Abstract
Viral protein 35 (VP35), encoded by filoviruses, are multifunctional dsRNA binding proteins that play important roles in viral replication, innate immune evasion and pathogenesis. The multifunctional nature of these proteins also presents opportunities to develop countermeasures that target distinct functional regions. However, functional validation and the establishment of therapeutic approaches toward such multifunctional proteins, particularly for non-enzymatic targets, are often challenging. Our previous work on filoviral VP35 proteins defined conserved basic residues located within its C-terminal dsRNA binding interferon (IFN) inhibitory domain (IID) as important for VP35 mediated IFN antagonism and viral polymerase co-factor functions. In the current study, we used a combination of structural and functional data to determine regions of Ebola virus (EBOV) VP35 (eVP35) to target for aptamer selection using SELEX. Select aptamers, representing two distinct classes, were further characterized based on their interaction properties to eVP35 IID. These results revealed that the aptamers bind to distinct regions of eVP35 IID with high affinity (10–50 nM) and specificity. These aptamers can compete with dsRNA for binding to eVP35 and disrupt the eVP35-nucleoprotein (NP) interaction. Consistent with the ability to antagonize eVP35-NP interaction, select aptamers can inhibit the function of the EBOV polymerase complex reconstituted by expression of select viral proteins. Taken together, our results support the identification of two aptamers that bind filoviral VP35 proteins with high affinity and specificity and have the capacity to potentially target filoviral VP35 proteins as a therapeutic target.
Keywords: RNA aptamers, filovirus, viral protein VP35, inhibitors, antivirals
Introduction
Negative stranded RNA viruses possess a limited number of open reading frames (ORFs), such that many of the encoded proteins function at multiple stages of the viral replication cycle. The multifunctional nature of these viral proteins potentially provides several opportunities to develop antivirals. However, target validation of non-enzymatic viral proteins is particularly challenging as inhibitors must be identified de novo. Innovative methods that combine structure/function information can provide critical target validation prior to initiation of more effective small molecule-based methods.
Filoviruses are single-stranded, negative sense RNA viruses that cause severe hemorrhagic fever in humans and non-human primates1 The family filoviridea includes genera the ebolaviruses and marburgviruses and a proposed genus of Cuevavirus2 The genus ebolavirus contains five species, which are Ebola virus (EBOV), Sudan virus (SUDV), Taï Forest virus (TAFV), Reston virus (RESTV), and Bundibugyo virus (BDBV), while the genus Marburg virus has two species, Marburg virus (MARV) and Ravn virus (RAVV)3 Among the five species of Ebolavirus, RESTV does not cause disease in humans, although it is pathogenic in non-human primates4, 5 However, RESTV was recently isolated from a swine population in the Philippines, whose animal handlers were seropositive suggesting the potential zoonotic nature of filoviruses5, 6 Together, these observations highlight the potential public health risk posed by filoviruses and the need to develop innovative countermeasures.
Filoviral viral protein 35 (VP35) is a multifunctional immune antagonist that functions at several stages in the viral replication cycle7– 11 VP35 is known to antagonize numerous components in the IFN induction and signaling pathway8– 17, including RIG-I-like receptors (RLRs)11, 18, 19, IFN kinases IKKε and TBK-120, and RNA-dependent protein kinase (PKR)13,21. VP35 also functions as an RNAi silencing suppressor14,22, a co-factor for the viral polymerase23, and a structural protein that is part of the viral nucleocapsid24–27 Some, but not all VP35 mediated functions depend on the ability of VP35 to bind double stranded RNA (dsRNA)10, 11, 17, 18, 23, 28 Recent structural and functional studies have identified two conserved basic patches, the first basic patch (FBP) and the central basic patch (CBP), within the C-terminal IFN inhibitory domain (IID) of EBOV VP35 (eVP35) (Figure 1A)17, 18, 29, 30 The CBP is critical for dsRNA binding and host immune suppression, likely by preventing RNA recognition by RLRs18 Viruses containing mutations in the CBP region are replication competent, but are attenuated in viral growth. Moreover, prior infection with a guinea pig adapted CBP-mutant EBOV protected guinea pigs against subsequent infection by wildtype EBOV, further supporting the importance of the VP35 CBP for viral pathogenesis28 In contrast, the VP35 FBP is not required for dsRNA binding or inhibiting host immune responses. The filoviral replication complex minimally consists of the viral polymerase L, nucleoprotein (NP), and VP35, with the addition of EBOV viral protein 30 (VP30) being required for viral transcription in EBOV31 Previous studies have shown that mutation of residues within the FBP as well as other basic residues that border the CBP (border basic residues) render VP35 defective in its polymerase co-factor function23 Mutation of FBP residues leads to loss of critical contacts with the NP protein, an interaction required for replication complex formation17, 18, 23, 30 In contrast, mutation of the border basic residues does not result in loss of NP binding23 Altogether, these studies highlight the importance of VP35 proteins in viral-mediated host immune suppression and viral RNA synthesis, two critical functions which contribute to filoviral pathogenesis.
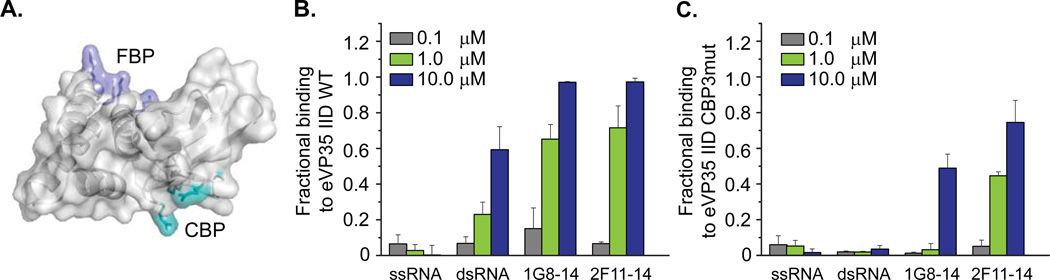
Figure 1. 1G8-14 and 2F11-14 aptamers bind eVP35 IID WT with high affinity.
(A) eVP35 IID contains two basic patches. The first basic patch (FBP) contains residues K222, R225, K251, and K248 (purple). The central basic patch (CBP) contains residues R312, K319, and R322 (cyan). eVP35 IID WT and eVP35 IID CBP3mut were used as targets for in vitro selection of 1G8-14 and 2F11-14 aptamers, respectively. Filter binding assays were used to assess the ability of (B) eVP35 IID WT and (C) eVP35 IID CBP3mut proteins to bind ssRNA, dsRNA, 1G8-14 aptamer, and 2F11-14 aptamer at protein concentrations of 0.1 µM (gray), 1 µM (green), and 10 µM (blue).
Despite increasing knowledge of filoviral pathogenesis, there are no approved treatments that target filoviral infections1, 32–37 Currently, there are several promising vaccine options under investigation38, 39; however, there are relatively few non-vaccine options available for treatment, particularly for post-exposure treatments38, 39 The multifunctional nature of filoviral VP35 and its relevance to viral pathogenesis makes VP35 a potential target for therapeutic development. Previous efforts using antisense oligonucleotides and small interfering RNAs have been shown to interfere with EBOV replication by targeting VP3512, 40–42 These studies coupled with recent results, which show that EBOVs containing mutant VP35 are highly attenuated and guinea pigs infected with these mutant EBOVs are protected against wildtype EBOV infections28, suggest that VP35 is a viable target for antiviral development.
Development of inhibitors that target key components in pathogens often requires detailed knowledge of structure and function or screening of large compound libraries with highly sensitive reporter(s). Nucleic acid aptamers provide an alternative method for inhibitor development as well as target validation43, 44 Aptamer identification and characterization does not require detailed knowledge of the target45–48 However, in situations where detailed structural and functional information are available, aptamer development can be tailored to enhance the impact of aptamer binding by targeting aptamer interactions to functionally important regions. Using such an approach, here we report the development of aptamers against eVP35 IID. Previous structural and functional results were utilized to identify functionally relevant regions of VP35 to target for aptamer development. Aptamers were identified using SELEX (Systematic Evolution of Ligands by EXponential enrichment49, 50) methods by targeting WT eVP35 IID and a mutant form of the protein. Two aptamers, representing distinct binding modes, were selected for further characterization. These studies revealed that one aptamers (1G8–14) preferentially binds EBOV and RESTV VP35 IID proteins, while the other (2F11-14) binds both EBOV and MARV VP35 IID proteins. Using in vitro studies, we demonstrate the utility of our selected aptamers as potential inhibitors, which can disrupt a critical protein-protein interaction in the filoviral replication complex and the activity of the minigenome system in cells. Taken together, our results suggest that aptamers identified here are valuable reagents for basic research and target validation of filoviral VP35 proteins for antiviral development.
MATERIALS AND METHODS
Protein expression and purification
Mutant eVP35 IID proteins were generated by overlap PCR and the plasmids were sequenced to confirm the mutation(s). Recombinant eVP35 IID WT and mutant proteins were expressed as previously described17, 51 The integrity of the proteins was assessed by 1H/15N-HSQC NMR spectra.
