The mitotic spindle checkpoint ensures accurate chromosome segregation and genomic integrity. Understanding the regulation of checkpoint protein Mad1 recruitment to the kinetochore has been an outstanding question in the field. Here, London and Biggins show that Bub1 is a receptor for Mad1, and the Bub1–Mad1 interaction at kinetochores is driven by Mps1-mediated phosphorylation of Bub1. This work reveals a long-sought mechanism that determines kinetochore activation of the spindle checkpoint.
Keywords: kinetochore, spindle checkpoint, Mps1 kinase, Mad1, Bub1 kinase
Abstract
The spindle checkpoint is a conserved signaling pathway that ensures genomic integrity by preventing cell division when chromosomes are not correctly attached to the spindle. Checkpoint activation depends on the hierarchical recruitment of checkpoint proteins to generate a catalytic platform at the kinetochore. Although Mad1 kinetochore localization is the key regulatory downstream event in this cascade, its receptor and mechanism of recruitment have not been conclusively identified. Here, we demonstrate that Mad1 kinetochore association in budding yeast is mediated by phosphorylation of a region within the Bub1 checkpoint protein by the conserved protein kinase Mps1. Tethering this region of Bub1 to kinetochores bypasses the checkpoint requirement for Mps1-mediated kinetochore recruitment of upstream checkpoint proteins. The Mad1 interaction with Bub1 and kinetochores can be reconstituted in the presence of Mps1 and Mad2. Together, this work reveals a critical mechanism that determines kinetochore activation of the spindle checkpoint.
Successful eukaryotic cell division requires accurate chromosome segregation, which relies on correct attachments of spindle microtubules to chromosomes. Defects in chromosome segregation result in aneuploidy, which is a hallmark of many cancers and birth defects. To ensure accurate segregation, a conserved signal transduction system called the spindle checkpoint prevents cell cycle progression until all chromosomes make correct attachments to spindle microtubules. The spindle checkpoint prevents the premature segregation of improperly attached chromosomes by inhibiting the activity of the anaphase-promoting complex (APC), a ubiquitin ligase that targets the anaphase inhibitor securin for destruction.
Spindle checkpoint function is mediated by the kinetochore, the macromolecular machine that assembles on the centromere of each chromosome and attaches to microtubules (Jia et al. 2013). The checkpoint and microtubule-binding functions of kinetochores are executed through a conserved set of subcomplexes termed KMN (Knl1/Spc105/Blinkin, Mis12/Mtw1, and Ndc80/Hec1) (Martin-Lluesma et al. 2002; McCleland et al. 2003; Cheeseman et al. 2006; Pagliuca et al. 2009). Checkpoint signaling further depends on kinetochore localization of the conserved checkpoint proteins Mps1, Bub1, Bub3, Mad1, and Mad2 as well as the Mad3 homolog BubRI in metazoans (Foley and Kapoor 2013; Jia et al. 2013). Mps1 is a protein kinase that interacts with Ndc80, and its kinase activity is required for checkpoint activity and kinetochore localization of all other checkpoint proteins in most organisms studied (Weiss and Winey 1996; Abrieu et al. 2001; Tighe et al. 2008; Kemmler et al. 2009; Heinrich et al. 2012; Nijenhuis et al. 2013; Zhu et al. 2013). Mps1 phosphorylation of Spc105 was recently shown to recruit Bub1 and Bub3 (the Bub1/3 complex) to the kinetochore (Kiyomitsu et al. 2007; London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012; Primorac et al. 2013), a requirement for the localization and activation of Mad1 and Mad2. However, Bub1/3 kinetochore localization is not sufficient for checkpoint activation without Mps1 activity, suggesting that Mps1 may have additional checkpoint targets (Rischitor et al. 2007; Ito et al. 2012; Yamagishi et al. 2012).
Mad1 and Mad2 form a complex (Mad1/2) that acts as a scaffold to activate soluble Mad2, which ultimately prevents cell cycle progression through APC inactivation (Luo et al. 2002; Sironi et al. 2002; Musacchio 2011). While Mad2 cycles rapidly on and off of the kinetochore, Mad1 is stably associated with kinetochores in the absence of microtubule attachments (Howell et al. 2004; Shah et al. 2004). Activated Mad2 signals the checkpoint downstream from kinetochores, and tethering Mad1 to kinetochores has been shown to constitutively stimulate checkpoint signaling in human cells (Maldonado and Kapoor 2011). Therefore, Mad1 kinetochore localization is the defining event in checkpoint activation, and elucidating the mechanism of this association is paramount in understanding checkpoint regulation. However, the direct binding partner and mechanism for the dynamic Mad1 kinetochore localization are unclear (Seeley et al. 1999; Brady and Hardwick 2000; Martin-Lluesma et al. 2002; McCleland et al. 2003; Gillett et al. 2004; Kiyomitsu et al. 2007; Yamamoto et al. 2008; Pagliuca et al. 2009; Ito et al. 2012; Kim et al. 2012). Here, we exploited our ability to isolate budding yeast kinetochores to demonstrate that Bub1 is a receptor for Mad1 and that the Bub1–Mad1 interaction at kinetochores is driven by Mps1-mediated phosphorylation of Bub1.
Results
Mad1 associates with purified kinetochores and requires Spc105
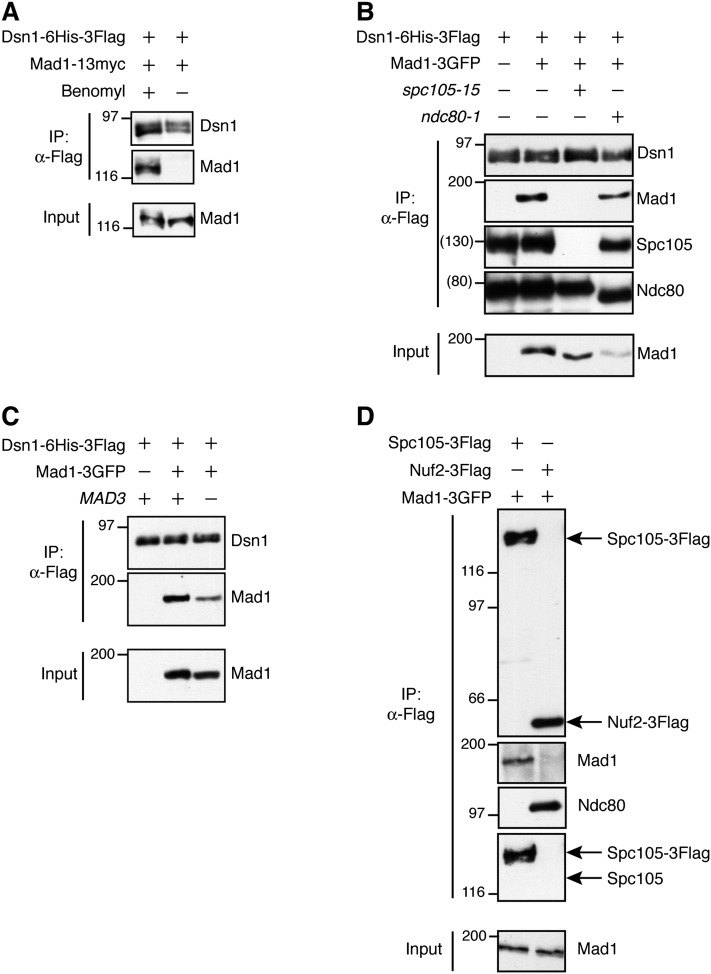
The outer kinetochore has been implicated in Mad1 recruitment, possibly through the conserved Ndc80/Hec1 subcomplex (Ndc80c) that is required for the spindle checkpoint (Martin-Lluesma et al. 2002; DeLuca et al. 2003; McCleland et al. 2003). To address which kinetochore components are required for Mad1 association, we first tested whether Mad1 associates with purified native kinetochore particles. Immunoprecipitation of the Mis12 subcomplex component Dsn1, in contrast to other kinetochore proteins, isolates native kinetochore particles containing approximately stoichiometric amounts of Spc105 and Ndc80 (Akiyoshi et al. 2010). Mad1 copurified with these kinetochore particles when cells were treated with the microtubule-destabilizing drug benomyl to activate the checkpoint (Fig. 1A), so we performed all subsequent copurification experiments using benomyl-treated cells.
