The nuclear envelope is a subdomain of the endoplasmic reticulum (ER). Here, Oegema and colleagues characterize the conserved lipin-activating protein phosphatase CNEP-1, which is specifically enriched on the nuclear envelope. The data show that CNEP-1 spatially controls lipin-dependent phospholipid flux to limit phosphatidylinositol levels and restrict ER sheet formation in the vicinity of the nuclear envelope. This study reveals coupling between control of phospholipid synthesis and ER subdomain structure.
Keywords: lipin, endoplasmic reticulum, phospholipid synthesis, phosphatidylinositol
Abstract
The nuclear envelope is a subdomain of the endoplasmic reticulum (ER). Here we characterize CNEP-1 (CTD [C-terminal domain] nuclear envelope phosphatase-1), a nuclear envelope-enriched activator of the ER-associated phosphatidic acid phosphatase lipin that promotes synthesis of major membrane phospholipids over phosphatidylinositol (PI). CNEP-1 inhibition led to ectopic ER sheets in the vicinity of the nucleus that encased the nuclear envelope and interfered with nuclear envelope breakdown (NEBD) during cell division. Reducing PI synthesis suppressed these phenotypes, indicating that CNEP-1 spatially regulates phospholipid flux, biasing it away from PI production in the vicinity of the nuclear envelope to prevent excess ER sheet formation and NEBD defects.
The endoplasmic reticulum (ER) is composed of sheets and tubules bounded by a membrane bilayer that faces the cytoplasm on one side and an internal lumen on the other. The ER is partitioned into subdomains specialized for different functions. The nuclear envelope subdomain is a spherical sheet perforated by nuclear pores that encases the chromatin. The outer membrane and lumen of the nuclear envelope are directly contiguous with other ER structural elements, whereas passage of proteins to the inner nuclear membrane (INM) is gated by the nuclear pores (Hetzer 2010; English and Voeltz 2013). The chromatin-facing INM is associated with the nuclear lamina, a dense filament meshwork that provides structural support (Hetzer 2010; Gerace and Huber 2012).
The ER also serves as a platform for de novo phospholipid synthesis (Fagone and Jackowski 2009; Lagace and Ridgway 2013). A central player is the conserved phosphatase lipin, which converts phosphatidic acid (PA) to diacylglycerol (DAG) (Carman and Han 2006; Siniossoglou 2013). In metazoans, lipin is at a branching point in the phospholipid synthesis pathway; the major building blocks of membrane bilayers (phosphatidylcholine [PC] and phosphatidylethanolamine [PE]) are synthesized from the lipin product DAG, whereas phosphatidylinositol (PI) is synthesized from the lipin substrate PA (Carman and Han 2006; Lagace and Ridgway 2013; Siniossoglou 2013). PI is converted to phosphoinositides (PIPs) when transported to other organelles that house specific PIP kinases and phosphatases (Balla 2013). Within the ER, PC and PE make up >70% of total phospholipid, whereas PI makes up <10% (van Meer et al. 2008). In metazoans, decreased lipin activity is predicted to shift phospholipid flux away from production of DAG and the major membrane phospholipids PC and PE toward the production of PI. This is in contrast to budding yeast, where a pathway that is not present in metazoans (the CDP–DAG pathway) (Supplemental Fig. S1A) converts PA to major membrane phospholipids (Carman and Han 2011; Siniossoglou 2013). Thus, lipin inhibition in budding yeast leads to an increase in all phospholipids and an abnormal expansion of the nuclear envelope that does not occur in metazoans (Han et al. 2006; Siniossoglou 2013).
Understanding lipin in metazoans is motivated by the fact that it is mutated in a number of human metabolic diseases and in a mouse model for lipodystrophy (Reue 2009). Lipin is recruited to the ER through a peripheral membrane-binding domain; however, soluble lipin also exists that transcriptionally coactivates genes involved in lipid metabolism (Siniossoglou 2013). Lipin activity and localization are regulated via multisite phosphorylation by Cdk1 and mTORC1 (Carman and Han 2011; Siniossoglou 2013), which negatively regulate lipin activity (Eaton et al. 2013). The conserved C-terminal domain (CTD) phosphatase CTDNEP1 (formerly Dullard) is an integral membrane protein enriched on the nuclear envelope that is predicted to activate lipin by dephosphorylating it (Siniossoglou et al. 1998; Kim et al. 2007; Han et al. 2012; Eaton et al. 2013).
Lipin inhibition in HeLa cells and Caenorhabditis elegans embryos slows nuclear envelope disassembly during mitosis (Golden et al. 2009; Gorjánácz and Mattaj 2009; Mall et al. 2012). This has been proposed to result from a reduction in DAG levels, which in turn could decrease the activity of DAG-dependent protein kinase Cs that phosphorylate lamins to promote their disassembly (Mall et al. 2012). Here, we investigate the role of lipin in nuclear envelope disassembly in the C. elegans embryo by characterizing the effects of inhibiting its nuclear envelope-localized regulator, CTDNEP1 (CNEP-1 [CTD nuclear envelope phosphatase-1] in C. elegans). CNEP-1 inhibition led to ectopic ER sheets that encased the nuclear envelope in an extra ER layer and interfered with nuclear envelope disassembly. CNEP-1 inhibition also led to elevated PI levels, and reducing PI synthesis suppressed ectopic ER sheet formation and nuclear envelope disassembly defects in the cnep-1 mutant. We conclude that CNEP-1 spatially controls lipin-dependent phospholipid flux to limit PI levels and restrict ER sheet formation in the vicinity of the nuclear envelope.