Crystallization and structure determination
Initial crystallization conditions for eVP35 IID CBP3mut (where CBP residues R312, K319, and R322 are mutated to alanine) and FBP4mut (where FBP residues K222, R225, K248, and K251 were mutated to alanine) were identified using commercially available crystallization screens (Hampton Research), and in-house optimized native crystals were grown at 25 °C using the hanging-drop vapor diffusion method.16.4 mg/mL eVP35 IID CBP3mut was diluted in a 1:1 ratio with reservoir solution containing 2.2 M Na/K phosphate (pH 4.5). 24 mg/mL eVP35 IID FBP4mut was diluted in a 1:1 ratio with reservoir solution containing 0.2 M MgCl2, 0.1 M HEPES (pH 7.5), 30% PEG 400. Crystals were soaked in reservoir solution containing 25% glycerol and vitrified in liquid nitrogen. Diffraction data was collected at Advanced Photon Source (Sector 19) at 100 K (Table S1). One hundred and eighty frames of data were collected with a frame width of 1.0° and detector-to-crystal distance of 300 mm for eVP35 IID FBP4mut. 450 frames of data were collected with a frame width of 0.2° and detector-to-crystal distance of 250 mm for eVP35 IID CBP3mut. Diffraction data were indexed, scaled and merged using HKL-300052. Intensities were converted to structure factors using CCP453. The structures were solved using molecular replacement, with chain B from eVP35 IID crystal structure (PDB ID 3FKE) as the search model using MOLREP53 The structures were refined using REFMAC554 and PHENIX55 Addition of solvent molecules and manual model building was performed using Coot56 TLS parameters were refined using the TLMSD server57 Final validation was performed using MOLPROBITY server58
In vitro selection of aptamers
In vitro SELEX selection of aptamers was carried out using eVP35 IID wildtype (WT) and eVP35 IID CBP3mut as targets59, 60. Oligonucleotide templates and primers were chemically synthesized with standard desalting by Integrated DNA Technology (Coralville, IA). The dsDNA library was generated from a ssDNA library (5`-GCCTGTTGTGAGCCTCCTGTCGAA(N45)TTGAGCGTTTATTCTTGTCTCCC-3`). Using the dsDNA library as a template, RNAs were in vitro transcribed following established protocols. The transcribed RNA was purified by phenol/chloroform extraction followed by ethanol precipitation and separated out on a 7M urea-8% PAGE. Purified RNAs were incubated with either eVP35 IID WT or eVP35 IID CBP3mut at 37°C for 0.5 to 1 hr in 10 mM HEPES (pH 7.0), 150 mM NaCl, 2mM TCEP (tris(2-carboxyethyl)phosphine), and 1 mM MgCl2 and filtered through nitrocellulose membrane (Millipore). The protein-bound RNA was eluted with 7M urea, precipitated by ethanol, and reverse transcribed using SuperScript™ reverse transcriptase (Invitrogen). The reverse transcribed cDNAs were amplified by polymerase chain reaction (PCR) using oligo-003 (5`-TAATACGACTCACTATAGGGAGACAAGAATAAACGCTCAA-3`) and oligo-004 (5`-GCCTGTTGTGAGCCTCCTGTCGAA-3`). In vitro transcription, RNA-protein binding, partition, and RT-PCR were repeated for 14 rounds and the resulting binders at rounds 10 and 14 were subcloned into the pCR-XL-TOPO vector (Invitrogen) for sequencing. Negative selection was carried out against the nitrocellulose membrane at rounds 3, 6, 9, and 12 to reduce non-specific interactions. The protein concentration for eVP35 IID WT was decreased from 0.4 µM at round 1 to 0.025 µM by round 14 in order to enhance selection stringency. For eVP35 IID CBP3mut, the protein concentration was decreased from 4 µM at round 1 to 0.05 µM by round 14. The concentration of protein used in each selection was adjusted so that the we can obtain about 1% retention of aptamers during each selection. Thus, the initial values of 0.4 µM for selection against eVP35 IID WT and 4 µM for selection against eVP35 IID CBP3mut were determined through this process.
End labeling of dsRNA and aptamers
32P 5`-end labeling of RNAs was carried out at 37°C for 1.5 hrs in a 20 µl reaction containing 70 mM Tris (pH 7.6 at 25°C), 10 mM MgCl2, 5 mM DTT, 1U/µl T4 polynucleotide kinase (New England Biolabs), 1 µM γ-ATP (Perkin Elmer), 5 µM RNA. 32P 3`-end labeling of RNA was carried out at 16°C for 18 hr in a 30 µl reaction of 50 mM Tris-HCl (pH 7.8 at 25°C), 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 10% DMSO, 0.67 U/µl T4 RNA ligase 1 (New England Biolabs), 2 µM 32P -cytidine-3`,5`-bis-phosphate (32pCp), 5 µM RNA (dsRNA or aptamer)61 Labeled RNAs were separated on a 7M urea-8% PAGE and eluted by crush soak method.
Filter binding assays for binding and competition assays for eVP35 IID-aptamer interaction
Filter binding assays were carried out as reported previously51 For dissociation constant (KD) measurements, the reaction mixtures contained 1–2 nM 32P-aptamer or 32P-dsRNA and eVP35 IID protein ranging from 0–100 µM. For single binding point assays, the reaction mixtures contained 10 nM 32P-aptamer and 0.5 µM eVP35 IID proteins. In the competition assay, the reaction mixtures included 10 nM 32P-aptamer, 0.5 µM eVP35 IID or 10 nM 32P-dsRNA, 10 µM eVP35 IID and unlabeled aptamer or dsRNA ranging from 0–20 µM. All reaction mixtures were incubated at 25°C for 15 min. Binding experiments were carried out in buffer containing 10 mM HEPES (pH 7.0), 150 mM NaCl, 2mM TCEP, 1 mM MgCl2 in 50 µl reactions in a 96-well vacuum filtration apparatus assembled with a nitrocellulose membrane on top of a positively charged nylon membrane. Membranes were washed and air-dried before exposure for 12–16 hrs along with a calibration standard, scanned on a Typhoon 9410 Variable Mode l Imager in phosphor storage mode, and quantified by ImageQuant (GE Healthcare). The ratio of the intensity on nitrocellulose membrane to that of the sum on nitrocellulose membrane plus nylon membrane was expressed as fractional binding. The binding curves were fit to the equation B=Bmax*[M] /(KD+[M]) using ORIGIN 7.0 software (OriginLab), where B is the fraction bound, Bmax is the maximum fractional binding, [M] is the protein concentration, and KD is the dissociation constant. Competition curves were fitted to the equation B/B0= 1/(1+[M]/IC50)n), where B is the fraction of 32P-labeled aptamers bound to nitrocellulose membrane at different concentrations of competitors (aptamer or dsRNA), B0 is fraction of 32P-labeled aptamers bound to nitrocellulose membrane without any competitors, [M] is the competitor concentration, IC50 is the concentration where 50% competition occurs, and n is a constant.
Isothermal titration calorimetry
Quantitative analysis of eVP35 IID protein-aptamer interactions were performed on a VP-isothermal titration calorimeter (VP-ITC) (Microcal). Protein samples were dialyzed against 500 mL of buffer containing 10 mM HEPES (pH 7.0), 150 mM NaCl, 1 mM MgCl2, and 2 mM TCEP for 12 h at 25 °C. Aptamer stocks were diluted >200-fold with dialysis buffer. The syringe contained eVP35 IID proteins at 100 to 150 µM, and the cell contained aptamers (1G8-14, 2F11-14) at 3 to 5 µM. ITC titrations were carried out using a reference power of 5 µcal/sec. The resulting ITC data were processed and fit to a one-site binding model or a two-site binding model to determine n (number of binding sites) and KD (dissociation constant) using ORIGIN 7.0 software. Both one and two site binding models were considered for each data set, but selection of the model was based on the quality of the fits. For two site binding, both sequential and independent binding modes were also considered. The two-site binding model used in this study assumes two thermodynamically independent binding events.
Pulldown assays
Pulldown assays were performed in buffer containing 10 mM HEPES (pH 7.0), 150 mM NaCl, 1 mM MgCl2, and 5 mM 2-mercaptoethanol at 25 °C. MBP-His tagged eVP35 IID WT or mutant proteins were immobilized on amylose resin. Resin bound MBP-His tagged eVP35 IID was incubated with purified His-tagged NP protein and subsequently washed. For aptamer competition assays, either 100µM aptamer or 300 µM dsRNA was incubated with MBP-His tagged eVP35 IID prior to incubation with NP. The level of protein used in the assay reflects our efforts to ensure proper detection of the pulldown experiment. Pulldown samples were separated on SDS-PAGE and analyzed by Western blot using mouse anti-His antibody (Santa Cruz biotechnology), followed by horseradish peroxidase (HRP) conjugated goat anti-mouse antibody (Bio-Rad). Membranes were developed using Millipore Immobilon Western Chemiluminescence HRP substrate and recorded on a ChemiDoc (Bio-Rad).
