is proposed to measure the separation performance as defined in equation(4).
| (4) |
where N is the number of PCs, IBOLD(k) is the MI between the feature projection on the kth PC and the canonical haemodynamic response function in task-related fMRI studies, and IPHYS(k) is the MI between the projection on the kth PC and synchronized cardiac or respiratory fluctuations, i.e., ICARD(k) or IRESP(k), which corresponds to SCARD or SRESP. If the feature projection on the kth PC has a high dependence on the expected haemodynamic response but low dependence on the physiological noise, or vice versa, then |IBOLD(k) – IPHYS(k)| is relatively large and indicates a good separation between the BOLD signal and noise. 1–e−|IBOLD (k)−IPHYS(k)| is used to map this difference to a range between 0 and 1, and a large |IBOLD(k) –IPHYS(k) |value results in a value of 1–e−|IBOLD(k)−IPHYS(k)| close to 1. The final S index is an average of all 1–e−|IBOLD(k)–IPHYS(k)| values calculated from the feature projections on N PCs. Therefore, a better signal-noise separation results in a higher S value. Since 0 < 1–e−|IBOLD (k)I−IPHYS(k)| < 1, the S value is also between 0 and 1. For resting state studies, there is no experimental paradigm, and we propose a different way to estimate IBOLD(k). First a seed is selected within a functional network of interest, and the average time course of the seed is used as the expected haemodynamic response to compute IBOLD (k). Although it is possible that the seed is contaminated by the physiological noise, the S value can provide a reliable evaluation of the separation performance because a kernel that can sufficiently separate the signal and noise to different PCs will generate large |IBOLD (k) – IPHYS (k)| values, leading to large S values.
Multiple data sets were used to identify a kernel and its parameter that outperform the others in terms of the signal-noise separation performance measured by equation(4). Due to the non-stationary nature of fMRI, the data collected from different subjects, or a same subject but at a different time may have very distinct signal and noise characteristics. Consequently, S values from different data sets are not directly comparable although they could be obtained using a same kernel and parameter, and only those computed from a same dataset with different kernels and parameters are comparable. To remove the effects from the non-stationarity, S values computed from each data set are normalized against the maximum S value from the same data set using the two kernels. Meanwhile, the difference between S values induced by different kernels and parameters using the same data set is preserved. After the normalization, the S values from all data sets are comparable and averaged together to increase the statistical power of the analysis. The kernel evaluation is not part of the block diagram as shown in Figure 1. It is implemented offline to provide guidance on the selection of a kernel function and its parameter for KPCA decomposition.
2.2.4. Component Analysis and Reconstruction
The KPCA-based noise removal is a dimensionality reduction problem that can be used to reveal low-dimensional representations of a data set and to reflect its underlying degrees of freedom. In existing KPCA-based approaches, the least significant PCs are supposed to characterize the noise and excluded from the reconstruction [31,34]. This might not be suitable for the physiological noise removal because the physiological noise could be at a similar or higher intensity level than BOLD signals [39,40]. Therefore, the noise could be captured by the most significant PCs. Prior information about the physiological noise is needed in order to identify PCs characterizing the noise, and is extracted from the synchronized physiological cycles.
The spectra of the synchronized physiological cycles were first calculated, and frequencies of interest (FOIs) showing peak power of the aliased cardiac and respiratory cycles were automatically detected and recorded. In our preliminary study [29], the spectrum of each feature projection was examined at FOIs to identify PCs capturing the greatest power of the noise. In this work, a more efficient way proposed by using the frequency information and the MI between the feature projections and physiological recording and expected BOLD signal. A set of feature projections are identified if they (i) exhibit high ICARD and IRESP but low IBOLD values, and (ii) have significant power at the FOIs. The analysis starts with a ranking of feature projections in terms of ICARD and IRESP , respectively. Since it was found that usually three to five feature projections showing much larger IBOLD values than other feature projections, in the rankings of ICARD and IRESP , feature projections that are among the top three showing the highest IBOL D are removed. The remained projections are examined at the FOIs. In each remained projection, the power at each FOI is estimated by summarizing the power of the closest frequency point to the FOI and the power of the two neighboring (one lower, one higher) frequency points. If the power at the FOI is greater than 0.5 times the average of power spectrum, then a finite impulse response band-stop filter is designed to attenuate the noise components at this FOI. Since most feature projections are not significant in terms of both BOLD signal and physiological noise, it might not be necessary to analyze each projection, and only those with ICARD or IRESP values greater than the average of ICARD or IRESP will be examined and filtered. This improves the computational efficiency without considerably affecting the noise removal performance.
KPCA reconstruction is not so straightforward as with linear PCA [34,41]. None of existing methods can provide a perfect reconstruction. In our work, the method proposed by Kwok was used [41]. It estimates the reconstructed data by utilizing distance constraints in input and feature spaces derived by the idea of multidimensional scaling (MDS) [42]. MDS states that the pairwise distance of two data points in an input space is preserved after projecting the data to a high dimensional feature space. The MDS-based KPCA reconstruction is an optimization problem that aims to minimize the difference between the pairwise distance of points in the input and high dimensional feature spaces.
2.2.5. Method Evaluation
Quantitative measures were used to evaluate the proposed method based on a comparison study with an adaptive filtering method [35], which was developed based on the widely used RETROICOR method [15]. For task-related data, a contrast-to-noise ratio (CNR) measurement was computed to assess the denoising performance [43]. It is defined as:
| (5) |
where ΔS is the change of signal intensity in response to a task stimulation, SE is the standard error of intensity in baseline or task stimulation period, and is defined as: SE = SD/M0.5, where SD is the standard deviation of the average intensity during the baseline or task period, and M is the number of images during the baseline or task period. The CNR values should increase or maintain at the same level after removing the noise in regions where BOLD activations appear during the stimulation, and to decrease in regions contaminated by the physiological noise. Temporal standard deviation maps were computed for the original and denoised data. For resting state data, MI values between a seed and part of the default mode network (DMN) was compared between the data denoised by the two methods. The MI between the synchronized physiological recording and fMRI data before and after the noise removal was also compared.