Results and Discussion
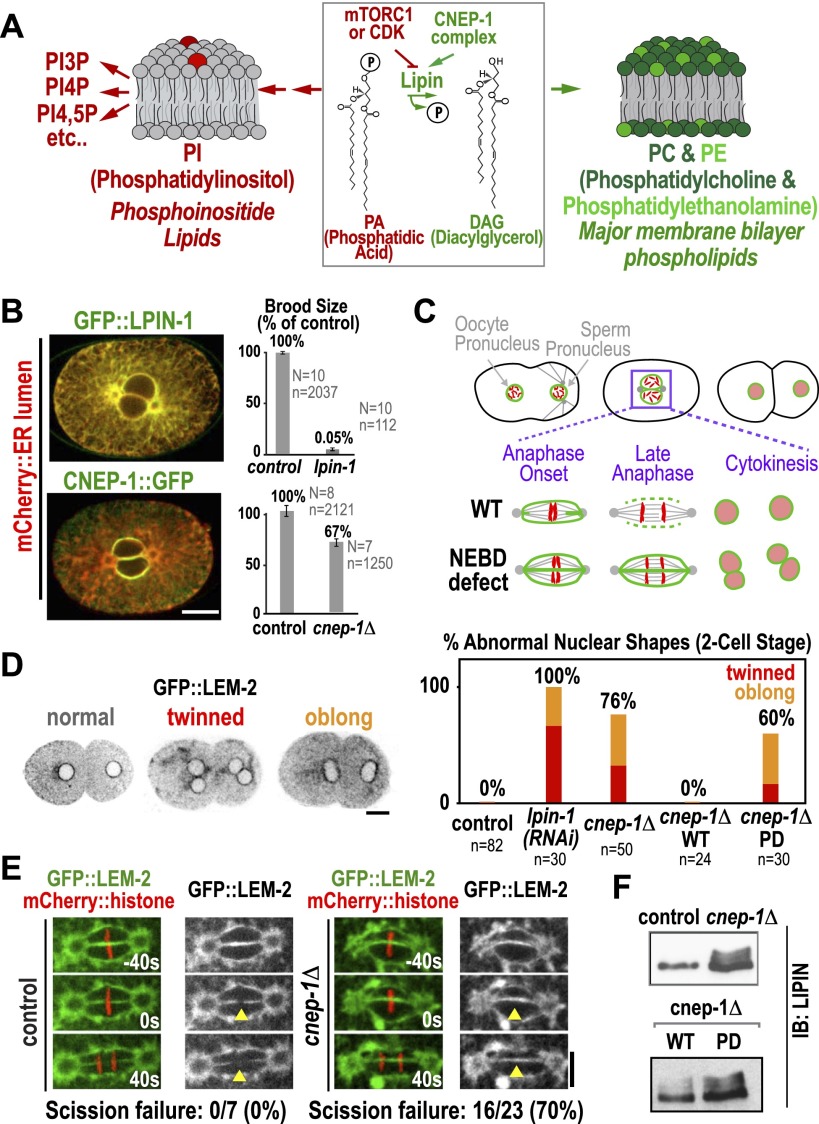
Inhibition of the C. elegans homolog of the lipin activator CTDNEP1, called CNEP-1, slows nuclear envelope disassembly during the first division of the C. elegans embryo (Han et al. 2012), a phenotype also observed following partial inhibition of lipin (LPIN-1 in C. elegans) (Golden et al. 2009; Gorjánácz and Mattaj 2009; Han et al. 2012). To investigate how CNEP-1 promotes nuclear envelope disassembly, we generated transgenes expressing GFP fusions with lipin and wild-type or phosphatase-defective (PD) CNEP-1 (CNEP-1WT or CNEP-1PD) (Fig. 1B; Supplemental Fig. S1B). In contrast to GFP∷LPIN-1, which localized throughout the ER (Fig. 1B; Gorjánácz and Mattaj 2009), CNEP-1∷GFP was specifically enriched on the nuclear envelope (Fig. 1B). Consistent with its central role in phospholipid synthesis (Fig. 1A), injection of dsRNA targeting lipin into adult worms led to the abrupt cessation of embryo production (Fig. 1B). In contrast, deletion of the cnep-1 gene (cnep-1Δ) (Supplemental Fig. S1C) led to only a modest reduction in brood size (Fig. 1B). The localization of CNEP-1 to the nuclear envelope, combined with the reduced severity of the cnep-1 phenotype, suggested that CNEP-1 activates a pool of lipin at the nuclear envelope to serve a specific function.
Figure 1.