EBOV transcription/replication assays
The plasmids used in the EBOV transcription/replication assays were described previously18, 23, 31 NP, VP35, VP30 and L proteins were cloned into pTM1 with only the cDNA sequence (no virus-derived UTRs were present). Aptamers were cloned into a modified pSilencer plasmid62 and the corresponding plasmids were sequenced to verify the accuracy of the aptamers.
Quantitation of total aptamer copy number in transfected cells by qRT-PCR
Genome equivalent (GEq) copy numbers (Total copies,) of aptamers in the minigenome assay were measured using primer/probes targeting the aptamers. Briefly, real-time PCR (RT-PCR) was performed using Superscript II RT-PCR kits (Invitrogen). All RT-PCR mixtures contained 5 µl of RNA eluate and master mixes were set up following the manufacturer’s protocols. Standards and test samples were assayed in triplicate using the CFX96 detection system (Bio-Rad) with the following cycle conditions: 50° C for 10 min, 95°C for 5 min, and 40 cycles of 95°C for 10 sec and 59°C for 30 sec. Threshold cycle (CT) values representing aptamers were used to determine Total copies. To create the GEq standard, DNA from aptamers stocks (100 ng) was used and the number of genomes was calculated using Avogadro’s number and the total molecular weight of the plasmid with aptamer.
Aptamer secondary structure prediction
The computational program RNAfold from Vienna package v.2.063 was used to predict the secondary structures for RNA aptamers 1G8-14 and 2F11-14. Default parameters on the RNAfold server were used except for the Turner Model 200464, which was used as the RNA parameter source. Results were assessed for the minimum free energy (MFE) structure prediction as well as thermodynamic ensemble predictions, including centroid structure65 and base-pairing probabilities using dot plot analysis66 as implemented in RNAfold. Resulting secondary structures for the MFE and the centroid were visualized with the RNA graphics program VARNA67
Circular dichroism (CD)
CD data were collected for 1G8-14 and 2F11-14 at 10 µM in a quartz cuvette with a 1 mm path-length using a Chirascan™ CD spectrometer. CD spectra were acquired from 200 to 320 nm at 5 and 95 °C at an interval of 1 nm and time-per-point of 2 sec. Temperature melting of aptamers was performed by monitoring the ellipticity at 260 nm from 5 to 95 °C with a 2 °C increment at a 1 °C/min gradient. ORIGIN 7.0 software was used to plot CD spectra and to calculate first derivatives from the temperature melting plots.
RESULTS
Mutation of conserved basic residues does not perturb the overall fold of eVP35 IID
The CBP and FBP contain highly conserved basic residues in VP35 IID that are functionally important as mutations of either the CBP or the FBP impair several VP35 mediated functions7 Therefore, a series of structural studies were carried out to characterize the impact of these mutations on the integrity of the structure of eVP35 IID. These studies resulted in crystal structures of eVP35 IID CBP3mut (PDB code 4IJE) and FBP4mut (PDB code 4IJF), which were solved to 1.9 and 2.5 Å, respectively using the eVP35 IID WT (PDB code 3FKE) structure as the search model for molecular replacement (Table S1). These structures revealed minimal changes in the overall protein fold when compared to eVP35 IID (Figure S1). The most notable difference between the structures was observed in a short helix (α5) located within the β-sheet subdomain, in which the CBP3mut and FBP4mut structures make one turn of the helix instead of two. Comparison of the eVP35 IID WT, CBP3mut, and FBP4mut structures revealed that eVP35 IID CBP3mut and eVP35 IID FBP4mut structures display RMSD values of 0.68 and 0.46 Å, respectively, which further suggest minimal structural changes upon mutating out the CBP and FBP. Analysis of surface electrostatics, however, showed significant differences among the three structures (Figure S2). As expected, the CBP3mut resulted in a loss of the basic charge at the CBP, but the FBP was relatively unaffected, while the structure of FBP4mut revealed that the basic charge at the FBP was lost with the CBP unaffected.
SELEX identifies RNA aptamers that binding eVP35 IID
In order to generate different classes of aptamers for eVP35 IID, aptamer selection was carried out against two targets, eVP35 IID WT and eVP35 IID CBP3mut proteins using SELEX (Figure 1A). By using eVP35 IID WT and CBP3mut proteins, we biased the aptamer selection to different conserved regions of eVP35 IID. We expected that the dsRNA binding site formed by CBP residues would dominate aptamer selection using eVP35 IID WT as a target due to the highly basic charge and dsRNA binding properties in this region. In contrast, by mutating the CBP (in eVP35 IID CBP3mut), we reduced the basic nature of the CBP region with the intent that other charged regions, preferably the FBP, could compete during aptamer selection. The initial RNA pool contained RNA species of approximately 90 bases, with 45 nucleotides in the variable region flanked on either side by a 5` and a 3` common region. This initial pool bound eVP35 IID WT with a KD of 2900 ± 200 nM and bound eVP35 IID CBP3mut with a KD of 36000 ± 8000 nM. After several rounds of SELEX, each aptamer pool displayed enhanced affinity and specificity for eVP35 IID WT and eVP35 IID CBP3mut proteins. Our progress curves show >10-fold affinity enhancement for the final aptamer pool, with an affinity of 300 ± 100 nM at round 14 for eVP35 IID WT (Figure S3A) and 2800 ± 600 nM at round 10 for eVP35 IID CBP3mut (Figure S3B). We also extended our selection against eVP35 IID CBP3mut up to round 14. Although this pool displayed a high degree of non-specific binding to the nitrocellulose membrane, we were able to identify select individual aptamers from rounds 10 and 14. As discussed below, we were able to further characterize these select aptamers for affinity measurements and ranking.
For our initial SELEX studies, we observed significant enrichment of select sequences from a total of 96 clones. Using ClustalW multiple sequence alignments, sequences with variations less than 5% of the total aptamer length (i.e. 4 nucleotides or less that includes the constant/primer regions and the variable region) were grouped together and represented by a single member. We first selected 9 aptamers from the 1-series and 3 aptamers from the 2-series (Figure S3C). Subsequently, we identified representative members from each group based on their affinity for eVP35 IID WT and their ability to inhibit the interaction between VP35 IID and NP (see below). These selected aptamers were subjected to further characterization and ranking based upon affinity measurements with the top binders from each series selected for further analysis. Aptamer 1G8-14 was chosen to represent the 1-series aptamers, which were selected against eVP35 IID WT, and aptamer 2F11-14 was chosen to represent the 2-series aptamers, which were selected against eVP35 IID CBP3mut. In order to further examine RNA structural determinants, we used ViennaRNA structural prediction software to predict the RNA secondary structure of 1G8-14 and 2F11-14. The resulting secondary structure predictions for the MFE and the centroid were visualized with the RNA graphics program VARNA67, and base-pairing probabilities for the MFE and ensemble are shown using dot plot analysis (Figure S4)54 Based on the structure predictions, the 1G8-14 MFE and centroid structure displayed 15.12 ensemble diversity and the MFE structure represented 5.12% of the ensemble compared to the 2F11-14 MFE and centroid structure with 13.46 ensemble diversity and 4.23% of the ensemble represented by the MFE. However, the free energy of the ensemble for 1G8-14 was −22.63 kcal/mol compared to −12.75 kcal/mol for 2F11-14 suggesting that the 1G8-14 aptamer may adopt a more stable structure. Our CD data show differences in the corresponding spectra providing some experimental evidence that indicate structural differences between 1G8-14 and 2F11-14 aptamers (Figure S5). However, additional studies are required to appropriately characterize the actual structural differences between 1G8-14 and 2F11-14 aptamers. As an initial step in our efforts to define the optimum aptamer, we carried out boundary determination studies and tested several shorter aptamers based on the results of our boundary determination experiments. These results are described in the section entitled “Primer binding region of 1G8-14 is important for high affinity VP35 IID binding” (see below). These studies supported the use of full length aptamers in our present studies as discussed below.
Aptamers 1G8-14 and 2F11-14 bound eVP35 IID WT with affinities higher than dsRNA. At 1µM protein, the fractional binding for both 1G8-14 and 2F11-14 was >0.5, while the fractional binding for dsRNA was <0.3 (Figure 1B). eVP35 IID CBP3mut bound to 2F11-14 with high affinity and to 1G8-14 only at higher protein concentrations (10 µM protein) (Figure 1C). These binding studies provided an initial indication that we had identified aptamers that bind to eVP35 IID differently, with the CBP residues contributing to 1G8-14 aptamer binding. The ability of eVP35 IID WT and eVP35 IID CBP3mut to bind ssRNA and dsRNA was also assessed. As previously shown, eVP35 IID WT binds to dsRNA, but not ssRNA (Figure 1C). eVP35 IID CBP3mut does not bind to dsRNA due to mutations at key dsRNA binding residues, R312, K319 and R322 (Figure 1C)16, 28 Previous studies have shown that eVP35 binds dsRNA containing a 5`ppp moiety with higher affinity than blunt end dsRNA18, 29 Therefore, we tested if a 5`ppp or 5`OH affected aptamer binding to eVP35 IID. The resulting data suggest that the 5` phosphorylation state of the aptamer does not influence eVP35 IID binding (Figure S6). Additionally, this observation supports the notion that the eVP35-aptamer interactions are distinct from those observed between eVP35 IID and dsRNA11, 18.