Figure 1.
Mad1 associates with purified kinetochores and requires Spc105. (A) Mad1 associates with kinetochore particles when cells are treated with benomyl. A Dsn1-6His-3Flag Mad1-13myc strain (SBY8412) was grown with or without benomyl for 2 h before harvest. Kinetochore particles were purified by α-Flag immunoprecipitation and analyzed by immunoblotting with α-Flag and α-myc antibodies. (B) Spc105 is required for Mad1 kinetochore association. Kinetochore particles were purified by immunoprecipitation of Dsn1-6His-3Flag from cells shifted to 37°C before harvest. Strains contain Dsn1-6His-3Flag and either untagged Mad1 (SBY8253), Mad1-3GFP (SBY8415), Mad1-3GFP spc105-15 (SBY8829), or Mad1-3GFP ndc80-1 (SBY8579). Immunoblots were probed with α-Flag, α-GFP, α-Spc105, and α-Ndc80 antibodies. Note that Ndc80-1 is a truncation and still associates with purified kinetochores. (C) Mad1-3GFP enrichment with Dsn1-6His-3Flag does not require a fully functional spindle checkpoint. Kinetochore particles were purified from the following strains after treatment with benomyl: untagged Mad1 (SBY8253), Mad1-3GFP (SBY8415), or Mad1-3GFP mad3Δ (SBY11497). Immunoblots were probed with α-Flag or α-GFP antibodies. (D) Mad1 copurifies with Spc105c but not with Ndc80c. α-Flag immunoprecipitations of Spc105-3Flag (SBY8970) or Nuf2-3Flag (SBY8971) from benomyl-treated cells containing Mad1-3GFP. Immunoblots were probed with α-Flag, α-GFP, α-Ndc80, or α-Spc105 antibodies. Arrows indicate the migration of Spc105-3Flag or Nuf2-3Flag or the expected migration of untagged endogenous Spc105 (shown at the bottom). Migration of molecular weight markers (in kilodaltons) is shown to the left of each blot. Approximate estimated molecular weights are indicated in parentheses where molecular weight markers were not present within the cropped region.
We next tested whether functional Ndc80 or Spc105 subcomplexes of KMN are required for Mad1 association by purifying kinetochore particles from strains containing the temperature-sensitive alleles ndc80-1 and spc105-15 (Wigge and Kilmartin 2001; Nekrasov et al. 2003; Akiyoshi et al. 2010). ndc80-1 cells trigger the spindle checkpoint due to defective kinetochore–microtubule attachments at the restrictive temperature, while spc105-15 cells do not have a functional checkpoint (Wigge and Kilmartin 2001; Pagliuca et al. 2009). Consistent with this, the Mad1 association with the kinetochore was preserved in ndc80-1 cells but completely abolished in spc105-15 cells (Fig. 1B). We therefore tested whether a functional checkpoint is required for Mad1 kinetochore association. Mad3 is required for checkpoint function downstream from the kinetochore but is not known to be involved in upstream checkpoint signaling (Hardwick et al. 2000; Gillett et al. 2004). Mad1 associated with kinetochores in a mad3Δ strain treated with benomyl (Fig. 1C), indicating that the loss of Mad1 association with spc105-15 mutant kinetochores is not solely due to a loss of checkpoint function.
To determine whether the Spc105 or Ndc80 subcomplexes specifically associate with Mad1, we immunoprecipitated Nuf2-3Flag (a component of Ndc80c) or Spc105-3Flag. While Mad1-3GFP copurified with Spc105, it was absent from the Nuf2 purification (Fig. 1D). The Spc105 immunoprecipitation did not contain significant levels of Ndc80, suggesting that Ndc80c does not have a direct role in mediating the Spc105–Mad1 interaction (Fig. 1D). Conversely, the Nuf2 purification did not contain detectable levels of the endogenous untagged Spc105 protein (Fig. 1D), consistent with the absence of Mad1. The association of Mad1 with Spc105 is supported by a recent report in which human Spc105/Knl1 was closely localized with Mad1 on the kinetochore and was required for Mad1 kinetochore localization (Varma et al. 2013).
The middle region of Bub1 recruits Mad1 to kinetochores
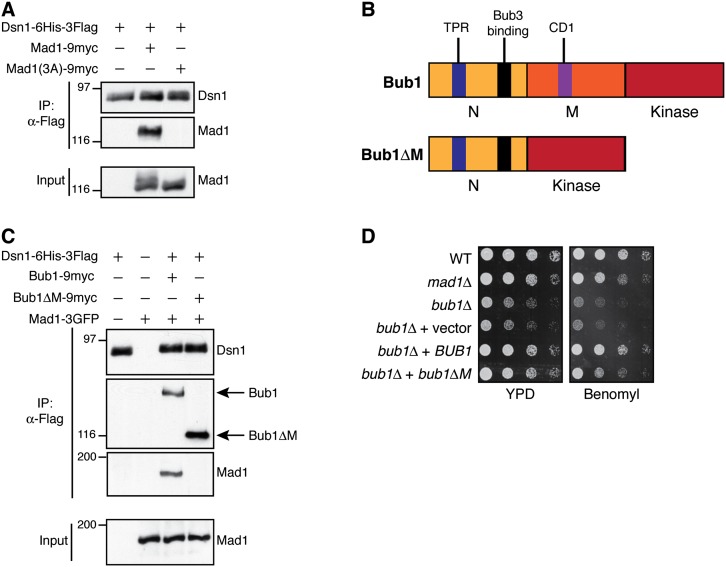
The Bub1/3 complex is required for Mad1 kinetochore localization and binds to kinetochores through Spc105 in numerous organisms (Kiyomitsu et al. 2007; London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012). Because Mad1 is present in Bub1 purifications from mitotic yeast extracts (Brady and Hardwick 2000), we hypothesized that Mad1 may interact with the kinetochore by binding through the Bub1/3 complex, as proposed in human cells and fission yeast (Heinrich et al. 2012; Kim et al. 2012). Mutation of a Mad1 conserved basic patch (RLK to AAA) prevents its association with Bub1 in yeast (Brady and Hardwick 2000) and reduces Mad1 kinetochore localization in human cells (Kim et al. 2012). Consistent with our hypothesis, this Mad1-3A mutant also failed to copurify with kinetochores (Fig. 2A).
Figure 2.
Mad1-3A and Bub1ΔM prevent Mad1 kinetochore association and checkpoint function. (A) The Mad1-3A mutant protein does not associate with kinetochores. Kinetochore particles were purified from cells containing untagged Mad1 (SBY8253), Mad1-9myc (SBY11840), or Mad1-3A-9myc (SBY11967). Immunoblots were probed with α-Flag or α-myc antibodies. (B, top) Bub1 schematic showing the N-terminal (1–368) and middle (369–608) regions and C-terminal kinase (609–1021) domain. The N-terminal region contains the conserved tetratricopeptide repeat (TPR) domain (residues 57–171) (Bolanos-Garcia et al. 2009) as well as the Bub3-binding/GLEBS domain (residues 315–350) (Larsen et al. 2007). Conserved domain 1 (CD1; residues 452–470 (Klebig et al. 2009), is present in the middle region. (Bottom) The middle region is deleted in Bub1ΔM. (C) The Bub1 middle region is required for Mad1 kinetochore association. Kinetochore particles were purified via Dsn1-6His-3Flag from cells with untagged Mad1 (SBY8253), Mad1-3GFP (SBY11392), Mad1-3GFP Bub1-9myc (SBY11317), or Mad1-3GFP Bub1ΔM-9myc (SBY11319). Immunoblots were probed with α-Flag, α-myc, or α-GFP antibodies. (D) bub1ΔM cells are benomyl-sensitive. Fivefold serial dilutions of the following strains were plated onto YPD or YPD + 5 μg/mL benomyl: wild type (WT) (SBY8412), mad1Δ (SBY8820), bub1Δ (SBY10596), bub1Δ + integrated 9myc control vector (SBY11232), bub1Δ + integrated BUB1-9myc (SBY11072), and bub1Δ + integrated bub1ΔM-9myc (SBY11083). Note that the cells for the immunoprecipitation experiments shown in this figure were treated with benomyl.