The lipin activator CNEP-1 concentrates on the nuclear envelope and is required for timely nuclear envelope disassembly during mitosis. (A) Schematic showing lipin at the branching point between the synthesis of major membrane phospholipids and PI. (B) Fluorescence confocal images of embryos expressing the ER lumen marker mCherry∷SP12 along with GFP∷LPIN-1 (top) (n > 10 embryos) or mCherry∷SP12 and CNEP-1∷GFP in a background deleted for the endogenous cnep-1 gene (bottom) (n > 10 embryos). Graphs plot normalized brood size. Error bars are SEM. (N) Number of worms; (n) total number of embryos. (C) Schematics highlight scission of the maternal and paternal pronuclear envelopes (green) during chromosome (red) segregation in the first division of control C. elegans embryos and failure of scission in embryos exhibiting a nuclear envelope breakdown (NEBD) defect. (D, left) Inverted contrast fluorescence confocal images of two-cell stage embryos expressing GFP∷LEM-2 illustrate nuclear morphology phenotypes. (Right) Graph plotting frequency of abnormal nuclear phenotypes for the indicated conditions. (E) Fluorescence confocal images of control and cnep-1Δ embryos expressing GFP∷LEM-2 and mCherry∷histone. Times are in seconds relative to anaphase onset. Yellow arrowheads mark the site of nuclear envelope scission in wild type. (F) Immunoblots of lysates from control and cnep-1Δ worms (top) or cnep-1Δ worms harboring the wild-type (WT) or PD transgenes (bottom) probed for LPIN-1. Bars, 10 μm.
To characterize the effects of the cnep-1 deletion on nuclear envelope dynamics, we filmed embryos expressing a GFP fusion with the INM protein LEM-2 (Galy et al. 2003). After fertilization, the maternal and paternal pronuclei form, increase in size, and migrate toward each other, meeting in the middle as the embryo enters mitosis (Fig. 1C). The nuclear envelopes become permeable, and microtubules penetrate to interact with the chromosomes. The nuclear envelopes continue to disassemble as the embryo progresses into anaphase, and the scission of maternal and paternal nuclear envelopes allows chromosome mixing approximately coincident with anaphase onset (Fig. 1C,E). When the nuclear envelopes reform in telophase, a single nucleus forms in each daughter cell (Fig. 1C,D). Perturbations that slow nuclear envelope disassembly result in a failure of scission (Audhya et al. 2007). In this case, two nuclei form in each daughter cell—one containing the maternal chromosomes, and one containing the paternal chromosomes (“twinned” nuclei) (Fig. 1C,D). Delayed scission can also result in a less severe phenotype characterized by misshapen, “oblong” nuclei (Fig. 1D).
In control embryos, 100% of nuclei at the two-cell stage exhibit a normal spherical morphology. In contrast, 76% of two-cell stage nuclei in cnep-1Δ embryos and 100% following partial LPIN-1 depletion were twinned or oblong, indicative of delayed nuclear envelope scission. The two-cell stage nuclear morphology defect in cnep-1Δ embryos was rescued by the wild-type, but not the PD, cnep-1 transgene (Fig. 1D); both transgenic proteins were expressed at similar levels (Supplemental Fig. S1B) and localized to the nuclear envelope (Fig. 1B; Supplemental Fig. S1D). Thus, the phosphatase activity of CNEP-1 is essential for its role in nuclear envelope disassembly. Consistent with the idea that CNEP-1 functions by dephosphorylating LPIN-1, levels of slower-migrating phosphorylated LPIN-1 species were increased in cnep-1Δ worms and restored to control levels by the wild-type, but not the PD, cnep-1 transgene (Fig. 1F). In contrast to the effect of the cnep-1 deletion on nuclear envelope scission, no defects were observed in nuclear envelope expansion following fertilization or in nuclear envelope permeabilization during mitotic entry (Supplemental Fig. S1E,F).