Aptamers recognize partially overlapping sites in eVP35 IID
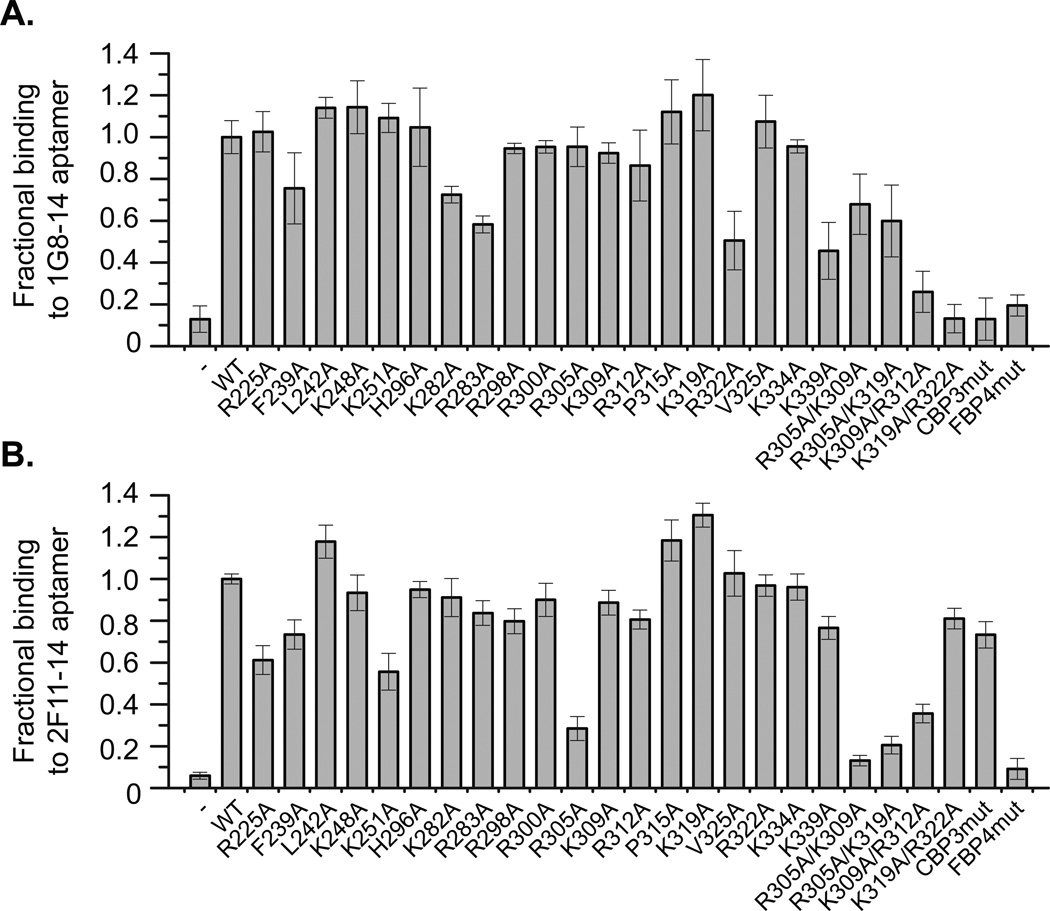
We and others have shown that eVP35-dsRNA interactions are mediated through the CBP region of eVP35 IID11, 16, 18, 29, 30 Therefore, a series of basic residues in eVP35 IID were mutated to determine the contribution of these residues, if any, to 1G8-14 and 2F11-14 aptamer binding. Representative single point binding data for 1G8-14 and 2F11-14 aptamers revealed distinct binding patterns. 1G8-14 aptamer binding primarily depends on a subset of residues that are also important for dsRNA binding and IFN inhibition. Single mutation of residues F239, R305, K309, R312, R322, and K339 to alanine had little effect on the 1G8-14 aptamer binding (<30% relative to WT) whereas double/triple mutations (K309A/R312A, K319A/R322A, CBP3mut) significantly diminished binding to 1G8-14 (>70% relative to WT) (Figure 2A). Mutations of the FBP residues (eVP35 IID FBP4mut) also impair binding to 1G8-14; however, further experiments revealed that this decrease in 1G8-14 binding to eVP35 IID FBP4mut is largely due to a decrease in the maximum fractional binding and not due to a substantial change in binding affinity (Figure S7). Moreover, these aptamers readily competed with dsRNA in competition assays (Figure S8). These results are different from those observed for dsRNA binding, where select single point mutation of residues in the CBP abolished dsRNA binding18 Collectively, these results suggest that aptamer binding involves multiple residues within a given eVP35 IID surface, with the CBP residues providing the largest contribution towards 1G8-14 aptamer binding.
Figure 2. Mutational analysis reveals differences in eVP35 IID-aptamer binding sites.
Fractional binding of aptamers (A) 1G8-14 and (B) 2F11-14 to eVP35 IID WT and mutant proteins measured by filter binding assay. Average fractional binding normalized to eVP35 IID WT are from two independent experiments, each with four repeats for individual eVP35 IID variants.
For the 2F11-14 aptamer, we observe a set of residues different from those important for 1G8-14 binding, including R225, K251, and F239, as mutation of these residues to alanine result in some loss of aptamer binding (<40% relative to WT) (Figure 2B). Most significantly, R305A mutant alone can drastically decrease binding to 2F11-14 (>70% relative to WT). In contrast to 1G8-14, eVP35 IID CBP3mut had limited impact on 2F11-14 binding, consistent with the single point data (Figure 1C). Residues R305 and K309 are important for both aptamers, suggesting that the aptamer binding sites are partially overlapping at or near these residues.
Residues in eVP35 IID CBP and FBP contribute differentially to aptamer binding
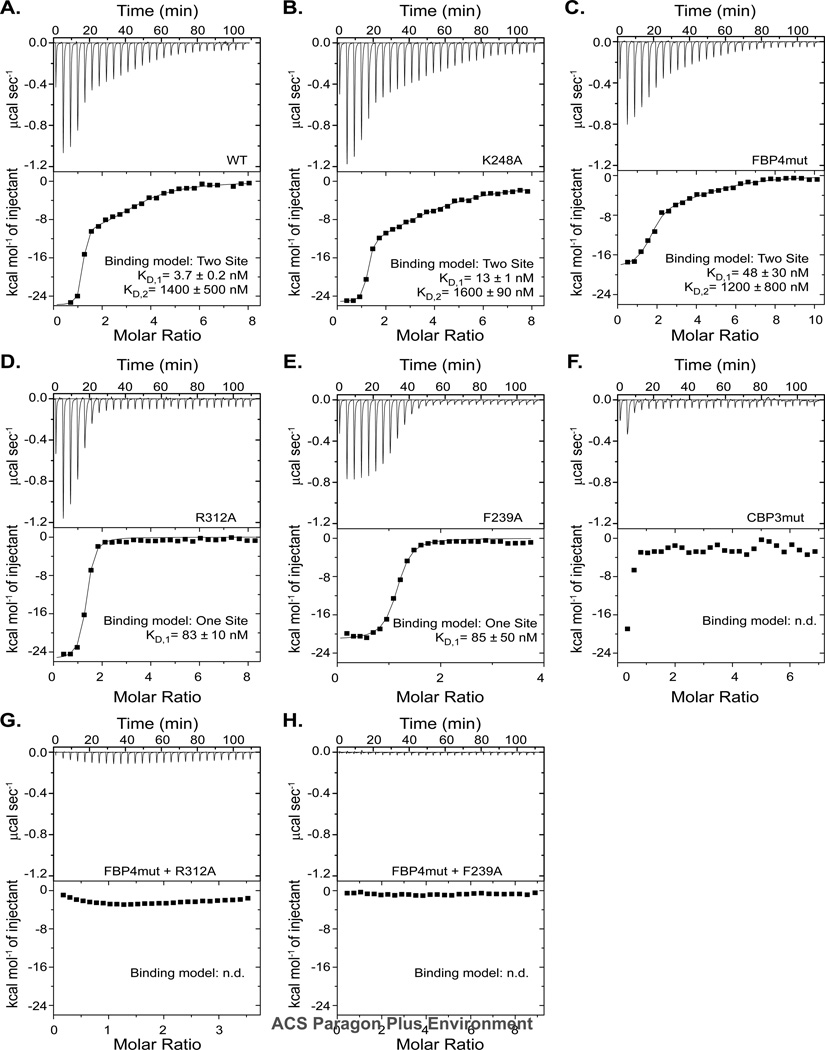
To further characterize eVP35-aptamer interactions, we measured the binding affinities of aptamer to eVP35 IID WT and mutant proteins using ITC. eVP35 IID WT bound 1G8-14 aptamer with high affinity compared to dsRNA (Figure 3A,18, 23). One-and two-site binding models were considered, ultimately resulting in the ITC data being fit using a two-site binding model, indicating that there are two independent binding modes for eVP35 IID on 1G8-14eVP35. The first binding mode is of high affinity (KD,1 = 3.7 ± 0.2 nM, n1 = 1.1 ± 0.1), which we term the “aptamer binding mode”, whereas the second binding mode showed moderate affinity (KD,2 = 1400± 500 nM, n2 = 2.7 ± 0.4) (Figure 3A, Table 1). The affinity and the multiple bindings displayed by the second binding mode suggest that this is the dsRNA binding mode. Mutation of residues in the FBP (e.g. K248A and FBP4mut) had little effect on binding (Figure 3B–C). When we test binding of aptamer to eVP35 IID proteins containing mutations of residues important for dsRNA binding (e.g. R312A and F239A), we find that the second binding mode is eliminated and the data can be fit to a one-site binding model (KD,1 values of 83 ± 10 nM and 85 ± 50 nM, respectively) (Figure 3D–E). Furthermore, mutation of three CBP residues together (i.e R312A/K319A/R322A triple mutant or CBP3mut) or in combination with mutations of the four FBP residues (i.e. FBP4mut/R312A and FBP4mut/F239A) in eVP35 IID results in complete loss of 1G8-14 aptamer binding (Figure 3F–H). For the second binding mode, these data are consistent with previously reported descriptions of the dsRNA-eVP35 interaction18 Altogether, our data suggest that the first binding mode is the high affinity aptamer-eVP35 IID interaction (aptamer mode) and the second binding mode is eVP35 IID binding to double stranded regions (dsRNA mode) in the aptamer (Table 2).