3. Results
3.1. Task-related Experiments
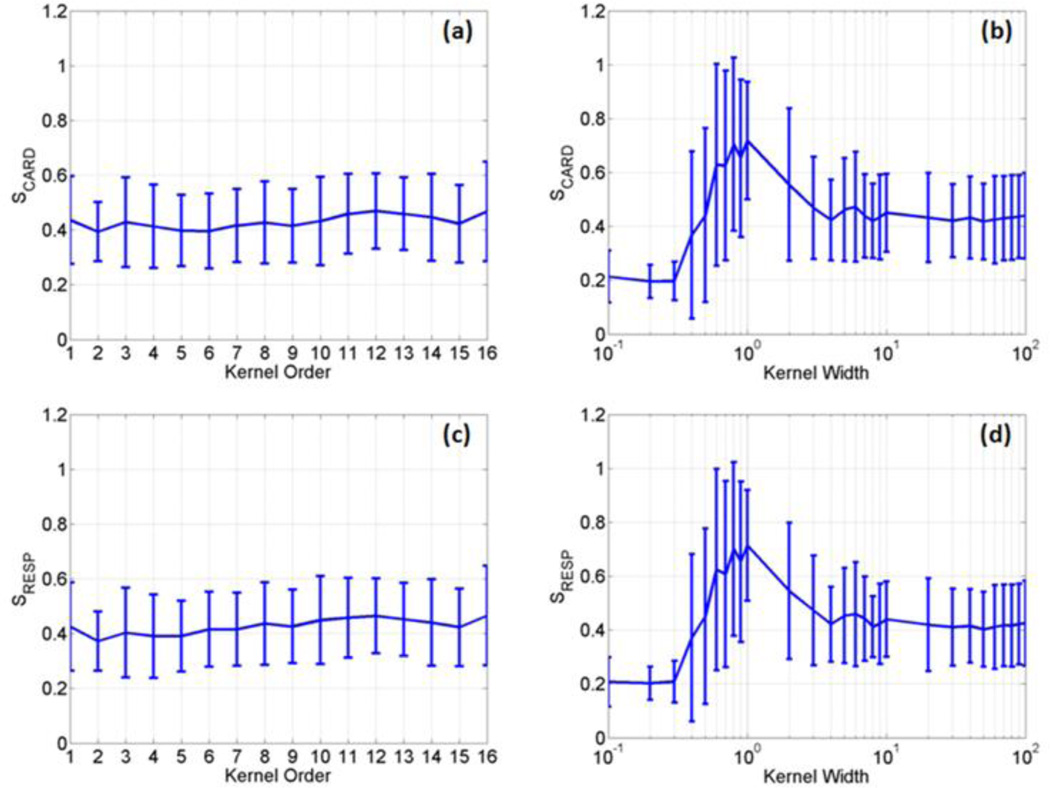
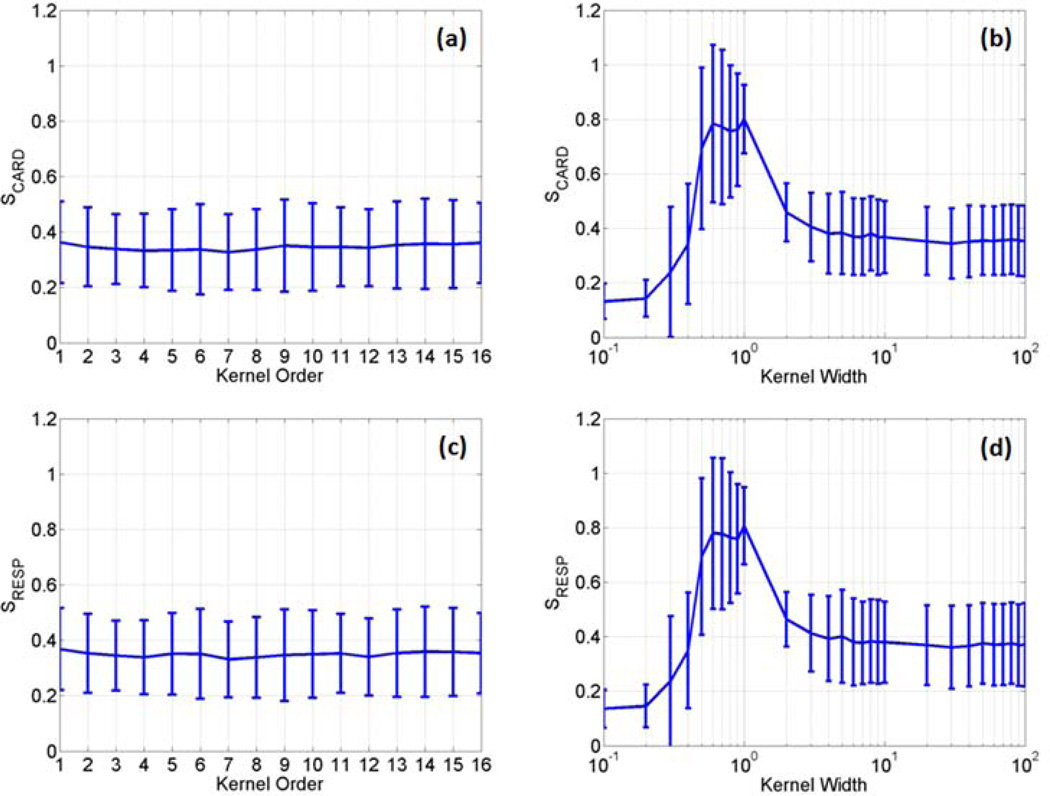
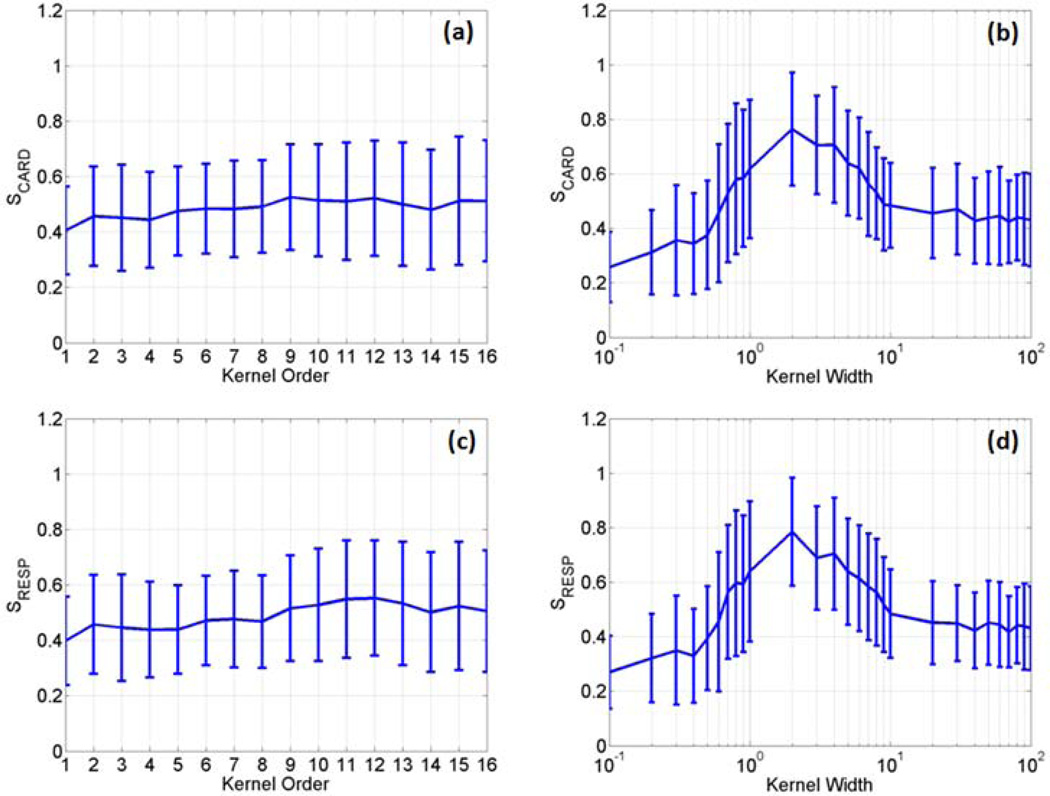
The two kernels described in equation(3) were first evaluated with different parameters using four sets of fMRI data acquired from the first task-related experiment. In each data set, three slices that contain significant BOLD responses were selected for the evaluation. Since each slice across time forms an image sequence, we had a total of twelve image sequences. For the polynomial kernel, the kernel order p ranged from 1 to 16 with an interval of 1, where order 1 corresponds to the linear discrimination. For the RBF kernel, the kernel width parameter < was set from 0.1 to 100, with an interval of 0.1 when 0.1 ≤ 1, with an interval of 1 when 1 ≤ 10, and with an interval of 10 when 10 ≤ σ ≤ 100. The average and standard deviation (SD) of S values of the two kernels were calculated using the twelve image sequences, as shown in Figure 2, where (a) and (b) are SCARD values as a function of p and < , respectively, and (c) and (d) are SRESP values as a function of p and < , respectively. When the polynomial kernel was used, there is no significant difference between S values as a function of p , as shown in Figure 2 (a) and (c). For the RBF kernel, there is a significant increase of SCARD and SRESP when 0.5 ≤ σ ≤ 10. When σ > 10, the S values are close to those obtained from the polynomial kernel.
Figure 2.
The average and standard deviation of SCARD and SRESP values calculated as a function of the polynomial kernel order (a, c) and the RBF kernel width (b, d) using four sets of data acquired from the first task-related experiment.