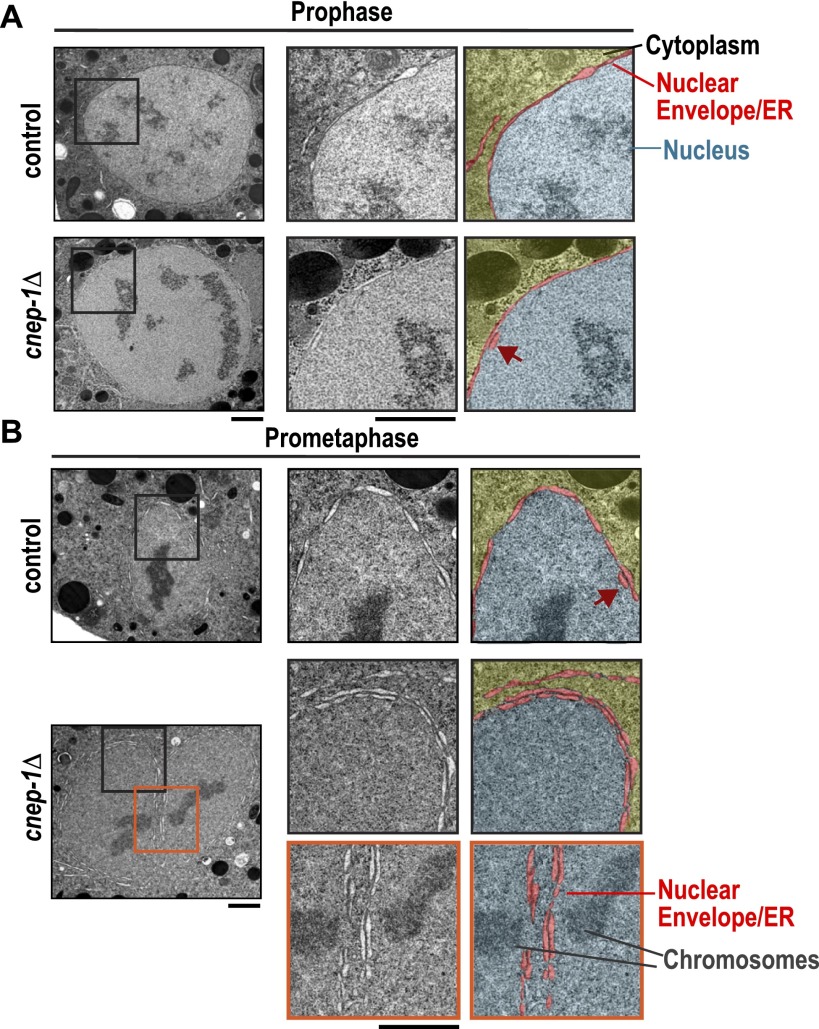
To understand how the defect in nuclear envelope disassembly arises in cnep-1Δ mutants, we used transmission electron microscopy (TEM) after high-pressure freezing and freeze substitution to examine the nuclear envelope at different stages of mitosis. In control embryos, prophase nuclei (judged by chromosome condensation state) were encased in a single double-membrane sheet (Fig. 2A). This was also the case for prometaphase–metaphase nuclei, although short isolated regions were observed where two stacked lumens could be detected along the nuclear envelope periphery (Fig. 2B, dark-red arrow). In cnep-1Δ embryos, short regions with two stacked lumens were observed in prophase nuclei (Fig. 2A), and prometaphase–metaphase nuclei appeared encased in a second ER layer; two parallel lumens could be traced along the majority of the nuclear periphery (Fig. 2B; Supplemental Fig. S2). This result suggests that the failure of nuclear envelope scission in cnep-1Δ embryos (Fig. 1E) is due to wrapping of the nuclear envelope in an extra ER layer, which places eight membrane bilayers and four lumens between the maternal and paternal genomes (Fig. 2B, bottom inset).
Figure 2.
Mitotic nuclei in cnep-1Δ mutants are wrapped in an additional ER layer. (A,B) Transmission electron micrographs of the nuclear regions of prophase (top) and prometaphase/metaphase (bottom) high-pressure-frozen embryos (eight to 16 cells) at the indicated mitotic stages (n = 13 mitotic cells for control; n = 11 mitotic cells for cnep-1Δ). Magnified images of boxed regions are paired with micrographs pseudocolored to highlight the nuclear envelope/ER lumens (red) that separate the cytoplasm (yellow) and nucleus (blue). Cinched-in regions along the nuclear envelope in A are where nuclear pore complexes are inserted. Arrows (dark red) point to isolated regions where two adjacent lumens are detected. One-hundred percent of cnep-1Δ nuclear envelopes during late stages of NEBD (n = 7), as compared with none in control (n = 6), displayed an additional double-membrane sheet encircling the nuclear envelope. Bars, 1 μm.
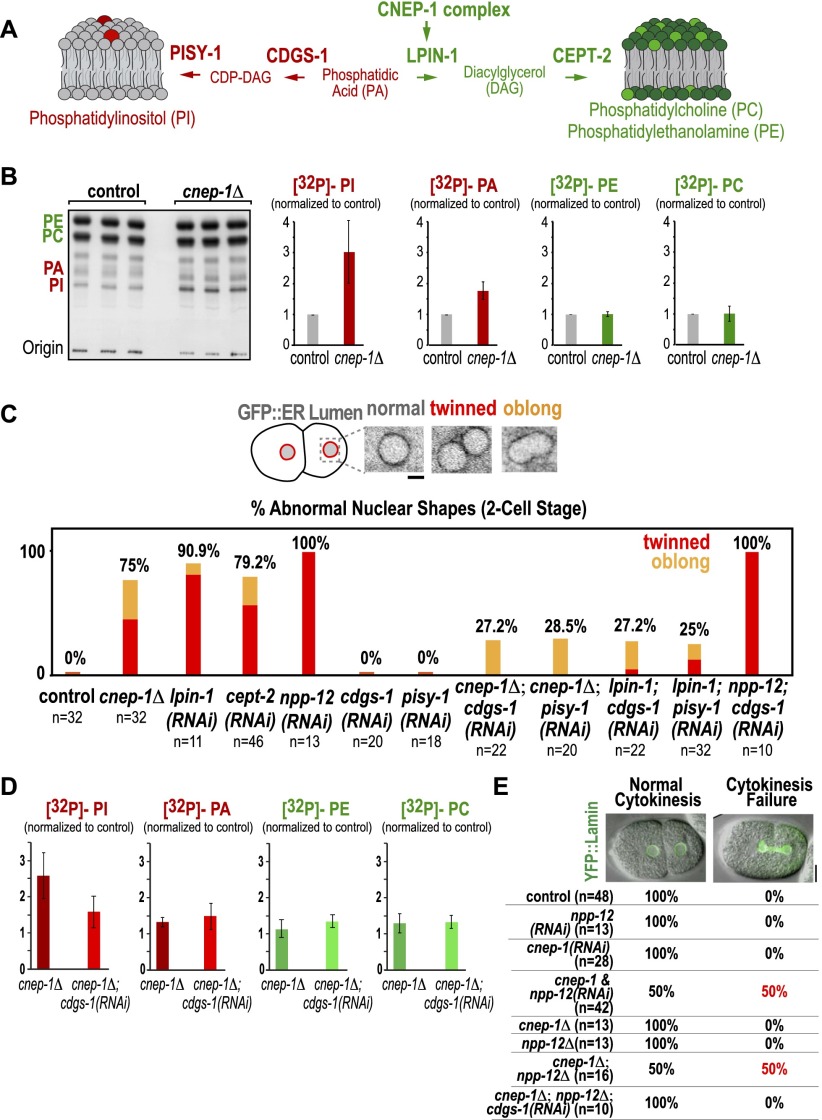
Wrapping of the nuclear envelope in an extra ER layer in the absence of CNEP-1 is likely due to reduced activation of lipin in the vicinity of the nuclear envelope. A reduction in lipin activity could exert such an effect by either reducing levels of its product, DAG, which in turn may reduce levels of the major membrane phospholipids PE and PC, or increasing levels of its substrate, phosphatidic acid (PA), which in turn may elevate production of PI (Fig. 3A). To distinguish between these possibilities, we directly measured phospholipid content by metabolically labeling control and cnep-1Δ worms and analyzing isolating lipids by thin-layer chromatography. This analysis revealed that PI and PA levels were increased in cnep-1Δ worms, with PI levels being elevated ∼2.5-fold compared with controls; in contrast, PE and PC levels were not significantly altered (Fig. 3B).