To identify the region of Bub1 responsible for mediating the Mad1 interaction with kinetochores, we deleted the middle region (residues 369–608) to generate Bub1ΔM (Fig. 2B). This region is implicated in the Bub1–Mad1 interaction (Warren et al. 2002) and includes a conserved patch (conserved domain 1 [CD1]) shown to be necessary for checkpoint function in human cells (Klebig et al. 2009). Mad1 did not copurify with these kinetochores or the Bub1ΔM protein even though Bub1ΔM still associated with kinetochores (Fig. 2C; Supplemental Fig. S1). Cells defective in the spindle checkpoint are sensitive to benomyl, so we analyzed the growth of bub1ΔM cells on medium containing benomyl (Fig. 2D). Consistent with a defect in the spindle checkpoint, bub1ΔM cells exhibited benomyl sensitivity comparable with mad1Δ cells (Fig. 2D). Deletions of BUB1 are even more sensitive to benomyl than mad1Δ cells because BUB1 has an additional chromosome segregation function in localizing the Sgo1 protein via histone H2A phosphorylation (Fernius and Hardwick 2007; Kawashima et al. 2010). Importantly, the viability of bub1ΔM cells was improved relative to bub1Δ cells, suggesting that the middle region mediates the checkpoint functions of Bub1 but does not affect its other functions in chromosome segregation.
We next tested whether the middle region of Bub1 is sufficient to promote kinetochore recruitment of Mad1. We previously generated an Spc105 phosphomutant allele, spc105-6A, that is defective in Bub1/3 binding to kinetochores because key Mps1 phosphorylation sites are mutated (London et al. 2012). As expected, kinetochore particles purified from spc105-6A cells also failed to copurify Mad1 (Fig. 3A; Supplemental Fig. S2A). We therefore fused Bub1(M) to the N terminus of Spc105-6A or wild-type Spc105. These fusions do not constitutively activate the checkpoint or inhibit the essential functions of Spc105 because viability is not reduced relative to nonfused SPC105 or spc105-6A strains (Supplemental Fig. S2B). Strikingly, the Bub1(M)-Spc105-6A fusion restored Mad1 purification with the kinetochore (Fig. 3A) even though the endogenous Bub1 and Bub3 proteins did not associate with these fusion kinetochores (Fig. 3B; Supplemental Fig. S2C). Additionally, and in contrast to wild-type kinetochores, Mad1 no longer required endogenous Bub1 to associate with the fusion kinetochore particles (Fig. 3C; data not shown). Fusion of Bub1(M) to wild-type Spc105 resulted in a strong increase in Mad1 levels associated with the kinetochore, consistent with Mad1 binding through both the fused Bub1(M) fragment and endogenous Bub1 (Fig. 3A).
Figure 3.
Bub1(M) mediates Mad1 kinetochore association and checkpoint function. (A) Bub1(M) is sufficient to recruit Mad1 to Spc105-6A mutant kinetochores. Kinetochore particles were purified by α-Flag immunoprecipitation of Dsn1-6His-3Flag from cells with endogenous Mad1 and Spc105-9myc (SBY10267) or cells with Mad1-3GFP and either Spc105-9myc (SBY11207), Bub1(M)-Spc105-9myc (SBY11208), Spc105-6A-9myc (SBY11210), or Bub1(M)-Spc105-6A-9myc (SBY11209). Immunoblots were probed with α-Flag or α-GFP antibodies. (B) Bub1(M)-Spc105-6A mutant kinetochores do not recruit endogenous Bub1/3. Kinetochores were purified as in A from the following strains: Spc105-9myc (SBY10267), Bub1-3GFP Spc105-9myc (SBY10347), Bub1-3GFP Spc105-6A-9myc (SBY10422), Bub1-3GFP Bub1(M)-Spc105-6A-9myc (SBY11725), Bub3-3GFP Spc105-9myc (SBY10351), Bub3-3GFP Spc105-6A-9myc (SBY10372), and Bub3-3GFP Bub1(M)-Spc105-6A-9myc (SBY11637). Immunoblots were probed with α-Flag or α-GFP antibodies. (C) Bub1(M) is sufficient to recruit Mad1 to Spc105-6A mutant kinetochores in the absence of endogenous Bub1. Kinetochore particles were purified as in A from strains containing Mad1-3GFP with Spc105-9myc (SBY11207), Bub1(M)-Spc105-6A-9myc (SBY11209), Spc105-6A-9myc (SBY11210), or Bub1(M)-Spc105-6A-9myc bub1Δ (SBY11303). Immunoblots were probed with α-Flag or α-GFP antibodies. (D) The middle region of Bub1 can restore partial checkpoint function in spc105-6A mutant cells. Cells were released from G1 into nocodazole-containing medium. Samples were taken at the indicated time points (in minutes) to analyze Pds1-18myc levels by immunoblotting with α-myc antibodies in SPC105-2V5 (SBY11892), spc105-6A-2V5 (SBY11893), and bub1(M)-spc105-6A-2V5 (SBY11894) cells. Pgk1 is shown as a loading control. Note that the cells for immunoprecipitation experiments were treated with benomyl, and protein levels for additional relevant panels are shown in Supplemental Figure S2.
To test whether the Mad1 that associates with Bub1(M)-Spc105-6A kinetochores is functional in vivo, we assayed checkpoint activity in response to nocodazole-induced microtubule destabilization by monitoring the protein levels of Pds1, the yeast Securin homolog (Cohen-Fix et al. 1996). Strikingly, Pds1 levels were significantly stabilized in bub1(M)-spc105-6A relative to spc105-6A cells (Fig. 3D). However, this stabilization still required endogenous Bub1 (data not shown), consistent with multiple requirements for Bub1 in the checkpoint (Elowe 2011). Together, these data indicate that the middle region of Bub1 can serve as a functional receptor for Mad1 at the kinetochore.
Phosphorylation of Bub1(M) regulates its interaction with Mad1
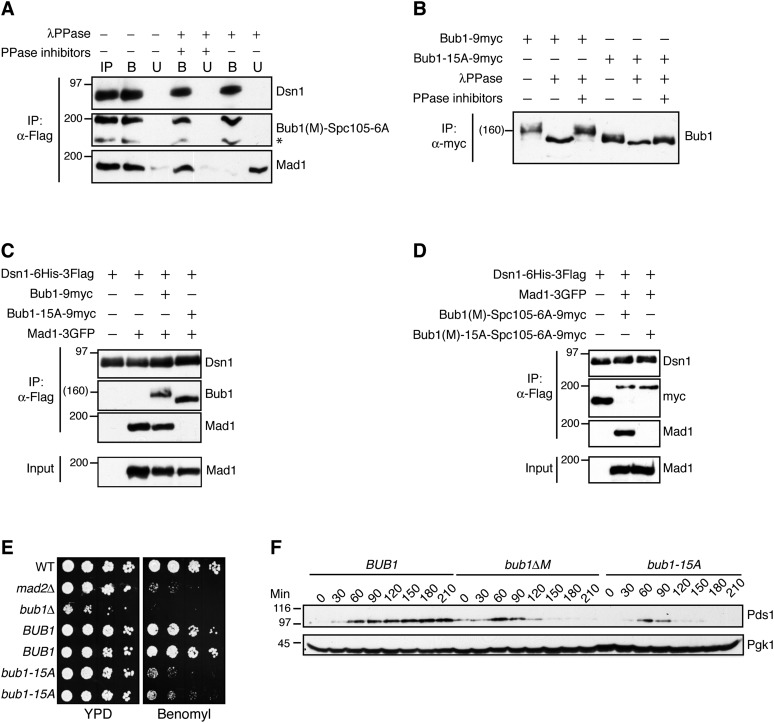
To address how Mad1 kinetochore localization is dynamically regulated, we tested whether phosphorylation is required for its association with kinetochores. Treatment of immunoprecipitated Bub1(M)-Spc105-6A kinetochores with λPPase released Mad1 from kinetochores (Fig. 4A), indicating that phosphorylation is required. Mad1 also dissociated from bead-bound Bub1 in a similar experiment (Supplemental Fig. S3).
Figure 4.