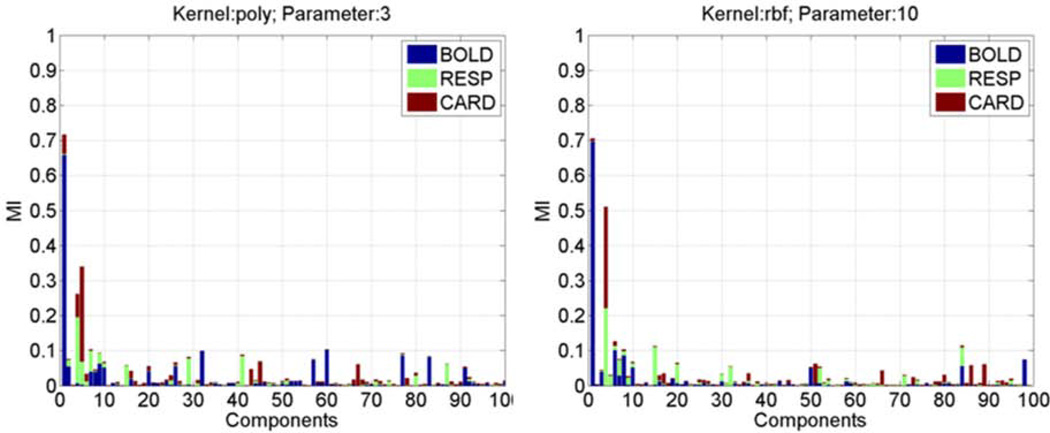
To illustrate the signal-noise separation in individual feature projections, stacked bars are used to represent the estimated IBOLD (blue), IRESP (light-green), and ICARD (red-brown) values of all feature projections, as shown in Figure 3, where (a) was obtained using the polynomial kernel with p = 3, and (b) was calculated by using the RBF kernel with σ = 10.0. Each stacked bar is corresponding to a feature projection, and the IBOLD , IRESP , ICARD values are represented by the lengths of associated color segments. It is expected that feature projections with large IBOLD values have small IRESP and ICARDvalues, which indicates a clear separation between the BOLD signal and physiological noise.
Figure 3.
The stacked bar representation of ICARD. IRESP, and IBOLD values of all PCs obtained from a set of fMRI data using (a) the third order polynomial kernel and (b) the RBF kernel with a kernel width σ=10.
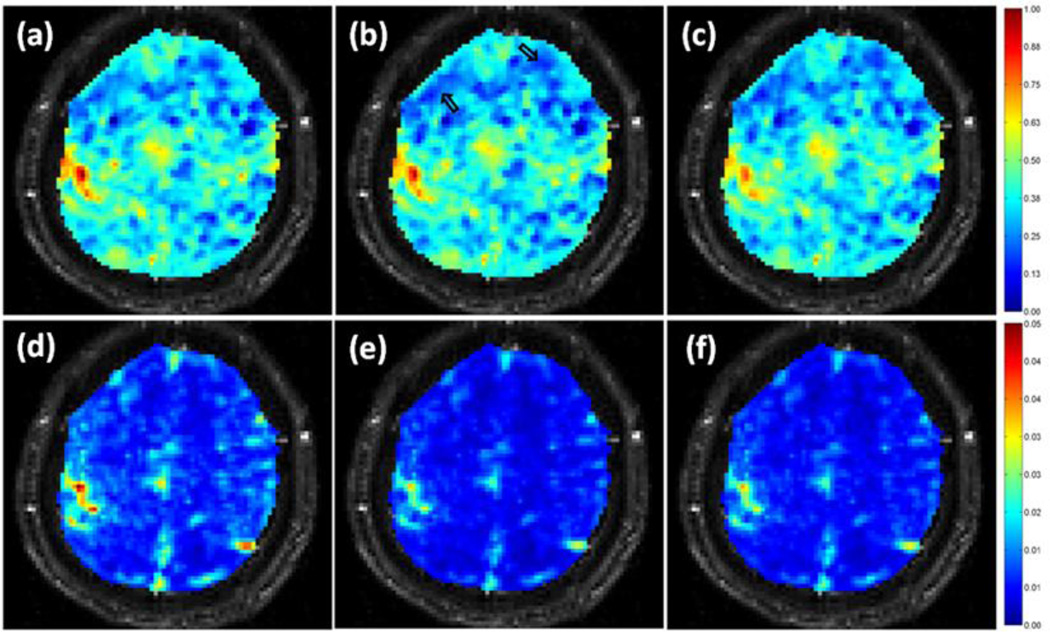
Since the RBF kernel provides a better signal-noise separation in terms of the measurement defined in equation(4) when 0.5 ≤ σ ≤ 10, the RBF kernel was used with σ = 6.0 in the proposed method to process the task-related data. For comparison, the adaptive filtering method was implemented for the same data. After the processing, CNR values were computed for the original and denoised data, and normalized between 0 and 1 based upon the maximum and minimum CNR values calculated from the original and denoised data from the two methods. Figure 4 (a)–(c) show a comparison of the normalized CNR values in a representing slice for one run of the first task-related experiment. Figure 4 (a) was obtained from the original data, (b) was calculated from the data processed by the proposed method, and (c) was from the data denoised by the adaptive filtering method. Note there are some voxels in regions not related to the task showing decreased CNR as indicated by the two arrows in (b). A similar decrease is observed in the adaptive filtering result in (c). A slight decrease of CNR in the left motor cortex is also observed in (c), implying possible attenuation of real BOLD activation by the adaptive filtering method. Figure 4 (d)–(f) are the temporal standard deviation maps, where (d) was computed from the original data, (e) and (f) were from the results of the proposed and adaptive filtering methods, respectively. The proposed method provides more attenuation on the temporal standard deviation than the adaptive filtering method without sacrificing CNR in the motor cortex.
Figure 4.
(a)–(c) Single slice CNR maps of a task-related experiment calculated from (a) the original data, (b) the data denoised by the proposed method, and (c) the data processed using the adaptive filtering method. The two arrows in (b) point to groups of voxels with decreased CNR. (d)–(f) Single slice temporal standard deviation maps obtained from (d) the original data, (e) the data denoised by the proposed method, and (f) the data smoothed using the adaptive filtering method.
So far the data used to evaluate the proposed method is the same data used to identify the kernel and its parameter. We further examined the method with the same kernel and parameter using different data sets. These data were obtained from different subjects with motor and visual stimuli. In addition, they have different spatial and/or temporal resolution, time length, and were acquired from different scanners. Figure 5 (a) is an EPI slice in a data set acquired from the second task-related experiment where a motor task stimulation was used. (b)–(d) are the normalized CNR maps overlaid on the slice calculated from the original data and the data denoised by using the proposed and adaptive filtering methods. Figure 5 (e) is another EPI slice in a data set collected from the third task-related experiment where a visual hemifield stimulation was implemented. (f)–(h) are the normalized CNR maps obtained from the original and denoised data. It might not be easy to visually identify the difference among these CNR maps. Table 1 shows the number of voxels with normalized CNR values above 0.6 and below 0.3 before and after the noise removal for the heretofore used three task-related fMRI data sets shown in Figures 4 and 5.
Figure 5.
(a) An individual slice from a data sets in the second task-related experiment where a motor task was performed, (b)–(d) are the CNR maps overlaid on this slice calculated from (b) the original data, (c) the data denoised by the proposed method, and (d) the data smoothed using the adaptive filtering method, (e) An individual slice from a data set in the third task-related experiment where a visual attention stimulation was applied, (f)–(h) are the CNR maps overlaid on the slice obtained from (f) the original data, (g) the data denoised by the proposed method, and (h) the data processed by the adaptive filtering method.
Table 1.