Figure 3.
The nuclear envelope disassembly defect in the cnep-1Δ mutant results from increased PI synthesis. (A) Schematic highlighting the C. elegans enzymes required to synthesize PI from the lipin substrate PA (CDGS-1 and PISY-1; red) and to synthesize PC and PE from the lipin product DAG (CEPT-2; green). (B) Autoradiograph of a chromatography plate after separation of isolated 32P-labeled phospholipids. Graphs plotting the mean band intensity for each phospholipid species from three independent replicates normalized against the mean value in control samples run in parallel. Error bars are the SD. (C, top) Inverted contrast confocal images of representative nuclear phenotypes in two-cell stage embryos expressing the ER lumen marker GFP∷SP-12. (Bottom) Plots quantifying abnormal nuclear phenotypes at the two-cell stage for the indicated conditions. Bar, 2 µm. (D) Graphs plotting the mean band intensity for each phospholipid species from three independent replicates normalized against the mean value in control samples run in parallel. Error bars are the SD. (E, top) Overlay of differential interference contrast and YFP∷lamin fluorescence confocal images for a control embryo (normal cytokinesis) and a cnep-1 and npp-12(RNAi) embryo failing cytokinesis (cytokinesis failure). (Bottom) Table showing the percentage of embryos exhibiting cytokinesis success or failure.
To assess the relationship of the alteration in lipid levels to the cnep-1Δ phenotype, we analyzed the consequences of depleting other enzymes in the phospholipid biosynthetic pathway (Fig. 3A) on nuclear envelope disassembly. Partial depletion of the germline choline/ethanolamine transferase CEPT-2 (choline/ethanolamine phosphotransferase-2), which acts downstream from lipin to convert DAG into PE/PC, phenocopied cnep-1 inhibition (Fig. 3C). Like inhibition of CNEP-1 and lipin, CEPT-2 inhibition is expected to bias biosynthetic flux toward PI. To test the prediction that elevated PI levels are the origin of the nuclear disassembly defect, we individually depleted the two enzymes required to convert PA to PI: CDGS-1 (CDP-DAG synthase-1), which converts PA to CDP-DAG, and PISY-1 (PI synthase-1), which converts CDP-DAG to PI. Neither depletion affected two-cell stage nuclear morphology (Fig. 3C). However, depletion of either PISY-1 or CDGS-1 significantly rescued the nuclear morphology defects resulting from inhibition of cnep-1 or partial inhibition of lpin-1 (Fig. 3C; Supplemental Movie S1). In contrast, neither depletion rescued the nuclear envelope disassembly defects observed following depletion of the transmembrane nucleoporin NPP-12 (the C. elegans gp210 homolog) (Fig. 3C; Audhya et al. 2007; Galy et al. 2008), indicating that the suppression is specific for the lipin pathway. Biochemical analysis revealed that cdgs-1 inhibition reduces the elevation of PI, but not of PA, in cnep-1Δ worms (Fig. 3D), consistent with the fact that CDGS-1 is between PA and PI in the lipid biosynthetic pathway (Fig. 3A). The suppression of the cnep-1 and lpin-1 phenotypes by inhibition of the enzymes that convert PA to PI, together with the direct biochemical analysis of lipid content, strongly suggest that the defect in nuclear envelope disassembly following CNEP-1 inhibition is caused by an increase in PI levels.