Figure 3. The CBP is important for high affinity binding of 1G8-14 aptamer to eVP35 IID.
ITC raw data and corresponding binding isotherms for 1G8-14 binding to eVP35 IID (A) WT, (B) K248A, (C) FBP4mut, (D) R312A, (E) F239A, (F) CBP3mut, (G) FBP4mut/R312A, and (H) FBP4mut/F239A.
Table 1.
Measured binding affinities between aptamers and WT or mutant eVP35 I ID proteins by ITC.
| 1G8-14 | 2F11-14 | |||||||
|---|---|---|---|---|---|---|---|---|
| n1 | KD,1(nM) | n2 | KD,2(nM) | n1 | KD,1(nM) | n2 | KD,2(nM) | |
| WT | 1.1±0.1 | 3.7±0.2 | 2.7±0.4 | 1400±500 | 1.2±0.0a | 7.1±0.1 | 5.1±0.1 | 380±40 |
| K248A | 1.1±0.0b | 13±1 | 3.3±0.4 | 1600±90 | 1.7±0.0c | 36±7 | 5.3±0.1 | 830±300 |
| FBP4mut | 1.4±0.2 | 48±30 | 3.2±0.6 | 1200±800 | n.d. | n.d. | n.d. | n.d. |
| R312A | 1.2±0.1 | 83±10 | n.d. | n.d. | 1.4±0.0d | 52±6 | n.d. | n.d. |
| F239A | 1.1±0.0e | 85±50 | n.d. | n.d. | 1.4±0.0f | 100±8 | n.d. | n.d. |
| CBP3mut | n.d. | n.d. | n.d. | n.d. | 1.3±0.1 | 56±7 | n.d. | n.d. |
| lFBP4mut/ R312A |
n.d. | n.d. | n.d. | n.d. | ||||
| CBP3mut/ K248A |
1.6±0.2 | 180±20 | n.d. | n.d. | ||||
= 1.2 ± 0.04
= 1.1 ± 0.03
= 1.7 ± 0.03
= 1.4 ± 0.01
= 1.1 ± 0.02
= 1.4 ± 0.04
n.d.: no binding
Table 2.
Summary of ITC results for aptamers binding to WT or mutant eVP35 IID proteins.
| dsRNA | 1G8-14 | 2F11-14 | |||
|---|---|---|---|---|---|
| Kd,1 | Kd,2 | Kd,1 | Kd,2 | ||
| WT | + + + + | + + + + | + + + + | + + + + | + + + + |
| K248A | + + + + | + + + + | + + + + | + + + + | + + + + |
| FBP4mut | n.a. | + + | + + + + | − | − |
| R312A | − | + | − | + + + | − |
| F239A | − | + | − | + + | − |
| CBP3mut | − | − | − | + + + | − |
| FBP4mut+R312A | n.a. | − | − | n.a. | n.a. |
| FBP4mut+F239A | n.a. | − | − | n.a. | n.a. |
| CBP3mut+ K248A | n.a. | n.a. | n.a. | + | − |
+ + + + = WT
+++ = 5–9 fold decrease in KD
++ = 10–19 fold decrease in KD
+ = >20 fold decrease in KD
− = no binding
n.a: not available
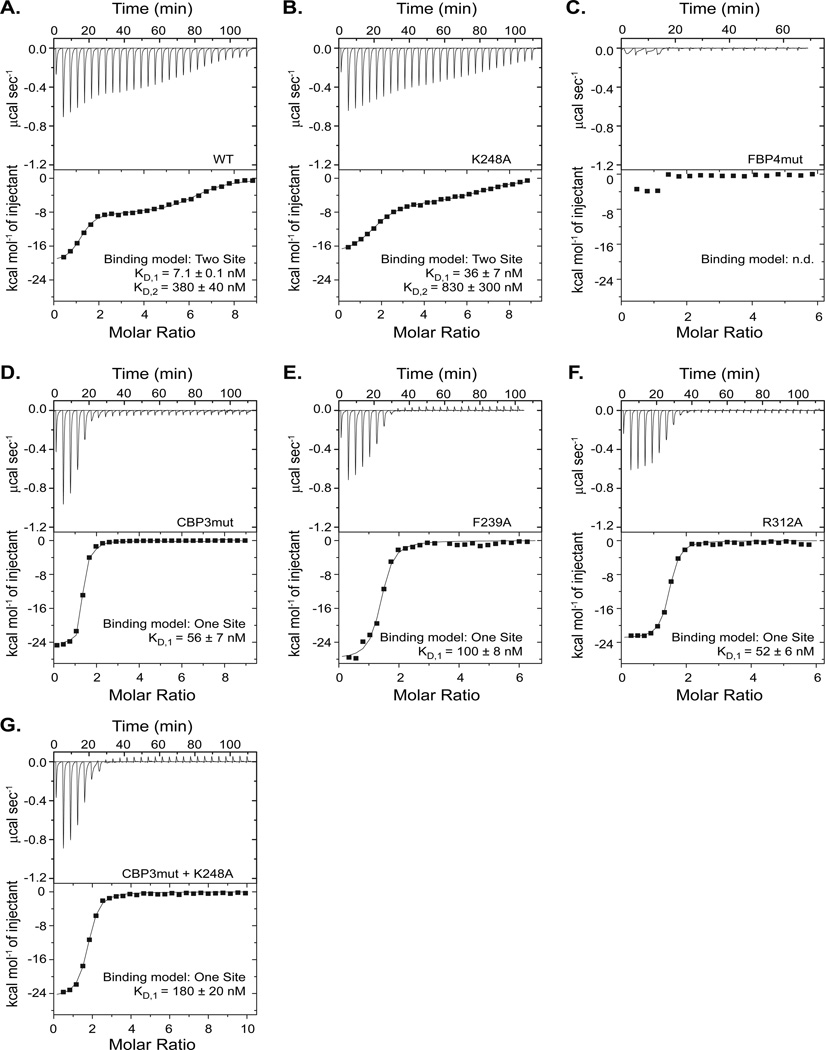
The ITC binding data for 2F11-14 aptamer to eVP35 IID were fit to a two-site binding model, where KD,1 = 7.1 ± 0.1 nM (n1 = 1.2 ± 0.0) and KD,2 = 380 ± 40 nM (n2 = 5.1 ± 0.1) (Figure 4A, Table 1). A single point mutation in the FBP, K248A, leads to slightly diminished binding with an approximately 5-fold increase in KD,1 and a 2.5-fold increase in KD,2 (Figure 4B). Mutation of four residues in the FBP (eVP35 IID FBP4mut) leads to near complete loss of 2F11-14 binding (Figure 4C), emphasizing a critical role for the FBP in 2F11-14 aptamer binding. Mutation of residues in the CBP eliminates the second binding event with KD,1 = 56 ± 7 nM, KD,1 = 100 ± 8 nM, and KD,1 = 52 ± 6 nM for eVP35 IID CBP3mut, F239A, and R312A, respectively (Figure 4D–F). Interestingly, mutation of K248A in combination with the CBP3mut leads to a ~25-fold increase in KD,1 compared to eVP35 IID WT (Figure 4G), but does not abolish binding altogether. Thus, in contrast to 1G8-14, residues in the FBP are critical for high affinity 2F11-14 binding. Similar to the results observed for 1G8-14 binding, residues in the CBP contribute to the second binding event, presumably through contacts with double stranded regions of the 2F11-14 aptamer. Our results are summarized in Table 2
Figure 4. The FBP is important for high affinity binding of 2F11-14 aptamer to eVP35 IID.