The number of voxels with high (>0.6) and low (<0.3) normalized CNR values before and after noise removal. The corresponding CNR maps are shown in Figures 4 and 5.
| Data set #1 | Data set #2 | Data set #3 | ||||
|---|---|---|---|---|---|---|
| High CNR (>0.6) |
Low CNR (<0.3) |
High CNR (>0.6) |
Low CNR (<0.3) |
High CNR (>0.6) |
Low CNR (<0.3) |
|
| No processing | 77 | 623 | 209 | 254 | 96 | 293 |
| Proposed method | 72 | 779 | 221 | 309 | 100 | 300 |
| Adaptive filtering | 63 | 757 | 183 | 303 | 87 | 280 |
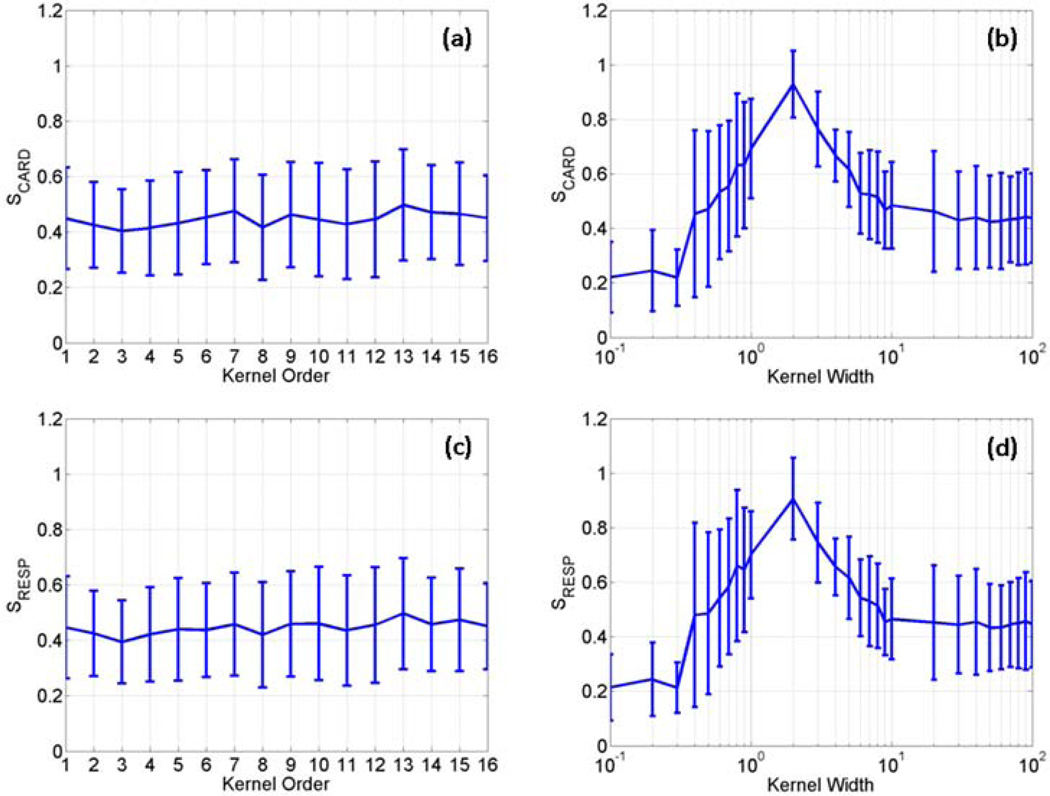
We further calculated S values using the data sets from the second and third task-related experiments. Figure 6 (a)–(d) are the average and SD of S values of the two kernels calculated using the data sets from the second and third task-related experiments. In the second task-related experiment, five slices showing BOLD activation over time in the motor cortex were identified in each data set, and totally ten image sequences were used to calculate the S values. In the data sets from the third experiment, fifteen image sequences were used to calculate S values with five image sequences from each data set showing BOLD activation in the primary visual cortex. It can be seen that all curves in Figures 2 and 6 exhibit a similar pattern.
Figure 6.
The average and standard deviation of SCARD and SRESP values calculated as a function of the polynomial kernel order (a, c) and the RBF kernel width (b, d) using two data sets collected from the second task-related fMRI experiment, and three data sets from the third task-related fMRI experiment.
3.2. Resting State Experiments
The proposed method was also evaluated using the resting state data. In the data from the first resting state experiment, a 3×3 voxels seed was manually selected in the posterior cingulate cortex (PCC) region that is part of the DMN. The average time course of this seed was used as an expected haemodynamic response, from which IBOLD was estimated. Figure 7 shows the average and SD of S values of the two kernels estimated using the four sets of resting state data. In each data set, three image sequences corresponding to three slices containing DMN regions were used to calculate S values, and there are total 12 image sequences used to obtain the results in Figure 7. Most S values are at a similar level, but we may see an increase in average value from the RBF kernel when 0.5 ≤ σ ≤ 10. Like the noise removal of task-related data, we also chose the RBF kernel with σ = 6.0 for the resting state physiological noise removal.
Figure 7.
The average and standard deviation of SCARD and SRESP values calculated as a function of the polynomial kernel order (a, c) and the RBF kernel width (b, d) using four sets of fMRI data collected from the first resting state fMRI experiment.
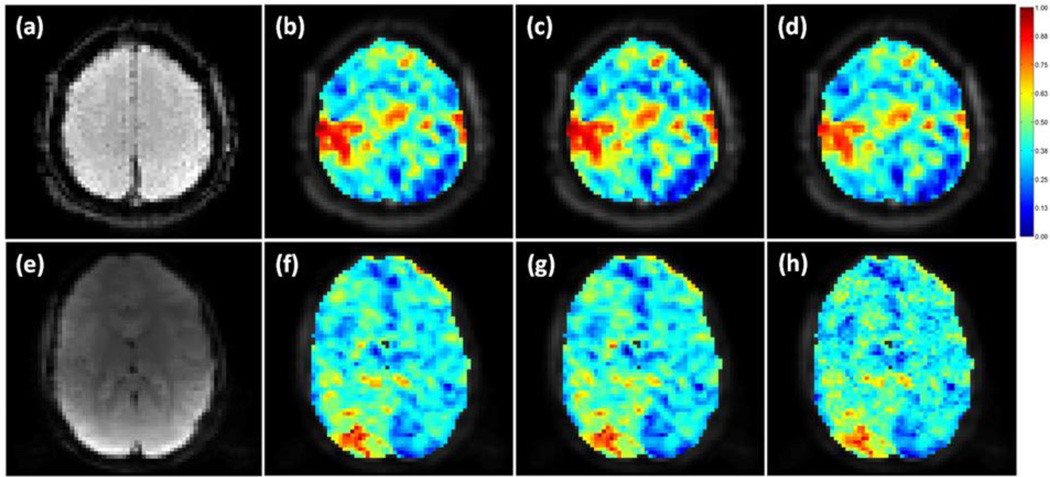
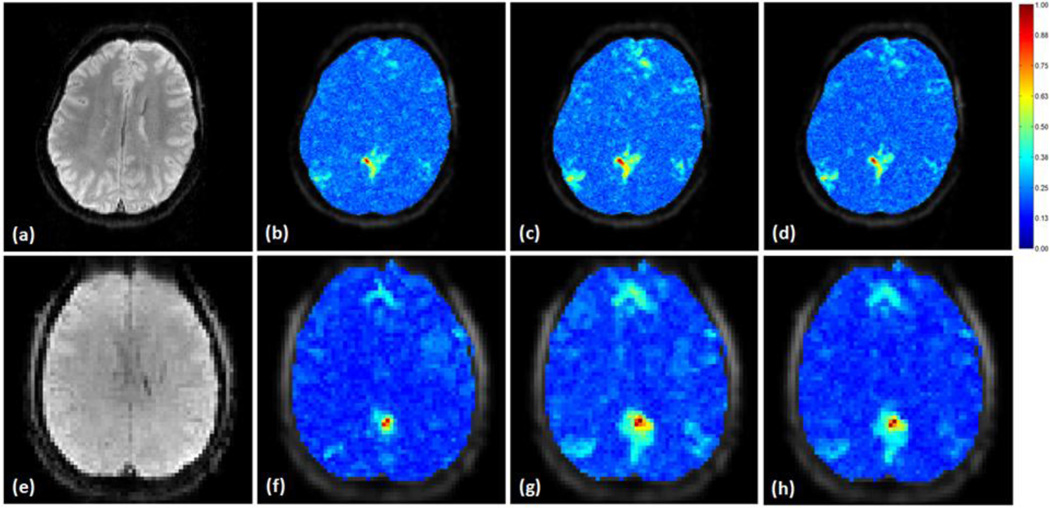
MI values were calculated between the seed’s and all voxels’ time courses using the original data, the data processed by the proposed and adaptive filtering methods. These MI values were normalized to the maximum MI value calculated from the original and denoised data, and overlaid on the corresponding image slice as shown in Figure 8 (a)–(c), where the encircled region in (a) is the seed based on which the S values in Figure 7 were obtained. Figure 8 (b) and (c) indicate that the proposed and adaptive filtering methods can increase the number of voxels showing high MI to the seed in PCC, medial prefrontal cortex (mPFC), and lateral parietal cortex (LPC). In order to examine the physiological noise distribution in this slice before and after the noise removal, the MI values between the fMRI data and the synchronized physiological cycles were computed and normalized to the maximum MI value computed from the original and denoised data, as shown in Figure 8 (d)–(i), where a MI value close to 1 means a high similarity between the data and noise. Specifically, Figure 8 (d)–(f) are MI maps between the synchronized cardiac recording and (d) original data, (e) data processed by the proposed method, and (f) the data smoothed by the adaptive filtering method. (g)–(i) are MI maps between the synchronized respiration recording and (g) original data, (h) data processed by the proposed method, and (i) the data denoised by the adaptive filtering method. It is observed that both methods can attenuate the dependence of fMRI data on the physiological fluctuations.