These findings argue that restricting PI synthesis in the vicinity of the nuclear envelope is important to prevent defects in nuclear envelope disassembly. The nuclear envelope disassembly defect resulting from cnep-1 and lpin-1 inhibition also appears to be distinct from that resulting from depletion of NPP-12, a transmembrane nucleoporin that controls lamin disassembly and INM protein dispersal (Audhya et al. 2007; Galy et al. 2008). Consistent with the idea that inhibition of CNEP-1 and NPP-12 disrupts nuclear envelope disassembly via distinct means, CNEP-1 inhibition slowed lamin disassembly and LEM-2 dispersal but to a reduced extent compared with NPP-12 depletion (Supplemental Fig. S3). In addition, simultaneous inhibition of CNEP-1 along with NPP-12 led to a greatly enhanced defect in nuclear envelope disassembly. In embryos in which CNEP-1 or NPP-12 alone was depleted by RNAi or in embryos homozygous for deletions of the cnep-1 or npp-12 genes, twinned nuclei were often observed at the two-cell stage, but cytokinesis was always successful. In contrast, simultaneous inhibition of cnep-1 and npp-12 led to a severe defect in which cytokinesis failed in ∼50% of cases due the inability of the cytokinetic furrow to cut through the remnants of the nuclear envelope (Fig. 3E; Supplemental Movie S2). Like the defects in nuclear envelope disassembly in the cnep-1Δ mutant, the cytokinesis defect in the double-deletion mutant was rescued by CDGS-1 depletion (Fig. 3E). We conclude that increased PI synthesis caused by CNEP-1 inhibition leads to wrapping of nuclei in an extra ER layer that slows nuclear envelope disassembly.
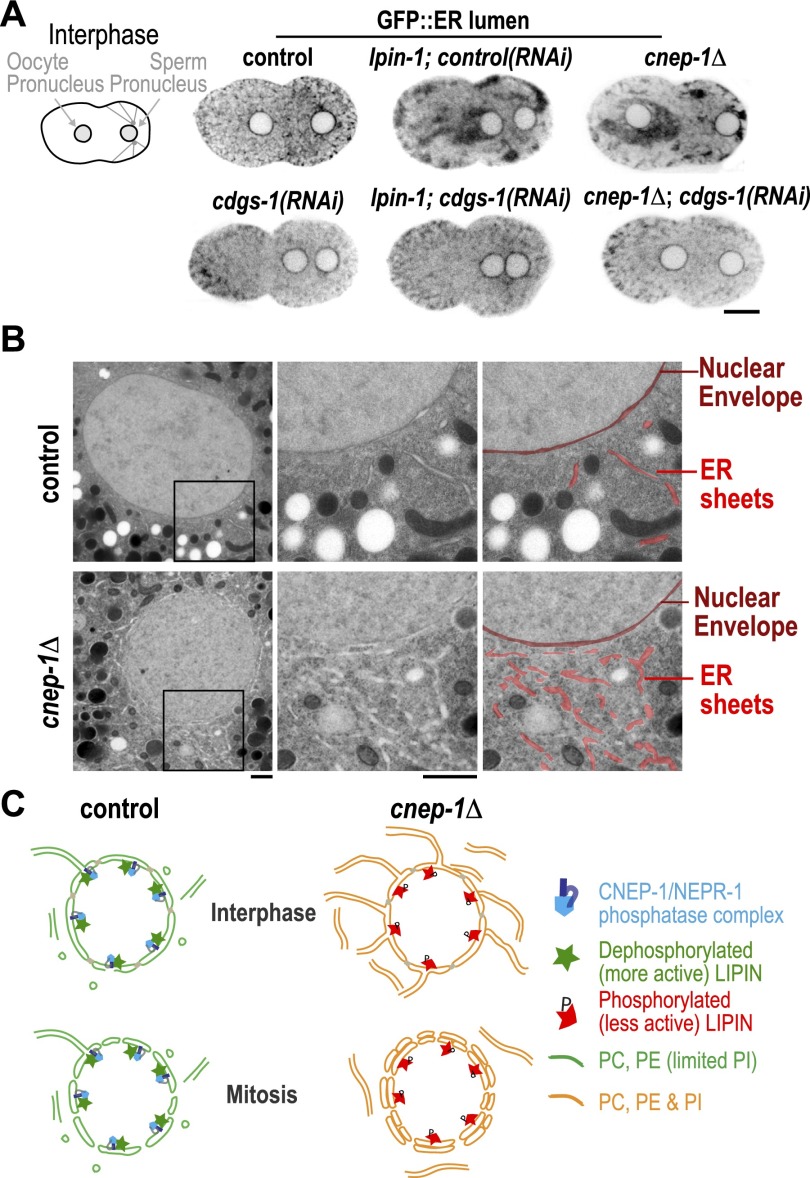
To identify the source of the extra ER layer that encases the nuclear envelope in cnep-1Δ embryos, we analyzed ER morphology in embryos expressing a GFP-tagged ER lumen marker (GFP∷SP12). The ER is normally relatively evenly distributed in interphase embryos and then condenses to form patches as embryos enter mitosis (Poteryaev et al. 2005; Audhya et al. 2007). ER morphology was noticeably altered in cnep-1Δ embryos with prominent ER patches apparent prior to the onset of mitosis (Fig. 4A); similar defects were seen following partial inhibition of lpin-1 or cept-2 (Fig. 4A; Supplemental Fig. S4A). Similar to our analysis of nuclear morphology, depletion of either of the enzymes required to convert PA to PI (PISY-1 or CDGS-1) did not lead to any detectable alteration in ER morphology on its own (Fig. 4A, Supplemental Fig. S4A); however, both inhibitions rescued the ER morphology defect resulting from deletion of cnep-1 or partial inhibition of lpin-1 (Fig. 4A; Supplemental Fig. S4; Supplemental Movie S3). In our fluorescence images, ER patches were often enriched near pronuclei (Fig. 4A; Supplemental Fig. S4A). Consistent with the idea that the ER patches detected at the light level are composed of ER sheets, examination of nuclei in cnep-1Δ embryos by TEM revealed a substantial increase in the number of ER sheets near the nuclear envelope (Fig. 4B). Unlike control embryos, where very few streaks of reduced electron density (consistent with an ER sheet in cross-section) were detected, many such sheets were present near nuclei in cnep-1Δ embryos. We therefore propose that the extra ER layer that encases the nucleus following permeabilization in cnep-1Δ embryos is composed of perforated ER sheets.