Identification of Bub1 phosphorylation sites required for the spindle checkpoint and Mad1 association. (A) The Mad1–kinetochore interaction requires Bub1 phosphorylation. λPPase treatment releases Mad1 from purified Bub1(M)-Spc105-6A kinetochore particles. Kinetochore particles (SBY11209) were immobilized on α-Flag beads and treated with λPPase with or without phosphatase inhibitors or without phosphatase. (IP) Starting input; (B) bound; (U) unbound. (*) An apparent degradation product of Bub1(M)-Spc105-6A corresponding to Spc105-6A migration. The immunoblots were probed with α-Flag, α-myc, or α-GFP antibodies. (B) The Bub1-15A mutant protein shows reduced phosphorylation in vivo. Bub1-9myc (SBY11764) or Bub1-15A-9myc (SBY11766) was immunoprecipitated from benomyl-treated cells and eluted directly or after treatment with λPPase with or without phosphatase inhibitors. The immunoblot was probed with α-myc. (C) Mad1 does not associate with kinetochore particles containing the Bub1-15A mutant protein. Kinetochore particles were immunoprecipitated from benomyl-treated cells with α-Flag beads. The strains contained untagged Mad1 (SBY8253), Mad1-3GFP (SBY8415), Mad1-3GFP Bub1-9myc (SBY11764), or Mad1-3GFP Bub1-15A-9myc (SBY11766). Immunoblots were probed with α-Flag, α-myc, or α-GFP antibodies. (D) Mad1 association with Bub1(M)-Spc105-6A kinetochores requires Bub1 phosphorylation sites. Dsn1-6His-3Flag was purified from a strain with untagged Mad1 (SBY10267) or strains with Mad1-3GFP and Bub1(M)-Spc105-6A-9myc (SBY11209) or Bub1(M)-15A-Spc105-6A-9myc (SBY11415). Immunoblots were probed with α-Flag, α-myc, or α-GFP antibodies. (E) bub1-15A mutant cells are benomyl-sensitive. Fivefold serial dilutions of the following strains were grown on YPD or YPD + 7.5 μg/mL benomyl: wild type (WT) (SBY8253), mad2Δ (SBY292), bub1Δ (SBY10596), and two independent transformants of the bub1Δ strain (SBY10596) with either BUB1-9myc (pSB1983) or bub1-15A-9myc (pSB1984). (F) bub1-15A mutant cells are defective in the spindle checkpoint. BUB1-6His (SBY11340), bub1ΔM-6His (SBY11342), and bub1-15A-6His (SBY11341) strains were released from a G1 arrest into medium containing nocodazole. Pds1-18myc levels were analyzed by α-myc immunoblotting at the indicted time points (in minutes). Pgk1 is shown as a loading control.
We next sought to identify the relevant phosphorylation sites by mass spectrometry. Because a basic patch on Mad1 is required for its association with Bub1 (Brady and Hardwick 2000), we looked for phosphorylation in the middle region of Bub1 that may promote an electrostatic interaction with Mad1. We identified phosphorylation sites and several adjacent patches of serines or threonines on Bub1 (Supplemental Fig S4A; Supplemental Table S1). We therefore mutated seven of these phosphorylation sites as well as eight neighboring serines or threonines to produce the bub1-15A allele. Bub1-15A migrated faster than wild-type Bub1, and λPPase treatment further enhanced its migration (Fig. 4B). Together, these data indicate that one or more phosphorylation sites in the middle region of Bub1 are eliminated in the Bub1-15A mutant protein, although the mutant protein is still phosphorylated in vivo.
Kinetochores purified from bub1-15A cells retained Bub1-15A but failed to associate with Mad1 (Fig. 4C), and the Bub1-15A protein did not copurify Mad1 (Supplemental Fig S4B). Mad1 binding to Bub1(M)-Spc105-6A kinetochores also required the phosphorylation sites on the fusion protein (Fig. 4D). As expected, cells harboring bub1-15A were less sensitive to benomyl relative to bub1Δ cells, consistent with this allele maintaining chromosome segregation functions but not checkpoint proficiency (Fig. 4E, cf. mad2Δ and bub1-15A). Indeed, bub1-15A cells exhibited a checkpoint defect because they failed to stabilize Pds1 upon nocodazole treatment, similar to bub1ΔM cells (Fig. 4F). Based on these data, we conclude that the Mad1 kinetochore interaction requires phosphorylation of key residues in the middle region of Bub1.
Mps1 phosphorylates Bub1 to recruit Mad1 to kinetochores
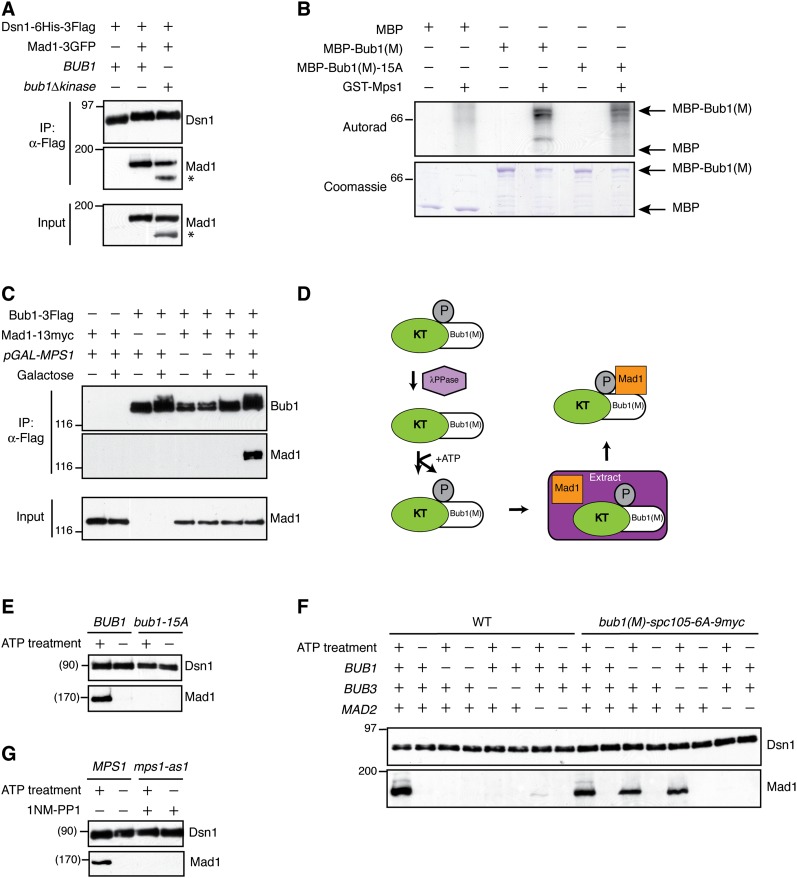
We next sought to identify the kinase responsible for Bub1 phosphorylation. While Bub1 is capable of autophosphorylation, its kinase activity is not required for the spindle checkpoint (Sharp-Baker and Chen 2001; Fernius and Hardwick 2007). Consistent with this, deletion of the Bub1 kinase domain did not affect Mad1 kinetochore association (Fig. 5A). Mps1 kinase activity is required for checkpoint activation under all conditions, so we tested whether Bub1 is an Mps1 substrate. We generated a recombinant fragment of the Bub1 middle region (residues 369–608 lacking the kinase domain) fused to maltose-binding protein, MBP-Bub1(M), and tested for radiolabeled phosphate incorporation in the presence of recombinant Mps1 (Weiss and Winey 1996). The Bub1 fragment was strongly phosphorylated by Mps1 in vitro, while the MBP tag alone did not exhibit detectable phosphorylation (Fig. 5B). MBP-Bub1(M)-15A showed reduced phosphorylation in this assay, indicating that the 15 sites contain one or more in vitro Mps1 target residues (Fig. 5B). To test for Mps1 phosphorylation of Bub1 in vivo, we overexpressed Mps1 in G1-arrested yeast cells. There was an apparent Bub1 phosphorylation shift, consistent with Bub1 phosphorylation in vivo being mediated by Mps1 activity and not requiring other mitotic events (Fig. 5C). Strikingly, Mps1 overexpression also led to the ectopic association of Bub1 with Mad1 in these G1-arrested cells (Fig. 5C).