ITC raw data and corresponding binding isotherms for 2F11-14 binding to eVP35 IID (A) WT, (B) K248A, (C) FBP4mut, (D) CBP3mut, (E) F239A, (F) R312A, and (G) CBP3mut/K248A.
In order to better define the binding interface for eVP35 IID-1G8-14 and eVP35 IID/2F11-14, and how these aptamers are oriented with respect to eVP35 IID, we used a filter binding competition assay to see if 1G8-14 could compete with 2F11-14 for eVP35 IID binding and vice versa. As expected, unlabeled 1G8-14 was able to compete off 32P-labeled 1G8-14. Unlabeled 1G8-14 can also compete off 32P-labeled 2F11-14 (Figure S9). However, unlabeled 2F11-14 could only compete off 32P-labeled 2F11-14, but not 32P-labeled 1G8-14. Since 1G8-14 was selected against WT eVP35 IID, it is conceivable that 1G8-14 aptamer would make contacts with both basic patches. This notion is supported by our ITC data (described above), showing that the CBP is the primary binding site for 1G8-14, but the FBP also contributes to 1G8-14 binding, and mutations to a CBP residue in combination with mutations to the FBP region (FBP4mut + R312A) results in loss of interaction. In contrast, 2F11-14 aptamer, which was selected against a mutant protein with only the FBP present, may predominantly interact with the FBP surface. Altogether, these data are consistent with the notion that distinct classes of high affinity aptamers can be developed using eVP35 WT and mutant eVP35 IID proteins as SELEX targets.
Primer binding region of 1G8-14 is important for high affinity VP35 IID binding
The structure predictions for the 1G8-14 aptamer (Figure S4) suggested that the primer binding regions may be important for secondary structure. To further evaluate the contributions from secondary structure of the 1G8-14 primer regions, we performed binding site boundary measurements to identify the nucleotides of 1G8-14 important for binding to eVP35 IID (Figure S10). The resulting data suggests that the region between nucleotides 36 and 65 is important. However, given the nature of the experiment, it was not clear if this is due to binding of the structural element shown in Figure S10B or if binding was primarily driven by the sequence between nucleotides 36 to 65 (indicated by arrows in Figure S10C). In this assay, we used partially digested aptamers under native conditions and isolated aptamer fragments that bound eVP35 IID binding. RNA bound eVP35 IID proteins were subsequently subjected to denaturing gel electrophoresis for sequence mapping. In order to differentiate between the two potential aptamer fragments that form the high affinity binding, we generated the minimum RNA sequence (nucleotides 36–65) and subjected that sequence to structure predictions and binding studies. In addition, we also tested RNA fragments corresponding to 1–65 and 36–90 nucleotides and the minimal free energy (MFE) structure predictions are shown in Figure S11 These predictions revealed that bases 36–65 forms a stem-loop (shaded region), which is not a sub-structure predicted for full length 1G8-14 structure (compare Figure S11A with Figure S11B). We performed ITC experiments in order to compare the full-length and truncated 1G8-14 constructs in their ability to bind eVP35 IID as well as mutant eVP35 IID proteins. Truncated 1G8-14 36–65 bound to eVP35 IID WT with a two-site binding mode similar to full-length 1G8-14. However, the truncated 1G8-14 36–65 aptamer was unable to bind eVP35 IID R312A (Figure S12; see Figure S12F vs Figure S12C). The sensitivity to the eVP35 IID R312A mutation was also observed previously for dsRNA binding. We also conducted ITC experiments for truncated 1G8-14 1–65 and 1G8-14 36–90 to assess the importance of the 5’ constant region and 3’ constant region, respectively (Figure S13). Truncated 1G8-14 1–65 bound eVP35 IID WT with a two-site binding mode, but similar to truncated 1G8-14 36–65 was unable to bind eVP35 IID R312A. Additionally, truncated 1G8-14 1–65 binding to eVP35 IID WT appear to be cooperative and this observation will require additional experiments in order to fully explore this behavior. However, it is evident that there are significant differences between WT and the 1–65 truncated aptamer. In contrast, truncated 1G8-14 36–90 bound eVP35 IID WT with a two-site binding mode similar to full length aptamer and more importantly, this truncated aptamer bound eVP35 IID R312A with high affinity (Figure S13I). Together these data suggest that the variable region of 1G8-14 is important for binding to eVP35 IID, but additional contributions from the constant regions are required for the optimal aptamer formation. Based on these observations, we used full length aptamers in our studies described here.
Aptamers disrupt eVP35 IID-NP interactions and inhibit EBOV replication/transcription activity
In order to test the utility of the 1G8-14 and 2F11-14 aptamers, we assessed their ability to inhibit interactions between eVP35 IID and NP proteins. This interaction is important for the function of the filoviral replication complex23, 31 Specifically, residues in the FBP of eVP35 IID are important for eVP35 IID-NP interactions and minigenome activity (Figure 5A)23 Mutation of all four residues in the FBP (eVP35 IID FBP4mut) leads to loss of eVP35 IID-NP binding in a pulldown assay. In contrast, mutation of residues in the CBP (eVP35 IID R305A/K309A and eVP35 IID CBP3mut) does not impact eVP35-NP binding. Since the FBP appears to be important for both 1G8-14 and 2F11-14 binding, we tested the ability of these aptamers to compete with NP for binding to eVP35. Both 1G8-14 and 2F11-14 aptamers were able to inhibit eVP35 IID-NP complex formation, while dsRNA and aptamers 2B3-10 and 2D1-10 are unable to disrupt eVP35 IID-NP interaction (Figure 5B). Aptamers 2B3-10 and 2D1-10, selected against eVP35 IID CBP3mut, are of similar length to 1G8-14 and 2F11-14 aptamers, and they display similar high affinity binding to eVP35 IID (Figure 5C). Together, our data demonstrate the ability of 1G8-14 and 2F11-14 aptamers to specifically disrupt the eVP35-NP interaction (Figure 6). We also tested the ability of aptamers 1G8-14 and 2F11-14 to inhibit RNA synthesis in a replication/transcription assay, where the viral replication complex was reconstituted by expression of select viral proteins31 Resulting data show that 1G8-14 and 2F11-14 can antagonize the replication/transcription activity in a dose dependent manner (Figure 6). In contrast to these two specific aptamers, additional aptamers with similar length and affinity show little to no inhibition, despite all six aptamers being present at comparable levels as judged by qRT-PCR (Figure S14). Altogether, these results demonstrate that 1G8-14 and 2F11-14 aptamers can potentially function as competitive inhibitors of the filoviral replication complex by disrupting eVP35 IID-NP interactions. However, it is worth noting that the aptamers can interact with NP at high protein concentrations (>100 µM); thus, it is not clear if additional rounds of aptamer optimization with negative selection against NP will be necessary prior to use of these aptamers in virally infected cells.
Figure 5. 1G8-14 and 2F11-14 aptamers disrupt eVP35 IID-NP interaction.
MBP-His tagged WT or mutant eVP35 IIDs were immobilized on amylose resin and incubated with His-tagged NP proteins in the (A) absence or (B) presence of dsRNA or aptamer. (A) Lanes 1, 3, 5, 7 are input (I) samples for MBP-His eVP35 IID WT, CBP3mut, 305A/309A, and FBP4mut, respectively. Lanes 2, 4, 6, and 8 are final bead (FB) samples for MBP-His eVP35 IID WT, CBP3mut, 305A/309A, and FBP4mut, respectively. (B) Lane 1 shows MBP-His tagged eVP35 IID WT bound to the amylose resin. Lane 2 shows the presence of both His-NP and MBP-His eVP35 IID WT, and lanes 3–8 are final bead samples for pulldowns done in the absence (lane 3) or presence of dsRNA (lanes 4) and aptamers 1G8-14 (lane 5), 2B3-10 (lane 6), 2D1-10 (lane7) and 2F11-14 (lane 8).
Figure 6. 1G8-14 and 2F11-14 aptamers inhibit EBOV replication/transcription activity in a dose dependent manner.
A replication/transcription assay was performed in which plasmids encoding the different aptamers (250, 500 ng, left and right, respectively) were co-transfected with the plasmids required for minigenome replication and transcription (plasmids expressing EBOV L, NP, VP30 and VP35 along with plasmid expressing T7 promoter dependent EBOV minigenome which encodes a Renilla luciferase reporter gene). Also co-transfected was a plasmid that expresses firefly luciferase from an RNA polymerase II promoter. Renilla luciferase activity was normalized firefly luciferase activity. Minigenome reporter activation was expressed as relative activity by setting the negative control (without VP35) to a value of 1. The error bars indicate standard deviation of three independent replicates, *p=0.001, **p=0.0005. The western blot shows expression of NP and eVP35.