Figure 4.
CNEP-1 suppresses ER sheet formation in the vicinity of the nuclear envelope. (A) Inverted contrast fluorescence confocal images of interphase embryos expressing the ER lumen marker GFP∷SP12. Bar, 10 μm. (B) Transmission electron micrographs of ultrathin cross-sections of high-pressure-frozen control and cnep-1Δ embryos. Magnified images of the boxed regions are paired with micrographs pseudocolored to highlight the location of the nuclear envelope (dark red) and ER sheets (red). Bars, 900 nm. (C) Model for spatial regulation of phospholipid synthesis by CNEP-1. In control embryos, the CNEP-1 complex activates lipin to limit the amount of PI on nuclear membranes and ER membranes in the vicinity of the nucleus (green nuclear envelope/ER membranes contain PC and PE with limited PI). In cnep-1Δ embryos, lipin is less active at the nuclear envelope, and PI levels are higher (orange nuclear envelope/ER membranes contain PC, PE, and PI). Increased PI levels lead to ectopic ER sheets in the vicinity of the nucleus that encase the nuclear envelope following mitotic permeabilization and inhibit its subsequent disassembly.
Wrapping of the nuclear envelope in an extra ER layer in cnep-1Δ embryos suggests that CNEP-1 prevents ER sheets from stacking onto the nuclear envelope by ensuring that the PI concentration on the nuclear envelope and ER membranes in the vicinity of the nucleus remains low (model in Fig. 4C). As the nuclear envelope and ER are a single contiguous membrane system, we expect that phospholipids diffuse between the ER and nuclear envelope while at the same time being acted on by localized biosynthetic enzymes that bias their distribution. In the absence of CNEP-1, higher PI levels would cause the ER near the nuclear envelope to form an extensive network of sheets that encases the permeabilized nuclear envelope, slowing its disassembly (Fig. 4C). The fact that the second ER layer in cnep-1Δ embryos only encases the nuclear envelope following its permeabilization suggests the existence of a mechanism that prevents ER sheets from stacking onto the nuclear envelope during interphase. This mechanism may be inactivated during mitosis when the nuclear envelope has been shown to invaginate and thus transiently form stacked membrane segments to aid in nuclear envelope disassembly (Calarco et al. 1972; Galy et al. 2008). It is also interesting that the nuclear envelope is uniformly encased in a single additional ER layer, suggesting that whereas stacking of ER sheets onto the nuclear envelope is permitted, stacking of ER sheets onto each other is not.
The mechanism that we propose to explain how lipin/CNEP-1 inhibition delays nuclear envelope disassembly is significantly different from the previously proposed mechanism, based on work in mammalian cells, in which lipin inhibition was suggested to decrease DAG-induced activation of protein kinase Cs, thereby slowing lamin phosphorylation and disassembly (Mall et al. 2012). Future work will be needed to determine whether this discrepancy is due to a difference between systems or whether a mechanism similar to the one that we propose also underlies the nuclear envelope disassembly defect in mammalian cells.
The idea that PI synthesis is spatially regulated to control ER structure is consistent with recent work showing that phospholipid synthesis enzymes, including phosphatidylinositol synthase, are enriched in a Rab10-dependent ER subdomain and that this compartmentalization is important to prevent the excessive formation of ER sheets (Kim et al. 2011; English and Voeltz 2012). How PI levels control the extent of ER sheet formation and whether the relevant lipid is PI itself or another PI-derived PIP are important future questions. Asymmetric distribution of lipids with curvature-favoring structures between the two membrane monolayers in a bilayer can generate membrane curvature. However, high curvature is thought to require levels of compositional asymmetry that are unlikely in vivo (Shibata et al. 2009). This, combined with recent work characterizing membrane-associated proteins that shape ER tubules and sheets, has led to the conclusion that proteins are the main players in controlling ER structure (Shibata et al. 2009; Goyal and Blackstone 2013). It is therefore attractive to think that higher PI concentrations could promote sheet formation by modulating the function of ER-shaping proteins; testing this hypothesis will be an important goal of future work.