Figure 8.
Mutual information (MI) based evaluation of resting state fMRI physiological noise removal. The MI values were normalized to the maximum MI computed from the original and denoised fMRI data, and overlaid on an image slice that covers part of the DMN. (a)–(c) are MI maps between a seed (encircled region in (a)) in the PCC area and (a) the original data, (b) the data denoised by the proposed method, and (c) the data processed using the adaptive filtering method. A MI value close to 1 means a high dependence on the seed. (d)–(f) are MI maps between the synchronized cardiac recording and (d) the original data, the data denoised by (e) the proposed method, and (f) the adaptive filtering method, (g)–(i) are MI maps between the synchronized respiration recording and the fMRI data in a same order as (d)–(e). The regions pointed by arrows in (d) partially overlap with the DMN but can be attenuated by the proposed and adaptive filtering methods without affecting the identification of voxels showing high MI with the seed.
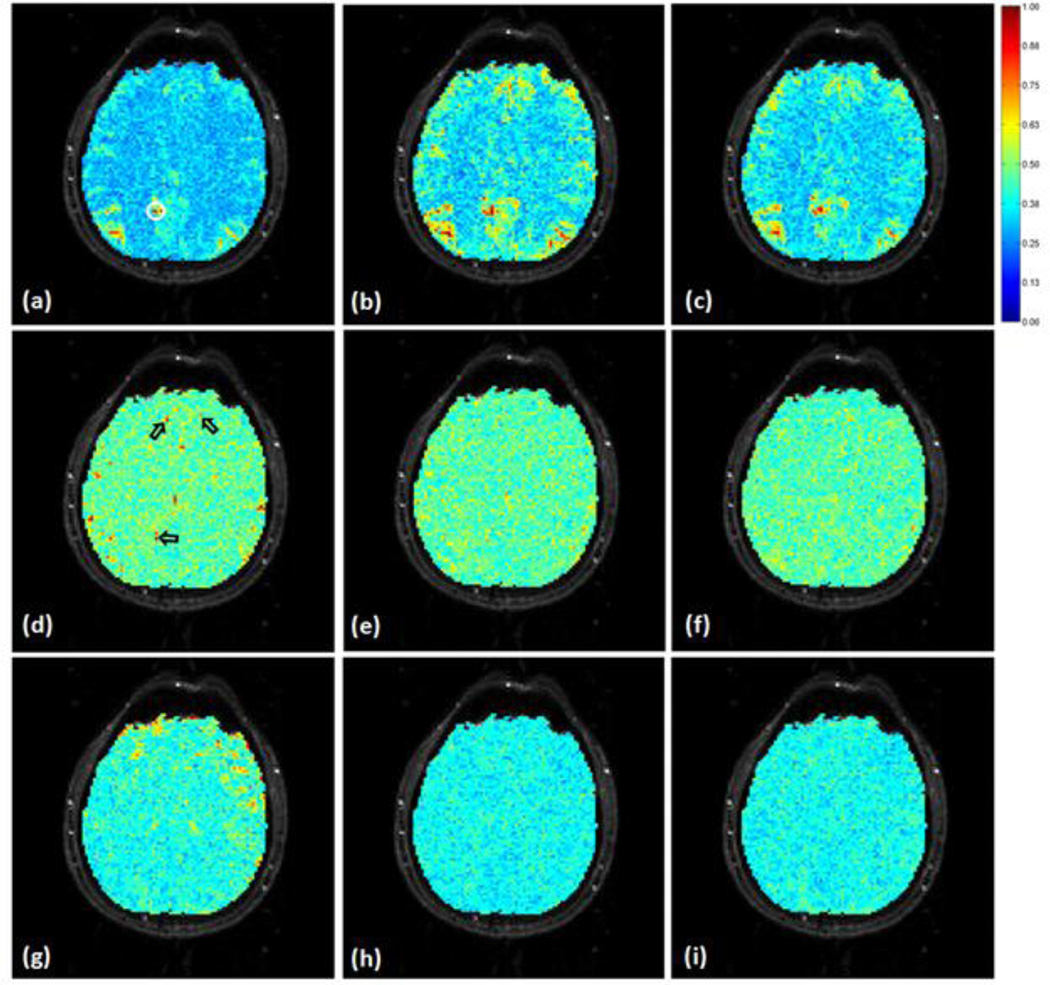
The same kernel and parameter were applied to the other data sets collected from the second and third resting state experiments. Figure 9 (a) shows an individual slice in a data set from the second resting state experiment. (b)–(d) are the MI maps between a 3×3 voxels seed in PCC and all voxels’ time courses overlaid on the slice using (b) the original data, (c) the data denoised by the proposed method, and (d) the data processed by the adaptive filtering method. Figure 9 (e) is an individual slice from another set of data collected from the third resting state experiment. (f)–(h) show the same type of MI maps as (b)–(d) using this data. The S values calculated using the data sets from the second and third resting state experiments are shown in Figure 10 (a)–(d). There are six sets of data from the second resting state experiment, and eighteen image sequences were used with three image sequences from each data set. There are two data sets from the third resting state experiment, with three image sequences from each data set. The average and SD of S values exhibit a similar pattern as those in Figure 8. This implies that the proposed signal noise separation measurement is independent of experiment paradigm, subjects, spatial and temporal resolution, and time length of experiments, and the identified kernel and its optimal parameter range are applicable to the physiological noise of different fMRI experiments.
Figure 9.
Mutual information (MI) between a seed in the PCC and the original and denoised fMRI data calculated from two sets of resting state data. The MI values were normalized to the maximum MI computed from the original and denoised fMRI data in each data set, and overlaid on the corresponding slice that covers the seed region, (a) shows an individual slice from one data set where the seed was selected, (b) is the MI map calculated using the original data, (c) is the MI map computed using the data denoised by the proposed method, and (d) was from the data processed by the adaptive filtering method, (e) is an individual slice from the other data set where the seed was selected in PCC. (f)–(h) are the MI maps following the same order as (b)–(d) calculated using the original and denoised slice in this data set.
Figure 10.