Aptamers differentially recognize closely related filoviral VP35 IID proteins
Filoviral VP35 proteins share high sequence similarity in the IID region (97.6% similar, 88.9% identical for RESTV aligned with EBOV; 81.7% similar, 22.9% identical for MARV aligned with EBOV) (Figure S15). Therefore, we tested the ability to the 1G8-14 and 2F11-14 aptamers to differentiate between closely related VP35 proteins. The resulting data showed that 1G8-14 aptamer preferentially binds to eVP35 IID proteins (EBOV and RESTV VP35 IID proteins) (Figure 7A). When binding was tested at protein concentrations <1 µM, EBOV and RESTV showed a fractional binding >0.6, while <0.1 fractional binding was observed for MARV. In contrast, 2F11-14 aptamer bound to all three filoviral VP35 IID proteins with >0.5 fractional binding at protein concentrations > 1µM. (Figure 7B). The ability of 1G8-14 to differentiate between eVP35 IID and MARV VP35 IID proteins is likely due to sequence differences between EBOV and MARV VP35 proteins near the CBP. For example, in EBOV VP35 IID positions 319 and 322 are K319 and R322, respectively, while corresponding positions in MARV VP35 IID are T309 and K312 (Figure 2 and Figure S15). Thus, it appears that the relative sensitivity to different filoviral species is entirely dependent on the relative contributions of the different basic patches.
Figure 7. Aptamers differentially bind to filoviral VP35 IID proteins.
eVP35 IID from EBOV, RESTV, and MARV were tested in their ability to bind (A) 1G8-14 and (B) 2F11-14 aptamers at varying protein concentrations shown as fractional binding.
DISCUSSION
In this study, we have identified and characterized two aptamers against eVP35 IID that bind with low nanomolar affinity using SELEX technology. These aptamers were generated by using two targets, eVP35 IID WT and eVP35 IID CBP3mut, with the goal of identifying different classes of aptamers that interact with distinct sets of residues within filoviral VP35 proteins. Target selection was in large part motivated by available structure-function data and our goal was to drive the selection process to different functionally important regions of this multifunctional non-enzymatic protein. Based on the data described above, our selected aptamers display binding affinities that are 20–100 fold higher than dsRNA and the 92 nucleotide long random RNA pool that was used in the initial SELEX, supporting the specific enrichment of aptamers. From the initial groups of aptamers, 1G8-14 and 2F11-14 aptamers were chosen to represent two aptamer classes as they bound eVP35 IID with high affinity, disrupted the eVP35IID-NP complex, and displayed significant differences at the nucleotide level (~40% difference in the aptamer variable region). Biochemical and functional mapping of select aptamers revealed that 1G8-14 and 2F11-14 aptamers bind eVP35 IID primarily through interacting with residues from the CBP and the FBP, respectively. Although it is apparent that additional interactions between eVP35 IID and aptamers are also formed due to the highly basic nature of the eVP35 IID surface30 Our ITC data revealed two independent binding modes: a high affinity aptamer binding mode and a the dsRNA binding mode (Figure 8). Each aptamer has a single high affinity binding site and multiple dsRNA binding sites. These two binding events are energetically independent, at least in the context of eVP35 IID. Interestingly, mutations to CBP residues, or key residues for dsRNA binding resulted in the presumable loss of the dsRNA binding mode. Therefore, at low eVP35 IID concentrations, we observe the high affinity aptamer binding event with a eVP35 IID:aptamer ratio of 1:1. As we increase eVP35 IID concentrations, eVP35 IID is able to make additional interactions with double stranded regions of the aptamers due to its inherent dsRNA binding function, therefore, allowing multiple eVP35 IID proteins to interact with a single aptamer.
Figure 8. 1G8-14 and 2F11-14 aptamers have multiple binding modes.
VP35 aptamers display two binding modes that include a single high affinity binding site and multiple dsRNA binding sites. At low eVP35 IID WTconcentrations or when binding to key dsRNA binding mutant R312A, only the high affinity site is occupied. At higher concentrations of eVP35 IID WT additional binding events occur via the dsRNA binding mode.
This work demonstrates how integrating available structural and functional data to tailor SELEX selection can result in aptamers that target different functional regions and potentially generate multivalent aptamers. 1G8-14 aptamer was selected against eVP35 IID WT and binds primarily to residues in the CBP with additional contacts to the FBP region. Despite the fact that the CBP is important for both 1G8-14 aptamer and dsRNA binding, there are significant differences in how eVP35 IID binds these two ligands. Most notably, a single point mutation within the CBP completely abolishes dsRNA binding16, 18, 28 However, single point mutations have only marginal effects on 1G8-14 aptamer binding (Figure 2). In contrast, double and triple mutation of CBP residues significantly affect the ability of 1G8-14 aptamer to bind eVP35 IID, which supports the notion that the aptamers establish multiple contact sites at the binding interface(s). In addition to the CBP, our data shows that mutations within the FBP affect 1G8-14 aptamer binding (increases KD,1). Mutation of the FBP along with a single mutation in the CBP region abolishes aptamer binding altogether. These results further support the view that 1G8-14 aptamer recognizes several surfaces across eVP35 IID. In contrast, the main site of interaction on eVP35 IID for the 2F11-14 aptamer appears to be near the FBP, with the CBP providing additional contact points. Our results clearly suggest that both 1G8-14 and 2F11-14 aptamers interact with multiple copies of eVP35 IID and therefore there is a high potential for these aptamers to inhibit multiple VP35-mediated functions in vivo.
In this work, we were able to target the CBP or the FBP of eVP35 IID and show that 1G8-14 and 2F11-14 aptamers have distinct binding footprints through biochemical mapping. Moreover, we are able to distinguish that the CBP is the primary binding site for 1G8-14, while 2F11-14 uses the FBP as its primary binding site. As a multifunctional protein, eVP35 IID inhibits IFN responses to viral infection, in part through dsRNA sequestration, and supports RNA viral synthesis through structural bridging of NP and the viral polymerase L. We have shown that aptamers bind eVP35 IID using multiple points of contact, a mode that is distinct from dsRNA binding. Furthermore, aptamers can compete with dsRNA for eVP35 IID binding, indicating that aptamers have the potential to disrupt the IFN inhibitory function of eVP35. Here we show that aptamers disrupt the eVP35-NP interaction and replication/transcription activity, indicating their potential to inhibit eVP35 polymerase cofactor function. It is interesting to note that selected aptamers can potentially inhibit both the eVP35 immune suppressor function and the polymerase cofactor function.
Aptamers are well known for their high affinity and specificity against their targets. The high specificity of aptamers can be used to distinguish among viral subtypes, which is relevant for the development of aptamers against viral proteins. On the other hand, aptamers with broad specificity can potentially serve as pan-antivirals. One of our selected aptamers, 1G8-14, can distinguish between EBOV and MARV VP35 IIDs, binding to EBOV and RESTV VP35 IIDs with KD’s 20-fold lower (at 1 µM concentration of protein, see Figure 5A) than MARV VP35 IID. The other selected aptamer, 2F11-14, can bind EBOV, RESTV, and MARV VP35 IIDs with comparable affinities. Thus, we have demonstrated that it is feasible to develop both highly specific and pan-antifiloviral aptamers using VP35 as the SELEX target molecule, and our success in selecting both highly specific and pan-antifiloviral aptamers indicates that there are highly conserved regions of VP35 which are recognized by 2F11-14, as well as species unique regions in VP35, which allows differential binding of 1G8-14 to EBOV and MARV VP35 IIDs.
Filoviruses cause high rates of fatalities during outbreaks and are potential agents of bioterrorism. Although filoviral vaccine options are under investigation, no approved vaccines are available at this time. Furthermore, other therapeutics such as small molecule or drug-like methods to counter disease are largely lacking at present. Work here, using available structure-function data, supports the development of VP35 specific aptamers that can facilitate validation of multifunctional VP35 as an antiviral target. Thus, inhibition of VP35 function using aptamers will not only provides a path to develop antivirals, but also provide novel target sites and mechanisms for antiviral development.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank P. Ramanan (Washington University School of Medicine) and Dr. M. Nilsen-Hamilton (Iowa State University) for reagents and helpful discussions and Dr. Vinayaka R. Prasad (Albert Einstein College of Medicine) for the pSilencer plasmid.
Funding:
This work is supported in part by NIH grants (R01AI081914 and MRCE Developmental Grant (U54AI057160-Virgin (PI) to G.K.A. and R01AI059536 to C.F.B.). Some reagents used in the project were generated under DOD/DTRA grant HDTRA1-12-1-0051. The project or effort depicted was or is sponsored by the Department of the Defense, Defense Threat Reduction Agency. The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred.