Figure 5.
Mps1 phosphorylation of Bub1 recruits Mad1 to the kinetochore. (A) Bub1 kinase activity is not required for Mad1 kinetochore association. Kinetochore particles were immunoprecipitated from benomyl-treated cultures from strains with untagged Mad1 (SBY8253), Mad1-3GFP (SBY8415), or Mad1-3GFP Bub1Δkinase (SBY11006). Note that a Mad1-3GFP degradation product is observed in this strain (*). Immunoblots were probed with α-Flag or α-GFP antibodies. (B) Mps1 phosphorylates the middle region of Bub1 in vitro. Kinase assays were performed with purified MBP or MBP fusions plus/minus recombinant GST-Mps1 bound to glutathione resin. Phosphorylation was detected by autoradiography (top), and total protein was detected by Coomassie staining (bottom). The 15A fragment phosphorylation signal was reduced 26% relative to wild type in this experiment. (C) Mps1 overexpression leads to Mad1–Bub1 association in G1-arrested cells. Bub1-3Flag was immunoprecipitated from cells arrested in G1 and treated with glucose or galactose. (Left to right) SBY11541 (pGAL-MPS1 MAD1-13myc), SBY11490 (pGAL-MPS1 BUB1-3Flag), SBY10532 (BUB1-3Flag MAD1-13myc), and SBY11488 (pGAL-MPS1 BUB1-3Flag MAD1-13myc). Immunoblots were probed with α-Flag or α-myc antibodies. (D) Schematic of experiment to determine whether Mps1 phosphorylation recruits Mad1 to kinetochores in vitro. Bub1(M)-Spc105-6A-9myc kinetochores [KT-Bub1(M)] bound to beads were treated with phosphatase and then incubated with ATP to allow Mps1 phosphorylation. The beads were then added to yeast extract containing Mad1-3GFP and washed, and the kinetochores were eluted. (E) Mps1 phosphorylation of kinetochore particles recruits Mad1 and depends on Bub1 phosphorylation sites. Bead-bound Bub1(M)-Spc105-6A-9myc (SBY11149) or Bub1(M)-15A-Spc105-6A-9myc kinetochores (SBY11325) were used for the experiment outlined in D. Immunoblots were probed with α-Flag or α-GFP antibodies. (F) Mad2, but not Bub1 or Bub3, is needed to enable Mad1 recruitment to Bub1(M)-Spc105-6A-9myc kinetochore particles phosphorylated by Mps1. Kinetochore particles were purified from wild-type (WT) (SBY8253) or bub1(M)-spc105-6A-9myc (SBY11149) strains and then treated as in D to analyze Mad1-3GFP binding from either wild-type (SBY8416), bub1Δ (SBY11464), bub3Δ (SBY11418), or mad2Δ (SBY11389) extracts. Immunoblots were probed with α-Flag or α-GFP antibodies. Binding of Mad1-3GFP to wild-type kinetochores in the +ATP condition is likely due to recruitment of preassociated Bub1–Mad1 complex through Spc105 phosphorylation. (G) Mad1 recruitment to kinetochore particles depends on Mps1 activity. Bub1(M)-Spc105-6A-9myc kinetochores were purified from either wild-type (SBY11149) or mps1-as1 (SBY11486) strains and used for the experiment outlined in D. mps1-as1 kinetochores were treated with the specific inhibitor 1NM-PP1. Immunoblots were probed with α-Flag or α-GFP antibodies.
To determine whether Mps1 phosphorylation of Bub1 is a signal for Mad1–kinetochore binding, we asked whether kinetochore-bound Bub1 recruits Mad1 upon Mps1 phosphorylation in vitro. Mps1 kinase activity specifically copurifies with kinetochores (London et al. 2012), so we tested whether ATP treatment of kinetochore particles enabled Mad1 binding from yeast extract (Fig. 5D). We first eliminated existing phosphorylation from the kinetochores with phosphatase treatment. Since phosphatase treatment dissociates Bub1 from kinetochores (London et al. 2012), we used Bub1(M)-Spc105-6A fusion kinetochores for these experiments. There was strong ATP-dependent binding of Mad1 to Bub1(M)-Spc105-6A kinetochores, and this also required the Bub1 phosphorylation sites (Fig. 5E). Because Bub1(M)-Spc105-6A kinetochores purified from yeast lack endogenous Bub1 or Bub3 (see Fig. 3B), the middle region of Bub1 mediates this interaction. Consistent with this, Mad1 from bub1Δ or bub3Δ lysates still associated with Bub1(M)-Spc105-6A kinetochores in vitro (Fig. 5F). Mad1 binding to wild-type kinetochores in this assay is expected to occur through the interaction of Bub1 in the extract with Spc105 on the kinetochore, resulting in high Mad1 levels from wild-type extract but no Mad1 binding from bub1Δ or bub3Δ extract (Fig. 5F). In contrast to Bub1 and Bub3, Mad2 was required for the association of Mad1 with the Bub1(M)-Spc105-6A fusion kinetochores (Fig. 5F), consistent with its requirement to localize Mad1 in vivo (Gillett et al. 2004).
To confirm that the phosphorylation that potentiates Mad1 binding is due specifically to Mps1, we used Bub1(M)-Spc105-6A kinetochores harboring the ATP analog-sensitive Mps1-as1 protein (Jones et al. 2005). These kinetochores exhibit kinase activity that is repressed by treatment with the specific inhibitor 1NM-PP1 (Jones et al. 2005; London et al. 2012). Mad1 did not bind to these kinetochores when they were treated with the inhibitor and ATP (Fig. 5G), indicating that Mps1 phosphorylation of Bub1(M) is a kinetochore recruitment signal for Mad1.
To further narrow down the relevant phosphorylation sites in Bub1(M), we made a series of mutants where the putative phosphorylation sites were mutated to alanine and analyzed for growth on benomyl medium. This analysis revealed that residues T485, T509, and T518 contribute to the benomyl sensitivity (Supplemental Table S2) and are required for Mad1 kinetochore binding (Supplemental Fig. S5A). In addition, we noticed a conserved phosphorylation site at T455 within the middle region that was also phosphorylated in our mass spectrometry data and in a data set of human mitotic phosphorylation (Daub et al. 2008). We therefore generated a bub1(T455A) mutant and found that it is also benomyl-sensitive (Supplemental Table S2) and abolishes the association of Mad1 with the kinetochore (Supplemental Fig. S5B). We additionally attempted to generate a phosphomimetic allele of Bub1 by substituting identified residues with aspartic or glutamic acid, but this has so far not proven successful (Supplemental Table S2; data not shown). Together, this suggests that phosphorylation of multiple residues in the N-terminal portion of the Bub1 middle region can mediate Mad1 binding, while the C-terminal residues do not contribute.