The average and standard deviation of SCARD and SRESP values calculated as a function of the polynomial kernel order (a. c) and the RBF kernel width (b. d) using six data sets acquired from the second resting state fMRI experiment, and two data sets from the third resting state fMRI experiment.
4. Discussion
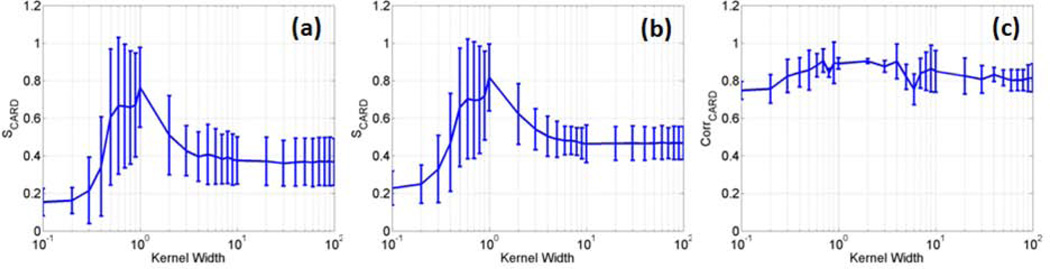
MI is used in this work to reveal high order dependencies between the KPCA feature projections and expected haemodynamic responses or synchronized cardiac/respiratory fluctuations. It is not easy to accurately estimate MI values, and two MI estimation methods were examined in this study [44,45]. It was found that the two methods generate slightly different MI estimates but provide almost same patterns for S values as a function of different kernel parameters. Therefore, they may be used to estimate relative increases/decreases of MI or MI-based measures, such as the proposed S index. The method proposed by Darbellay et al. was finally used for this study because it is computationally more efficient [45]. The MI measures in equation (4) could be replaced by the absolute value of Pearson's correlation coefficient, which can characterize the second order dependencies between feature projections and signal/noise. A comparison between the MI- and correlation-based S indices was performed using all task-related fMRI experimental data presented in this work, and part of results is shown in Figure 11, where (a) and (b) are S values as functions of the RBF kernel width obtained using the two MI estimation methods, and (c) was obtained by using the correlation coefficient in equation (4). The variation of S values in (a) and (b) are quite similar to each other except for their values after the normalization. The S values obtained using the correlation coefficient also show an increase when 0.5 ≤ σ ≤ 10, but the increase is not so significant as those in (a) and (b) calculated using MI. A possible reason of this difference is that MI can provide a more complete characterization of dependencies between the feature projections and haemodynamic responses or synchronized cardiac/respiratory cycles than the correlation coefficient.
Figure 11.
The average and standard deviation of SCARD values calculated as a function of the RBF kernel width based on all task-related fMRI data presented in this work, (a) and (b) were obtained by using the MI estimation methods proposed by Darbellay et al. [45], and Beirlant et al. [44]. respectively, (c) was obtained by replacing MI with the absolute values of Pearson’s correlation coefficient in the calculation of S values.
Under the task stimulation, there is no significant difference in S values when different polynomial kernel order is used, as shown in Figure 2 (a), (c), Figure 6 (a), (c), (e), and (g). When the RBF kernel is used, the average S values are usually less than those of the polynomial kernel if < 0.5, as shown in Figure 2 (b), (d), Figure 6 (b), and (d). However, when 0.5 ≤ σ ≤ 10, the average S values are greater than those obtained from the polynomial kernel. When σ> 10, the average S values are close to those calculated from the polynomial kernel. A similar observation was made from the S values computed from the resting state experiments, as shown in Figures 7 and 10. When the polynomial kernel is used with p = 1, the KPCA becomes the linear PCA. When the RBF kernel is used with a large kernel width, the KPCA is close to the linear PCA. Figures 2, 6, 7, and 10 also indicate that the nonlinear KPCA can provide better signal-noise differentiation than the linear PCA if the kernel and its parameter are properly chosen. For instance, the S values shown in these figures tell us that the RBF kernel with 0.5 ≤ σ ≤ 10 may be used. This observation is reasonable from the machine learning aspect. When the RBF kernel width is small (i.e., σ< 0.5) , the feature space distance between different feature points is close to zero, and PCA analysis in the feature space becomes unstable and meaningless [46]. On the other hand, if the RBF kernel width is large (i.e., σ> 10), KPCA is close to the conventional linear PCA, which is not sufficient to characterize the nonlinear fMRI data structure. A moderate kernel width (i.e., 0.5 ≤ σ ≤ 10) not only can introduce sufficient nonlinearity to characterize fMRI data structure, but also can avoid the unstable PCA computation in the feature space. A further examination of Figures 2, 6, 7, and 10 indicates that when 0.5≤ σ ≤2, the standard deviation of S is relative large in some data sets, implying an inconsistent signal-noise separation performance. The reason of this inconsistency is that these kernel width values are still small and cannot guarantee to provide reliable PCA analysis for all data sets. They may yield more scattering of the feature variance into the PCs, resulting in a significant number of PCs showing either high ICARD, IRESP, or high IBOLD values, which are not expected for the analysis. An expected KPCA decomposition generates only a few PCs that are significant in terms of either BOLD signal or the physiological noise. Therefore, in order to obtain a reliable decomposition, we suggest setting the RBF kernel width between 2 and 10. In the experiments, we examined a kernel width of 6.0 for the noise removal of both task-related and resting state data.
The IBOLD, ICARD, and IRESP values of all PCs shown in Figure 3 (a) were obtained after the KPCA decomposition of a task-related fMRI time series using the third order polynomial kernel. The third order polynomial kernel has been widely used in various pattern recognition tasks because it could provide sufficient nonlinearity without a significant increase of computational load. From the length of each color segment in the stacked bar, it is observed that the most significant PC computed by using this kernel shows a significant dependence on the BOLD signal. It also has an apparent dependence on the cardiac cycles, and little dependence on the respiration. Within the first ten most significant PCs, there are two PCs showing significant dependence on the physiological noise and little dependence on the BOLD signal. From the 11th significant PC to the least significant PC, there are a small number of PCs showing significant dependence on the BOLD activation and little dependence on the physiological noise, or the opposite situation. Most other PCs exhibit either similar or little dependence on both the BOLD signal and physiological noise. When the RBF kernel with σ = 10 was used for the same data, the first most significant PC shows a significant dependence on the BOLD signal, but little dependence on either cardiac or respiration cycles, as shown in Figure 3 (b). Within the first ten most significant PCs, there is one PC showing significant dependence on the physiological noise and little dependence on the BOLD signal. If we compare the PCs from the 11th significant to the least significant to those obtained from the 3rd order polynomial kernel, we may find that the polynomial kernel generates more PCs showing apparent dependence on the BOLD activation than the RBF kernel. These observations indicate that the RBF kernel with σ = 10 has a better characterization of the BOLD signal and physiological noise than the third order polynomial kernel, and provides a better signal-noise separation for this data. None of existing techniques, such as ICA, PCA, and KPCA, can completely separate the BOLD signal and physiological noise. But they can facilitate the identification of the majority of physiological fluctuations. As long as KPCA can provide a reasonable decomposition with a properly selected kernel and parameter, the most significant part the physiological noise can be characterized by PCs that are different from those characterizing BOLD signal, and a frequency analysis performed on feature projections of these PCs can better differentiate the physiological noise than the conventional frequency analysis performed on the original fMRI data.