ABBREVIATIONS
- EBOV
Ebola virus
- RESTV
Reston virus
- MARV
Marburg virus
- LLOV
Lloviu virus
- IFN
interferon
- IID
IFN inhibitory domain
- VP35
viral protein 35
- eVP35 IID
EBOV VP35 IID
- RLRs
RIG-I-like receptors
- CBP
central basic patch
- FBP
first basic patch
- SELEX
Systematic Evolution of Ligands by EXponential enrichment
- NP
nucleoprotein
- ITC
isothermal titration calorimetry
- dsRNA
double stranded RNA
- IKKε
IkappaB kinase epsilon
- TBK-1
TANK-binding kinase 1
- PKR
RNA-dependent protein kinase
- CD
circular dichroism
- TCEP
tris(2-carboxyethyl)phosphine
- HRP
horseradish peroxidase
- MFE
minimum free energy
- RMSD
root-mean-square deviation
Footnotes
Supporting Information
Data collection, structure solution, and refinement statistics for eVP35 IID CBP3mut and eVP35 IID FBP4mut; alignment of crystal structures and electrostatic surface representation for zVP35 IID WT, CBP3mut, and FBP4mut; enrichment of aptamer populations using SELEX; secondary structure prediction for 1G8-14 and 2F11-14; CD spectra show that 1G8-14 and 2F11-14 have different secondary structures; aptamer binding is not affected by the 5`-ppp moiety; mutations in the FBP of zVP35 IID result in overall lower maximum fractional binding; aptamers compete with dsRNA for eVP35 IID binding; 1G8-14, but not 2F11-14, can compete with 1G8-14 for binding to eVP35 IID; determination of the minimal binding region of 1G8-14 with eVP35 IID; structure predictions and binding studies for truncated 1G8-14; similar amounts of aptamer RNA in transfected cells as judged by qRT-PCR; sequence alignment of EBOV, RESTV, and MARV IIDs. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bray M, Murphy FA. Filovirus research: knowledge expands to meet a growing threat. J Infect Dis. 2007;196(Suppl 2):S438–S443. doi: 10.1086/520552. [DOI] [PubMed] [Google Scholar]
- 2.Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martinez M, Herrera JE, Pizarro M, Hutchison SK, Echevarria JE, Lipkin WI, Tenorio A. Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog. 2011;7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groseth A, Stroher U, Theriault S, Feldmann H. Molecular characterization of an isolate from the 1989/90 epizootic of Ebola virus Reston among macaques imported into the United States. Virus Res. 2002;87:155–163. doi: 10.1016/s0168-1702(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 5.Jahrling PB, Geisbert TW, Dalgard DW, Johnson ED, Ksiazek TG, Hall WC, Peters CJ. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet. 1990;335:502–505. doi: 10.1016/0140-6736(90)90737-p. [DOI] [PubMed] [Google Scholar]
- 6.Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh WJ, Batten B, Sealy TK, Carrillo C, Moran KE, Bracht AJ, Mayr GA, Sirios-Cruz M, Catbagan DP, Lautner EA, Ksiazek TG, White WR, McIntosh MT. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 7.Leung DW, Prins KC, Basler CF, Amarasinghe GK. Ebolavirus VP35 is a multifunctional virulence factor. Virulence. 2010;1:526–531. doi: 10.4161/viru.1.6.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, Klenk HD, Palese P, Garcia-Sastre A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enterlein S, Warfield KL, Swenson DL, Stein DA, Smith JL, Gamble CS, Kroeker AD, Iversen PL, Bavari S, Muhlberger E. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob Agents Chemother. 2006;50:984–993. doi: 10.1128/AAC.50.3.984-993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. J Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Leung DW, Shabman RS, Farahbakhsh M, Prins KC, Borek DM, Wang T, Muhlberger E, Basler CF, Amarasinghe GK. Structural and functional characterization of Reston Ebola virus VP35 interferon inhibitory domain. Journal of molecular biology. 2010;399:347–357. doi: 10.1016/j.jmb.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, Basler CF, Amarasinghe GK. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF. Mutual Antagonism between the Ebola Virus VP35 Protein and the RIG-I Activator PACT Determines Infection Outcome. Cell Host Microbe. 2013;14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prins KC, Cardenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumann M, Gantke T, Muhlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J Virol. 2010;84:10581–10591. doi: 10.1128/JVI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi W, Huang Y, Sutton-Smith M, Tissot B, Panico M, Morris HR, Dell A, Haslam SM, Boyington J, Graham BS, Yang ZY, Nabel GJ. A filovirus-unique region of Ebola virus nucleoprotein confers aberrant migration and mediates its incorporation into virions. J Virol. 2008;82:6190–6199. doi: 10.1128/JVI.02731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, Basler CF. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimberlin CR, Bornholdt ZA, Li S, Woods VL, Jr, MacRae IJ, Saphire EO. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci U S A. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, Amarasinghe GK. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci U S A. 2009;106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson JA, Bray M, Bakken R, Hart MK. Vaccine potential of Ebola virus VP24, VP30, VP35, and VP40 proteins. Virology. 2001;286:384–390. doi: 10.1006/viro.2001.1012. [DOI] [PubMed] [Google Scholar]
- 34.Geisbert TW, Jahrling PB. Towards a vaccine against Ebola virus. Expert Rev Vaccines. 2003;2:777–789. doi: 10.1586/14760584.2.6.777. [DOI] [PubMed] [Google Scholar]
- 35.Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V, Johnson JC, de Jong S, Tavakoli I, Judge A, Hensley LE, Maclachlan I. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hensley LE, Jones SM, Feldmann H, Jahrling PB, Geisbert TW. Ebola and Marburg viruses: pathogenesis and development of countermeasures. Curr Mol Med. 2005;5:761–772. doi: 10.2174/156652405774962344. [DOI] [PubMed] [Google Scholar]
- 37.Hartman AL, Towner JS, Nichol ST. Ebola and marburg hemorrhagic fever. Clin Lab Med. 2010;30:161–177. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Fausther-Bovendo H, Mulangu S, Sullivan NJ. Ebolavirus vaccines for humans and apes. Curr Opin Virol. 2012;2:324–329. doi: 10.1016/j.coviro.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warfield KL, Aman MJ. Advances in virus-like particle vaccines for filoviruses. J Infect Dis 204 Suppl. 2011;3:S1053–S1059. doi: 10.1093/infdis/jir346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geisbert TW, Hensley LE, Kagan E, Yu EZ, Geisbert JB, Daddario-DiCaprio K, Fritz EA, Jahrling PB, McClintock K, Phelps JR, Lee AC, Judge A, Jeffs LB, MacLachlan I. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J Infect Dis. 2006;193:1650–1657. doi: 10.1086/504267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groseth A, Hoenen T, Alimonti JB, Zielecki F, Ebihara H, Theriault S, Stroher U, Becker S, Feldmann H. In vitro evaluation of antisense RNA efficacy against filovirus infection, by use of reverse genetics. J Infect Dis 196 Suppl. 2007;2:S382–389. doi: 10.1086/520604. [DOI] [PubMed] [Google Scholar]
- 42.Warfield KL, Swenson DL, Olinger GG, Nichols DK, Pratt WD, Blouch R, Stein DA, Aman MJ, Iversen PL, Bavari S. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2006;2:e1. doi: 10.1371/journal.ppat.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang KN, Lee YS. RNA Aptamers: A Review of Recent Trends and Applications. Adv Biochem Eng Biotechnol. 2012;131:153–169. doi: 10.1007/10_2012_136. [DOI] [PubMed] [Google Scholar]
- 44.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Bunka DH, Platonova O, Stockley PG. Development of aptamer therapeutics. Curr Opin Pharmacol. 2010;10:557–562. doi: 10.1016/j.coph.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Soontornworajit B, Wang Y. Nucleic acid aptamers for clinical diagnosis: cell detection and molecular imaging. Analytical and bioanalytical chemistry. 2011;399:1591–1599. doi: 10.1007/s00216-010-4559-x. [DOI] [PubMed] [Google Scholar]
- 47.Wang T. Function and dynamics of aptamers: A case study on the malachite green aptamer. IOWA STATE UNIVERSITY; 2009. p. P156. [Google Scholar]
- 48.Binning JM, Leung DW, Amarasinghe GK. Aptamers in virology: recent advances and challenges. Front Microbiol. 2012;3:29. doi: 10.3389/fmicb.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 50.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 51.Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, Amarasinghe GK. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc Natl Acad Sci U S A. 2012;109:20661–20666. doi: 10.1073/pnas.1213559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscilation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.Collaborative Computational. Project N. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 58.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jhaveri SD, Ellington AD. In vitro selection of RNA aptamers to a protein target by filter immobilization. Curr Protoc Mol Biol Chapter. 2001;24:23. doi: 10.1002/0471142727.mb2403s52. Unit 24. [DOI] [PubMed] [Google Scholar]
- 60.Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- 61.Peters GA, Li S, Sen GC. Phosphorylation of specific serine residues in the PKR activation domain of PACT is essential for its ability to mediate apoptosis. J Biol Chem. 2006;281:35129–35136. doi: 10.1074/jbc.M607714200. [DOI] [PubMed] [Google Scholar]
- 62.Ramalingam D, Duclair S, Datta SA, Ellington A, Rein A, Prasad VR. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J Virol. 2011;85:305–314. doi: 10.1128/JVI.02626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. Journal of molecular biology. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 65.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 67.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.