Bub1 and Mad1(C)/Mad2 interact directly
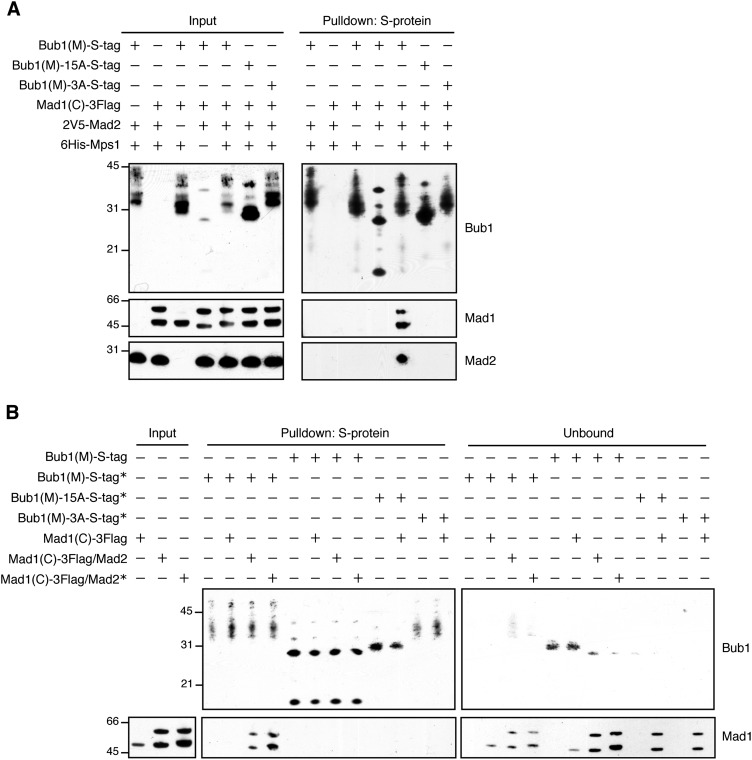
We next tried to reconstitute the minimal Bub1–Mad1 interaction given that unknown factors may be present in lysate that promote the interaction. To do this, we asked whether the coexpression of Mps1 with Mad1 and Bub1 in bacteria was sufficient for their association. We used the middle region of Bub1 and a C-terminal Mad1 fragment [residues 318–749, Mad1(C)] that was previously shown to retain kinetochore-binding function (Scott et al. 2005). Because Mad2 is required for Mad1 kinetochore localization in vivo (Gillett et al. 2004) and the Mad1–Bub1 interaction (Brady and Hardwick 2000; see above), we also coexpressed Mad2. Coexpression of all of these proteins was necessary and sufficient for the association of Mad1(C) with Bub1(M) (Fig. 6A). Importantly, this interaction was essentially abolished upon mutation of the phosphorylation sites on Bub1(M) (Fig. 6A). This is also consistent with the requirement for Mad2 (but not other checkpoint proteins) to promote Mad1 association with Bub1(M)-Spc105-6A fusion kinetochores (Fig. 5F). As Mad2 also associated with Bub1(M) in a Mad1(C)-dependent manner (Fig. 6A), the Mad1–Mad2 interaction appears to be necessary for Mad1(C) to bind to Bub1(M). We also note that Mad2 and Mad1 interacted when coexpressed in bacteria without Mps1 or Bub1(M) (Supplemental Fig. S6A), consistent with the reported phosphorylation-independent, constitutive nature of Mad1–Mad2 binding (Chen et al. 1999; Sironi et al. 2001). Because Mps1 is not present in the Bub1(M) pull-down at significant levels under these conditions (Supplemental Fig. S6B), we conclude that Mps1 association does not directly mediate the interaction between Bub1(M) and Mad1(C)/Mad2.
Figure 6.
Recombinantly expressed Bub1 and Mad1 interact and require Mps1 and Mad2. (A) Recombinantly expressed Bub1 and Mad1 interact. The indicated proteins were coexpressed in bacteria and purified using S-protein resin. Note that there are multiple Mad1 and Bub1 species, presumably due to degradation, and the phosphorylation of the Bub1 fragments by Mps1 leads to a large mobility shift that is difficult to resolve by SDS-PAGE. The immunoblots were probed with α-S-tag, α-Flag, or α-V5 antibodies. (B) Bub1 phosphorylation, but not Mad1 phosphorylation, is required for in vitro binding of Bub1 and Mad1. Mad1(C)-3Flag was purified by itself or together with 2V5-Mad2 in the presence or absence of Mps1 expression. (*) Mps1 coexpression. The indicated Bub1(M)-S-tag fragments were immobilized on S-protein beads and incubated with the Mad1 proteins in vitro to assay for binding. The immunoblots were probed with α-S-tag or α-Flag antibodies.
We next wanted to ascertain whether Bub1(M) phosphorylation is sufficient for Mad1(C) binding or whether there is also a requirement for Mps1 phosphorylation of Mad1(C) or Mad2. To do this, we purified recombinant Mad1(C) alone or together with Mad2 with or without Mps1 coexpression. We then attempted to bind this in vitro to the wild-type Bub1(M) protein prepared with or without Mps1 coexpression or to phosphomutant Bub1(M) proteins prepared with Mps1 coexpression. While Mps1 expression appeared to phosphorylate Mad1(C) because it migrated more slowly, this phosphorylation did not promote the association with unphosphorylated Bub1(M) (Fig. 6B). In contrast, phosphorylated Bub1(M) bound to Mad1(C) with or without Mad1(C) phosphorylation, and the binding was abolished by mutation of the Bub1 phosphorylation sites. These results indicate that Mps1 phosphorylation on Bub1 is necessary and sufficient for Mad1(C)–Mad2 binding in vitro, whereas Mad1(C) phosphorylation is not sufficient for the interaction.
Discussion
The identity of the Mad1 kinetochore receptors has been a long-standing question that has been difficult to address in vivo (Jia et al. 2013). Previously, Ndc80 was implicated as a receptor because it is required for Mad1 localization in various organisms (Martin-Lluesma et al. 2002; DeLuca et al. 2003; McCleland et al. 2003). Recently, Bub1 was also proposed to be a potential Mad1 receptor in human cells (Kim et al. 2012). In Caenorhabditis elegans, the kinetochore protein Spindly interacts with Mad1 by coimmunoprecipitation (Yamamoto et al. 2008), although this may be species-specific, and yeast does not have a known SPINDLY homolog (Griffis et al. 2007; Maresca and Salmon 2010). However, a direct interaction between Mad1 and these proteins has never been demonstrated at the kinetochore. We determined that a central region of Bub1 is required for Mad1 association with kinetochores. The interaction requires Bub1 phosphorylation by Mps1, which interacts with Ndc80 (Kemmler et al. 2009; Nijenhuis et al. 2013). We propose that the requirement for Ndc80c in Mad1 localization is due to Mps1 recruitment rather than the direct binding of Mad1. The yeast Ndc80-1 mutant protein that we tested still associates with kinetochores (see Fig. 1B), so it likely retains Mps1 activity. Consistent with this, ndc80-1 mutant cells are competent to activate the spindle checkpoint, while cells with a complete depletion of NDC80 are not (McCleland et al. 2003). Although metazoan Mad1 may have multiple receptors, our data are consistent with Bub1 as the sole Mad1 kinetochore receptor in yeast.
Bub1 is also a target of Cdk1 as well as autophosphorylation (Goto et al. 2011). In fission yeast, mitotic phosphorylation of Bub1 by Cdk1 was found to be important for robust maintenance of mitotic arrest (Yamaguchi et al. 2003), consistent with a possible role in Mad1 recruitment. However, we found that Mps1 phosphorylation is sufficient for Bub1–Mad1 binding in vitro, indicating that neither Cdk1 nor Bub1 phosphorylation is required for the interaction. Consistent with this, none of the critical phosphorylation sites that we identified match the Cdk1 consensus (see Supplemental Table S2), and Bub1 kinase activity is not required for the checkpoint in budding yeast (Fernius and Hardwick 2007).
Our discovery that the Bub1–Mad1 interaction is mediated by Mps1 phosphorylation on Bub1 allowed us to reconstitute this association. Although Mad1 phosphorylation by Mps1 has been implicated in its kinetochore localization (Hardwick et al. 1996), the interaction between Bub1 and Mad1 does not require Mad1 phosphorylation in vitro (Fig. 6B). However, Bub1-associated Mad1 is phosphorylated in vivo, suggesting that Mad1 phosphorylation may mediate other functions of Mad1 (Cairo et al. 2013). The interaction between Bub1 and Mad1 also requires Mad2, as was previously reported in budding yeast in vivo (Brady and Hardwick 2000; Gillett et al. 2004). Although the contribution of Mad2 is currently unclear, Mad1/2 forms stable tetramers that may promote Bub1 association (Sironi et al. 2002). Concordantly, Mad2 coexpression appears to stabilize Mad1(C) when coexpressed in bacteria (Supplemental Fig. S6B). In contrast, Mad2 is not required for Mad1 kinetochore localization in other organisms (Seeley et al. 1999; Hewitt et al. 2010; Kim et al. 2012), so this requirement may not be broadly conserved. The ability to reconstitute the Bub1–Mad1 interaction will facilitate structural studies that should reveal the requirement for Mad2 in the future.