There are many methods that can easily reduce the temporal standard deviation of voxels. But as a consequence, CNR of truly activated brain voxels is also attenuated. From the results in Figure 4, it is interesting to find that the proposed method can provide a more significant attenuation of the temporal standard deviation than the adaptive filtering without sacrificing CNR of truly activated brain voxels. This implies that the decreased standard deviation likely represents the physiological fluctuations, and the BOLD CNR can be maintained or enhanced by the proposed method. This is further verified by the results from the other two data sets collected from different subjects, different experiments with different tasks, as shown in Figure 5. The numerical results in Table 1 were computed to help the comparison of CNR maps shown in Figures 4 and 5. Compared to the number of voxels showing high and low CNR values in the original data, the data denoised by the proposed method have a similar or greater number of high CNR voxels, and more low CNR voxels. There are less high CNR voxels in the data processed by the adaptive filtering method. This indicates that the attenuation of the physiological noise also lead to the reduction of BOLD signal when the adaptive filtering method was used. This also implies that part of BOLD frequencies overlap with the aliased physiological noise, and the proposed method can differentiate the frequency-overlapping signal and noise.
The evaluation of the denoising performance on resting state data is challenging because it is difficult to judge if there would be greater or fewer functionally connected regions after the processing. There are two types of errors caused by the physiological noise in resting state studies [47]. One is the false positive when there is no functional connection but the dependence between noise components is significant enough with respect to a predefined threshold. The other is false negative when the true functional connection is obscured by the physiological noise and cannot be detected. The first type of error leads to more voxels showing significant dependence to the seed, and the noise removal should reduce the number of connected voxels. The second type of error results in less voxels connected to the seed, and the noise removal should increase the connected area. In practice, it is difficult to know if the number of voxels connected to the seed should increase or decrease after the noise removal. But according to the existing knowledge about the spatial distribution of physiological noise, and results from previous studies, it is possible to determine if the noise removal is effective or not.
DMN is widely used to evaluate the physiological noise effect in resting state fMRI studies. In this study, MI was computed to identify DMN based on a seed in PCC as shown in Figure 8 (a). Regions with red color indicate significant connections with the seed. Since all MI values were normalized against a same base, the comparison of MI maps in each row of Figure 8 are meaningful. For the data shown in Figure 8, both the proposed and adaptive filtering methods increase the number of functionally connected voxels in PCC, mPFC, and LPC, as shown in Figure 8 (b) and (c). In addition, the proposed processing results in more connected voxels than the adaptive filtering method. It is also observed from Figure 8 (b) and (c) that some regions that are not considered as part of DMN show increased MI with the seed region. In order to check the significance of these increases, we manually selected four DMN regions covering PCC (288 voxels), mPFC (180 voxels), left and right LPC (321 voxels), and calculated the ratio between the average MI within these regions and the average MI of all other regions (3372 voxels). The ratio is 1.18 in the original data, 1.23 after the adaptive filtering, and 1.33 after the proposed processing. Therefore, the increases of MI in DMN regions are more significant than those in other regions. An examination of the noise distribution before and after the noise removal in Figure 8 (d)–(i) may give us more information about the performance of two methods. It is observed that both methods can considerably attenuate the dependence of the fMRI data to the synchronized physiological cycles, implying that the methods can effectively reduce the physiological noise. It is also observed that there is more attenuation to the respiration-induced noise than to the cardiac-induced noise, as shown in Figure 8 (e)–(f), and (h)–(i). There are several regions indicated by arrows in Figure 8 (d). These regions partially overlap with the DMN, and show significant dependence on the synchronized cardiac cycles. Both methods can reduce the cardiac noise in these regions, and facilitates the identification of more functionally connected voxels, as indicated by the voxels that show low MI to the seed in Figure 8 (a), but show high MI values in Figure 8 (b) and (c). The performance of the proposed method can be further verified by the identified DMN from two different data sets shown in Figure 9, where each row starts with an EPI image slice on which the denoising was performed. Figure 9 (b) and (f) are the normalized MI maps between a seed in PCC and the original data. (c) and (g) are the normalized MI maps between the seed and the data denoised by the proposed method. (d) and (e) are the normalized MI maps between the seed and the data processed by the adaptive filtering method. Both the proposed and adaptive filtering approaches can increase the MI with the seed in PCC, mPFC, and LPC regions, and the results from the proposed method show higher MI values in these regions than those from the adaptive filtering method.
The proposed KPCA based method also has limitations. First, like ICA- or PCA-based methods, it is not guaranteed that signal and noise can be completely separated into different components. Secondly, the frequency analysis is implemented on each PC across the entire recording period, and short time changes of noise frequencies may not be sufficiently characterized. The simultaneous physiological recording is not mandatory for the study. Approaches that use either ICA components [48,49], or data from a noise region in white matter or ventricle, or regions around large vessels can also be used to provide an estimation of the physiological noise [50].
5. Conclusion
The challenge of fMRI physiological noise removal is addressed in this study based on a KPCA based physiological noise removal method. The polynomial and RBF kernels were examined for the KPCA using a signal-noise separation measurement that was proposed to evaluate the kernels’ performance in differentiating the physiological noise and BOLD signal. Experimental study shows that the RBF kernel is a better choice for the separation of the physiological noise when the kernel width is properly set. Based on the RBF kernel, the KPCA-based physiological noise removal method was developed and evaluated using both task-related and resting state fMRI data acquired from multiple human subjects. A comparison study with an adaptive filtering method was also performed. The results indicate that the proposed method can provide comparable or better physiological noise removal performance than the adaptive filtering method. Particularly, a major advantage of the proposed method is its potential capability to differentiate frequency-overlapping signal and noise. This distinguishes it from most existing methods that use adaptive filtering and/or regression models to smooth the data, which could simultaneously attenuate the noise and signal.
Acknowledgements
This research was partially supported by NIH R01-NS074045 grant (to N.-K. Chen).
Reference
- 1.Ugurbil K, Hu X, Chen W, Zhu X, Kim SG, Georgopoulos A. Functional mapping in the human brain using high magnetic fields. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1999;354:1195–1213. doi: 10.1098/rstb.1999.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruger G, Glover GH. Physiological noise in oxygenation-sensitive magnetic resonance imaging. Magn Reson Med. 2001;46:631–637. doi: 10.1002/mrm.1240. [DOI] [PubMed] [Google Scholar]
- 3.Di Salle F, Esposito F, Elefante A, Scarabino T, Volpicelli A, Cirillo S, Elefante R, Seifritz E. High field functional MRI. Eur. J. Radiol. 2003;48:138–145. doi: 10.1016/j.ejrad.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Voss HU, Zevin JD, McCandliss BD. Functional MR imaging at 3.0 T versus 1.5 T: a practical review. Neuroimaging Clin. N. Am. 2006;16:285–297. doi: 10.1016/j.nic.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 6.Fox M, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 8.Birn RM, Murphy K, Bandettini PA. The effect of respiration variations on independent component analysis results of resting state functional connectivity. Hum Brain Mapp. 2008;29:740–750. doi: 10.1002/hbm.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJ. Default-mode brain dysfunction in mental disorders: A systematic review. Neurosci. Biobehav. Rev. 2009;33:279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Zollei L, Panych L, Grimson E, Wells IIIWM. Exploratory identification of cardiac noise in fMRI images. Proc. International Society and Conference on Medical Image Computing and Computer-Assisted Intervention. 2003 [Google Scholar]