Materials and methods
C. elegans strains and RNA-mediated interference
C. elegans strains and dsRNAs are listed in the Supplemental Material. L4 or young adult hermaphrodites were injected with dsRNA and incubated for 12–48 h at 20°C before dissection to obtain embryos for filming (for details, see the Supplemental Material).
Immunoblotting
Affinity-purified antibodies against amino acids 25–161 of LPIN-1 were generated using standard procedures. Antibody signals were detected using an HRP-conjugated secondary antibody (GE Healthcare) and the WesternBright Sirius detection system (Advansta).
Light microscopy
Embryos were imaged at 20°C with a 60× 1.4 NA PlanApochromat lens on an Andor Revolution XD Confocal System (Andor Technology) mounted on an inverted microscope (TE2000-E, Nikon) equipped with a confocal scanner unit (CSU-10, Yokogawa), solid-state 100-mW lasers, and a high-resolution interline CCD camera (Clara or iXon; Andor Technology). Images of the ER lumen marker were acquired using an inverted Zeiss Axio Observer Z1 system with a Yokogawa spinning-disk confocal head (CSU-X1), a 63× 1.4 NA Plan Apochromat objective, and a QuantEM:512SC EMCCD camera (Photometrics).
Phospholipid analysis
Worms were labeled overnight with 1 mCi/mL phosphorus-32 orthophosphate (PerkinElmer), and lipids were extracted following standard protocols (Bligh and Dyer 1959). Total phospholipids were spotted in triplicate, separated by thin-layer chromatography (60:35:10 solution of chloroform:methanol:20% methylamine), and detected by exposing onto film (for details, see the Supplemental Material).
High-pressure freezing and electron microscopy
Whole animals were high-pressure-frozen and freeze-substituted. Samples were embedded, and images of post-stained ultrathin sections were taken using an FEI Tecnai Spirit TEM at 80 kV (Fig. 2) or an FEI Tecnai 12 TEM operated at 100 kV (Fig. 4). For details, see the Supplemental Material.
Acknowledgments
We thank the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources (NCRR), and Shohei Mitani (Tokyo Women's Medical University, Japan) for strains; Dr. Marilyn Farquhar for the use of the Electron Microscopy Core Facility; Ying Jones for the semi- and ultrathin sectioning of samples; and Gregory S. Taylor for assistance with lipid analysis. S.B. was supported by fellowships from the A.P. Giannini Foundation and The Hartwell Foundation. K.O. and A.D. received salaries and additional support from the Ludwig Institute for Cancer Research. This work was funded by grants from the Deutsche Forschungsgemeinschaft (MU 1423/3-2 and MU 1423/4-1) and Human Frontier Science Program (RGP 0034/2010) to T.M.-R. and from the NIH to J.E.D (DK18024 and DK18849) and A.A. (GM088151).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.230599.113.
References
- Audhya A, Desai A, Oegema K 2007. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol 178: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T 2013. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev 93: 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Calarco PG, Donahue RP, Szollosi D 1972. Germinal vesicle breakdown in the mouse oocyte. J Cell Sci 10: 369–385 [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS 2006. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci 31: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS 2011. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu Rev Biochem 80: 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JM, Mullins GR, Brindley DN, Harris TE 2013. Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association. J Biol Chem 288: 9933–9945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Voeltz GK 2012. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol 14: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Voeltz GK 2013. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol 5: a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P, Jackowski S 2009. Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res 50: S311–S316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P 2003. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell 14: 5104–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I 2008. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci 121: 317–328 [DOI] [PubMed] [Google Scholar]
- Gerace L, Huber MD 2012. Nuclear lamina at the crossroads of the cytoplasm and nucleus. J Struct Biol 177: 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O 2009. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci 122: 1970–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjánácz M, Mattaj IW 2009. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci 122: 1963–1969 [DOI] [PubMed] [Google Scholar]
- Goyal U, Blackstone C 2013. Untangling the web: Mechanisms underlying ER network formation. Biochim Biophys Acta 1833: 2492–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 281: 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM 2012. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J Biol Chem 287: 3123–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW 2010. The nuclear envelope. Cold Spring Harb Perspect Biol 2: a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Dixon JE 2007. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci 104: 6596–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Balla T 2011. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell 21: 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Ridgway ND 2013. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim Biophys Acta 1833: 2499–2510 [DOI] [PubMed] [Google Scholar]
- Mall M, Walter T, Gorjánácz M, Davidson IF, Nga Ly-Hartig TB, Ellenberg J, Mattaj IW 2012. Mitotic lamin disassembly is triggered by lipid-mediated signaling. J Cell Biol 198: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A 2005. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell 16: 2139–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue K 2009. The lipin family: Mutations and metabolism. Curr Opin Lipidol 20: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, Rapoport TA 2009. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 25: 329–354 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S 2013. Phospholipid metabolism and nuclear function: Roles of the lipin family of phosphatidic acid phosphatases. Biochim Biophys Acta 1831: 575–581 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J 17: 6449–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW 2008. Membrane lipids: Where they are and how they behave. Nature Publishing Group 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]