Mps1-mediated phosphorylation has at least one additional role in the checkpoint, which is to recruit Bub1/3 to kinetochores via phosphorylation of Spc105/Knl1 (London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012). This is due to the direct binding of the Bub3 protein to phosphorylated MELT motifs within Spc105/Knl1 (Primorac et al. 2013). Strikingly, tethering the middle region of Bub1 to a phospho-deficient Spc105-6A mutant protein is sufficient to recruit Mad1 to kinetochores and mediate the spindle checkpoint in the complete absence of a kinetochore-bound endogenous Bub1/3 complex. This suggests that the only requirement for Bub3 kinetochore localization in spindle checkpoint initiation is to localize Bub1. However, fusion of the middle region of Bub1 to the mutant Spc105-6A protein did not result in constitutive checkpoint activation, presumably because the recruitment of Mad1/2 is still regulated normally by Mps1 phosphorylation. In contrast, tethering Mps1 to the kinetochore through the kinetochore scaffold protein Mis12 promoted prolonged Mad1 kinetochore localization and spindle checkpoint activity in human cells (Jelluma et al. 2010). Similarly, tethering of fission yeast Mps1 to kinetochores by fusion to Ndc80 resulted in a checkpoint-dependent cell cycle arrest (Ito et al. 2012). These apparently contradictory findings can be reconciled by proposing that Mps1 is not regulated normally when tethered to Ndc80 or an exogenous kinetochore receptor, such as Mis12–Mps1, which leads to constitutive checkpoint activation. An intriguing possibility is that Mps1 kinase activity removes Mps1 from the kinetochore upon microtubule binding, thus facilitating checkpoint silencing (Hewitt et al. 2010; Jelluma et al. 2010; Nijenhuis et al. 2013). While a previous report suggested that Ndc80 phosphorylation is important for Mps1 activation of the checkpoint (Kemmler et al. 2009), this phosphorylation was not required for the Mad1 kinetochore interaction (data not shown).
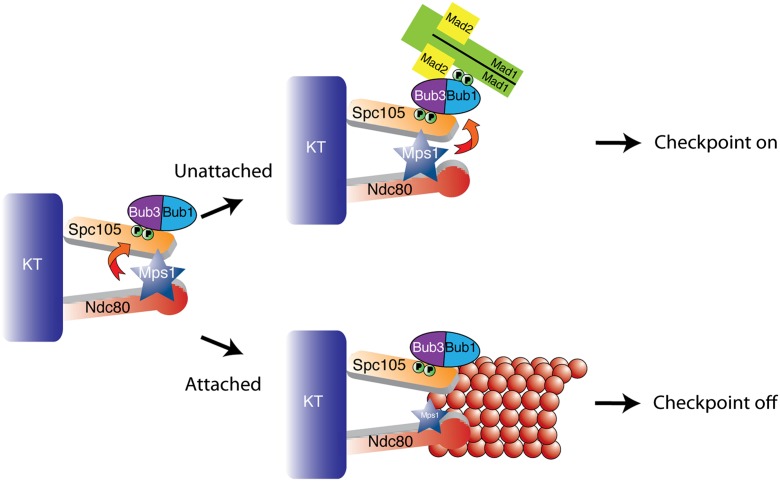
Our findings are consistent with the following model (Fig. 7). Mps1 phosphorylates Spc105 to recruit Bub1/3 during mitosis. When kinetochores lack proper microtubule attachments or tension, kinetochore-bound Mps1 phosphorylates Bub1 to recruit Mad1, thus activating the checkpoint. Because the Mad1–Mad2 interaction is constitutive (Chen et al. 1999), the Bub1–Mad1 interaction at kinetochores must specifically facilitate the catalytic conversion of Mad2. Upon formation of microtubule attachments, Mad1 is removed from the kinetochore, and the checkpoint is silenced. Microtubule binding could dissociate Mad1 from the kinetochore through a variety of mechanisms, such as active stripping of Mad1 and/or activation of a protein phosphatase. However, Mps1 overexpression results in Mad1 kinetochore localization even in the presence of normal kinetochore–microtubule attachments (Hardwick et al. 1996; Jelluma et al. 2010; Cairo et al. 2013), suggesting that Mps1 activity toward Bub1 must also be silenced to release Mad1. Activity of Mps1 toward Bub1 could be altered by spatial separation that occurs upon tension-induced stretching or conformational changes within Bub1/3 or Mps1 upon microtubule attachment. Proper microtubule attachments may also oppose Mps1 localization to kinetochores. Additionally, Glc7/PP1 activity promotes checkpoint silencing (Pinsky et al. 2009; Vanoosthuyse and Hardwick 2009) at least in part by removing Bub1/3 from the kinetochore (London et al. 2012). Future work will be necessary to establish precisely how phosphatase activity opposes the Bub1–Mad1 interaction in vivo.
Figure 7.
Model. Mps1 activity during mitosis phosphorylates Spc105 to recruit the Bub1/3 complex (London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012). Mps1 then phosphorylates Bub1 at unattached kinetochores to recruit Mad1/2 and activate the downstream checkpoint response. Microtubule attachment opposes Mps1 activity, possibly by promoting Mps1 dissociation from kinetochores, and results in Mad1 dissociation from kinetochores to silence the checkpoint (Jelluma et al. 2010).
Bub1 phosphorylation is likely to be a conserved mechanism of Mad1 recruitment. Bub1 is necessary for Mad1 recruitment in other organisms (Abrieu et al. 2001; Heinrich et al. 2012), and regions of Bub1 and Mad1 that are required for the checkpoint are conserved. Checkpoint activation in Xenopus egg extracts requires Bub1 phosphorylation (Chen 2004), and we identified a conserved phosphorylation site required for Mad1 localization to kinetochores in yeast that is also a mitotic phosphorylation target in human cells (Daub et al. 2008). Together, our work provides a foundation for future structural, biophysical, and biochemical investigations aimed at understanding how phosphorylation-driven Mad1 binding and activation are coupled to kinetochore–microtubule attachment.
Materials and methods
Strain and plasmid construction
All strains are isogenic with the W303 background, and standard genetic crosses were performed to introduce temperature-sensitive kinetochore alleles. All strains and plasmids used in this study are listed in the Supplemental Tables. Additional information on strain and plasmid construction is in the Supplemental Material.
Yeast methods and purifications
Media and microbial techniques were as described (Rose et al. 1990), and additional details about experimental conditions are in the Supplemental Material. Kinetochore particle and Flag-tagged protein purifications from yeast were performed essentially as described (Akiyoshi et al. 2010). For kinetochore particle purifications, cells expressing Dsn1-6His-3Flag were cultured to OD600 of ∼2.0. Lysis was performed by grinding of yeast pellets with dry ice followed by resuspension in immunoprecipitation lysis buffer (25 mM HEPES at pH 8.0, 150 mM KCl, 0.1% NP-40, 2 mM MgCl2, 0.1 mM EDTA at pH 8.0, 0.5 mM EGTA at pH 8.0, 15% glycerol) supplemented with protease and phosphatase inhibitors (Akiyoshi et al. 2010) and clarification by ultracentrifugation. Cells used in Pds1 stability, pGAL-MPS1 induction, and Bub1 in vivo phosphorylation shift experiments were lysed by bead beating (Ranjitkar et al. 2010). Immunoprecipitated proteins were washed three times in lysis buffer with protease and phosphatase inhibitors, twice in lysis buffer alone, and then eluted in SDS sample buffer prior to analysis.
Phosphatase and kinase assays
Phosphatase and kinase assays are described in the Supplemental Material.
Protein-binding assays
Kinetochore–Mad1-binding assays using lysates were performed essentially as described with additional details in the Supplemental Material (London et al. 2012). Recombinant protein-binding experiments were performed by incubating resin-bound Bub1-S-tag proteins with purified Mad1-3Flag proteins. In vitro binding buffer consisted of PBS with 0.1% Tween-20, 150 mM NaCl, 300 ng/μL bovine serum albumin (BSA), and approximately equivalent amounts of each test protein. Binding reactions were performed for 1–2 h at 4°C. Beads were washed twice in an excess of binding buffer with phosphatase inhibitors, and proteins were eluted in sample buffer.