- 11.Guimaraes AR, Melcher JR, Talavage TM, Baker JR, Ledden P, Rosen BR, Kiang NY, Fullerton BC, Weisskoff RM. Imaging subcortical auditory activity in humans. Hum. Brain Mapp. 1998;6:33–41. doi: 10.1002/(SICI)1097-0193(1998)6:1<33::AID-HBM3>3.0.CO;2-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenger VA, Peltier S, Boada FE, Noll DC. 3D spiral cardiac/respiratory ordered fMRI data acquisition at 3 Tesla. Magn. Reson. Med. 1999;41:983–991. doi: 10.1002/(sici)1522-2594(199905)41:5<983::aid-mrm18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Hu X, Kim SG. Reduction of signal fluctuation in functional MRI using navigator echoes. Magn. Reson. Med. 1994;31:495–503. doi: 10.1002/mrm.1910310505. [DOI] [PubMed] [Google Scholar]
- 14.Hu X, Le TH, Parrish T, Erhard P. Retrospective estimation and correction of physiological fluctuation in functional MRI. Magn. Reson. Med. 1995;34:201–212. doi: 10.1002/mrm.1910340211. [DOI] [PubMed] [Google Scholar]
- 15.Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn. Reson. Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Wowk B, McIntyre MC, Saunders JK. k-Space detection and correction of physiological artifacts in fMRI. Magn. Reson. Med. 1997;38:1029–1034. doi: 10.1002/mrm.1910380625. [DOI] [PubMed] [Google Scholar]
- 17.Frank LR, Buxton RB, Wong EC. Estimation of respiration-induced noise fluctuations from undersampled multislice fMRI data. Magn. Reson. Med. 2001;45:635–644. doi: 10.1002/mrm.1086. [DOI] [PubMed] [Google Scholar]
- 18.Biswal B, DeYoe AE, Hyde JS. Reduction of physiological fluctuations in fMRI using digital filters. Magn, Reson, Med. 1996;35:107–113. doi: 10.1002/mrm.1910350114. [DOI] [PubMed] [Google Scholar]
- 19.Meyer FG. Wavelet-based estimation of a semiparametric generalized linear model of fMRI time-series. IEEE Trans. Med. Imaging. 2003;22:315–322. doi: 10.1109/TMI.2003.809587. [DOI] [PubMed] [Google Scholar]
- 20.Song X, Murphy M, Wyrwicz AM. Spatiotemporal Denoising and Clustering of fMRI Data. Proc. IEEE International Conference on Image Processing. 2006:2857–2860. [Google Scholar]
- 21.Harvey A, Pattinson K, Brooks J, Mayhew S, Jenkinson M, Wise R. Brainstem functional magnetic resonance imaging: disentangling signal from physiological noise. J Magn Reson Imag. 2008;28:1337–1344. doi: 10.1002/jmri.21623. [DOI] [PubMed] [Google Scholar]
- 22.Beall E. Adaptive cyclic physiological noise modeling and correction in functional MRI. Journal of Neuroscience Methods. 2010;187:216–228. doi: 10.1016/j.jneumeth.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 23.McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Hum. Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen AH, Gash DM, Avison MJ. Principal component analysis of the dynamic response measured by fMRI: a generalized linear systems framework. Magn. Reson. Imaging. 1999;17:795–815. doi: 10.1016/s0730-725x(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 25.Thomas CG, Harshman RA, Menon RS. Noise reduction in BOLD-based fMRI using component analysis. Neuroimage. 2002;17:1521–1537. doi: 10.1006/nimg.2002.1200. [DOI] [PubMed] [Google Scholar]
- 26.Churchill W, Yourganov G, Spring R, Rasmussen M, Lee W, Ween E, Strother C. PHYCAA: data-driven measurement and removal of physiological noise in BOLD fMRI. NeuroImage. 2012;59(2):1299–1314. doi: 10.1016/j.neuroimage.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Schölkopf B, Smola A, Müller K. Nonlinear Component Analysis as a Kernel Eigenvalue Problem. Neural Computation. 1998;10:1299–1319. [Google Scholar]
- 28.Thirion B, Faugeras O. Dynamical components analysis of fMRI data through kernel PCA. NeuroImage. 2003;20:34–49. doi: 10.1016/s1053-8119(03)00316-1. [DOI] [PubMed] [Google Scholar]
- 29.Song X, Ji T, Wyrwicz A. Baseline drift and physiological noise removal in high field fMRI data using kernel PCA. Proc. IEEE International Conference on Acoustics, Speech, and Signal Processing. 2008:441–444. [Google Scholar]
- 30.López M, Ramírez J, Górriz J, Álvarez I, Salas-Gonzalez D, Segovia F, Chaves R. SVM-based CAD system for early detection of the Alzheimer's disease using kernel PCA and LDA. Neuroscience Letters. 2009;464(3):233–238. doi: 10.1016/j.neulet.2009.08.061. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen P, Abrahamsen T, Madsen K, Hansen L. Nonlinear denoising and analysis of neuroimages with kernel principal component analysis and pre-image estimation. NeuroImage. 2012;60(3):1807–1818. doi: 10.1016/j.neuroimage.2012.01.096. [DOI] [PubMed] [Google Scholar]
- 32.Friston K, Phillips J, Chawla D, Büchel C. Revealing interactions among brain systems with nonlinear PCA. Hum. Brain Mapp. 1999;8:92–97. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<92::AID-HBM4>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friston K, Phillips J, Chawla D, Büchel CNonlinear PCA. characterizing interactions between modes of brain activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:135–146. doi: 10.1098/rstb.2000.0554. 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mika S, Schölkopf B, Smola A, Miller K, Scholz M, Ratsch G. Kernel PCA and De-noising in Feature Spaces; Advances in Neural Information Processing Systems 11. Morgan Kaufmann. 1998 [Google Scholar]
- 35.Deckers R, Gelderen P, Ries M, Barret O, Duyn J, Ikonomidou V, Fukunaga M, Glover GH, Zwart J. An adaptive filter for suppression of cardiac and respiratory noise in MRI time series data. NeuroImage. 2006;33:1072–1081. doi: 10.1016/j.neuroimage.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkinson M, Bannister P, Brady J, Smith S. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 37.Woolrich M, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, Beckmann C, Jenkinson M, Smith S. Bayesian analysis of neuroimaging data in FSL. NeuroImage. 2009;45:S173–S186. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 38.Ben-Hur A, Ong CS, Sonnenburg S, Schölkopf B, Ratsch G. Support vector machines and kernels for computational biology. PLoS Computational Biology. 2008;4(10):1–10. doi: 10.1371/journal.pcbi.1000173. e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandettini P, Wong E, Jesmanowicz A, Prost R, Cox R, Hinks R. MRI of human brain activation at 0.5T, 1.5T and 3.0T: comparison of DR2 and functional contrast to noise ratio. Proc. ISMRM. 1994;1:434. [Google Scholar]
- 40.Triantafyllou C, Hoge R, Krueger G, Wiggins C, Potthast A, Wiggins G, Wald L. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. NeuroImage. 2005;26:243–2505. doi: 10.1016/j.neuroimage.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Kwok JT, Tsang IW. The pre-image problem in kernel methods. IEEE Trans. Neural Netw. 2004;15:1517–1525. doi: 10.1109/TNN.2004.837781. [DOI] [PubMed] [Google Scholar]
- 42.Williams CKI. Advances in Neural Information Processing Systems. Vol. 13. MIT Press; 2001. On a connection between kernel PCA and metric multidimensional scaling. [Google Scholar]
- 43.Menon RS, Thomas CG, Gati JS. Investigation of BOLD contrast in fMRI using multi-shot EPI. NMR Biomed. 1997;10:179–182. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<179::aid-nbm463>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 44.Beirlant J, Dudewicz E, Gyorfi L, Van der Meulen E. Nonparametric entropy estimation: An overview; International Journal of Mathematical and Statistical. Sciences. 1997;6(1):17–39. [Google Scholar]
- 45.Darbellay G, Vajda I. Estimation of the information by an adaptive partitioning of the observation space. IEEE Trans. on Information Theory. 1999;45(4):1315–1321. [Google Scholar]
- 46.Hoffmann H. Kernel PCA for novelty detection. Pattern Recognition. 2007;40(3):863–874. [Google Scholar]
- 47.Chang C, Glover GH. Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. NeuroImage. 2009;47(4):1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beall E, Lowe M. Isolating physiologic noise sources with independently determined spatial measures. NeuroImage. 2007;37:1286–1300. doi: 10.1016/j.neuroimage.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Perlbarg V, Bellec P, Anton JL, Pelegrini-Issac M, Doyon J, Benali H. CORSICA: correction of structured noise in fMRI by automatic identification of ICA components. Magn, Reson, Imaging. 2007;25:35–46. doi: 10.1016/j.mri.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 50.Behzadi Y, Testom K, Liau J, Liu T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37(1):90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]