Recombinant protein purification and coexpression
GST-Mps1 was expressed and purified as described (Holinger et al. 2009), with glutathione-sepharose (GE Healthcare) resin. Resin-bound Mps1 was washed several times in wash buffer (0.5 mM DTT, 250 mM KCl in PBS) before addition of substrate. All proteins were expressed in freshly transformed codon-optimized BL21-Codonplus Escherichia coli by induction of log-phase cultures. Induction of MBP-tagged proteins was performed for 3 h at 37°C with 1 mM isopropyl β-D-thiogalactoside (IPTG). MBP proteins were purified as GST-Mps1 using amylose resin (New England Biolabs), eluted into elution buffer (10 mM maltose, 50 mM Tris at pH 8.0, 250 mM KCl), and then dialyzed against PBS with 30% glycerol.
Coexpression experiments used the pET-Duet and pCDF-Duet expression system (Novagen). Proteins were induced with 100 μM IPTG for 1 h at 37°C, and purifications were performed with S-protein agarose resin (EMD Millipore) or α-Flag-conjugated Dynabeads. Bacterial cell pellets were resuspended in bacterial lysis buffer (2 mM EDTA, 2 mM EGTA, in PBS with 1 mM PMSF and 10 μg/mL each leupeptin, pepstatin, and chymostatin), including phosphatase inhibitors for in vitro binding experiment proteins, and then sonicated three times for 20 sec. Triton X-100 was added to 1%, and samples were incubated for at least 30 min at 4°C with gentle mixing. Extract was separated from cellular debris by centrifugation and then incubated with S-protein agarose resin (Millipore) for at least 1 h at 4°C with gentle agitation. Resin was then washed six times with an excess of wash buffer including 1% Triton X-100 and then eluted into SDS sample buffer (for coexpression experiments) or resuspended in binding buffer (for in vitro binding experiments). Mad1-3Flag proteins were eluted as above with 0.5 μg/μL 3Flag peptide in wash buffer lacking DTT and then dialyzed into PBS with 30% glycerol.
Immunological methods
Immunoblotting was performed using the following antibodies diluted in PBS-Tween buffer: α-myc 9E10 (Covance) at 1:10,000, α-Flag M2 (Sigma-Aldrich) at 1:3000, α-V5 (Thermo/Pierce) at 1:5000, α-GFP (Roche) at 1:1000 or α-GFP JL-8 (Clontech) at 1:5000, α-Pgk1 (Invitrogen) at 1:10,000, and α-S-tag (Thermo/Pierce) at 1:2000; α-Spc105 and α-Ndc80 antibodies were both used at 1:10,000 and were kind gifts from Arshad Desai (Akiyoshi et al. 2010). Immunoprecipitations were performed according to the kinetochore purification conditions using these antibodies conjugated to Protein G Dynabeads. Note that the images shown are cropped from original immunoblots for clarity.
Mass spectrometry
Bead-bound kinetochore particles were phosphorylated in vitro and incubated with lysate, similar to Figure 6D. Samples were purified and prepared essentially as described (Akiyoshi et al. 2009, 2010; London et al. 2012). Details are included in the Supplemental Material.
Acknowledgments
We are grateful to Arshad Desai, Kevin Hardwick, Johannes Lechner, Kevan Shokat, and Mark Winey for sharing reagents, and Bungo Akiyoshi, Chip Asbury, and the Biggins laboratory for critical comments on the manuscript. We also thank Jeff Ranish and Phil Gafken for help with the mass spectrometry, and the Asbury and Tsukiyama laboratories for discussions. This work was supported by a National Institutes of Health (NIH) center interdisciplinary training grant (T32 CA080416) to N.L., and an NIH grant (GM064386) to S.B.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.233700.113.
References
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106: 83–93 [DOI] [PubMed] [Google Scholar]
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev 23: 2887–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S 2010. Tension directly stabilizes reconstituted kinetochore–microtubule attachments. Nature 468: 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Kiyomitsu T, D'Arcy S, Chirgadze DY, Grossmann JG, Matak-Vinkovic D, Venkitaraman AR, Yanagida M, Robinson CV, Blundell TL 2009. The crystal structure of the N-terminal region of BUB1 provides insight into the mechanism of BUB1 recruitment to kinetochores. Structure 17: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady DM, Hardwick KG 2000. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol 10: 675–678 [DOI] [PubMed] [Google Scholar]
- Cairo LV, Ptak C, Wozniak RW 2013. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol Cell 49: 109–120 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Chen RH 2004. Phosphorylation and activation of Bub1 on unattached chromosomes facilitate the spindle checkpoint. EMBO J 23: 3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell 10: 2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10: 3081–3093 [DOI] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 31: 438–448 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Howell BJ, Canman JC, Hickey JM, Fang G, Salmon ED 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol 13: 2103–2109 [DOI] [PubMed] [Google Scholar]
- Elowe S 2011. Bub1 and BubR1: At the interface between chromosome attachment and the spindle checkpoint. Mol Cell Biol 31: 3085–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Hardwick KG 2007. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet 3: e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK 2004. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J Cell Biol 164: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto GH, Mishra A, Abdulle R, Slaughter CA, Kitagawa K 2011. Bub1-mediated adaptation of the spindle checkpoint. PLoS Genet 7: e1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273: 953–956 [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol 148: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S, Windecker H, Hustedt N, Hauf S 2012. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J Cell Sci 125: 4720–4727 [DOI] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS 2010. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J Cell Biol 190: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holinger EP, Old WM, Giddings TH Jr, Wong C, Yates JR III, Winey M 2009. Budding yeast centrosome duplication requires stabilization of Spc29 via Mps1-mediated phosphorylation. J Biol Chem 284: 12949–12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 14: 953–964 [DOI] [PubMed] [Google Scholar]
- Ito D, Saito Y, Matsumoto T 2012. Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1. Proc Natl Acad Sci 109: 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJ 2010. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J Cell Biol 191: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Kim S, Yu H 2013. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci 38: 302–311 [DOI] [PubMed] [Google Scholar]
- Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, Bloom K, Winey M 2005. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol 15: 160–165 [DOI] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327: 172–177 [DOI] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J 28: 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sun H, Tomchick DR, Yu H, Luo X 2012. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci 109: 6549–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13: 663–676 [DOI] [PubMed] [Google Scholar]
- Klebig C, Korinth D, Meraldi P 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J Cell Biol 185: 841–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NA, Al-Bassam J, Wei RR, Harrison SC 2007. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci 104: 1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, Biggins S 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol 22: 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell 9: 59–71 [DOI] [PubMed] [Google Scholar]
- Maldonado M, Kapoor TM 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol 13: 475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED 2010. Welcome to a new kind of tension: Translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci 123: 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 17: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A 2011. Spindle assembly checkpoint: The third decade. Philos Trans R Soc Lond B Biol Sci 366: 3595–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell 14: 4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W, von Castelmur E, Littler D, De Marco V, Tromer E, Vleugel M, van Osch MH, Snel B, Perrakis A, Kops GJ 2013. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol 201: 217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P 2009. Roles for the conserved Spc105p/Kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE 4: e7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, Biggins S 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol 19: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife 2: e01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischitor PE, May KM, Hardwick KG 2007. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS ONE 2: e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Heiter P 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell 16: 4362–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley TW, Wang L, Zhen JY 1999. Phosphorylation of human MAD1 by the BUB1 kinase in vitro. Biochem Biophys Res Commun 257: 589–595 [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 14: 942–952 [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol 153: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol 22: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A 2001. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J 20: 6371–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A 2002. Crystal structure of the tetrameric Mad1–Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J 21: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Staples O, Taylor S 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol 181: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Hardwick KG 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol 19: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Wan X, Cheerambathur D, Gassmann R, Suzuki A, Lawrimore J, Desai A, Salmon ED 2013. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J Cell Biol 202: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CD, Brady DM, Johnston RC, Hanna JS, Hardwick KG, Spencer FA 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell 13: 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M 1996. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol 132: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Yang CH, Tanno Y, Watanabe Y 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol 14: 746–752 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Decottignies A, Nurse P 2003. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J 22: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto TG, Watanabe S, Essex A, Kitagawa R 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J Cell Biol 183: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Dou Z, Qin B, Jin C, Wang X, Xu L, Wang Z, Zhu L, Liu F, Gao X, et al. 2013. Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J Biol Chem 288: 36149–36159 [DOI] [PMC free article] [PubMed] [Google Scholar]