The progressive bone marrow failure syndrome dyskeratosis congenita (DC) is often caused by mutations in telomerase or factors involved in telomerase biogenesis and trafficking. However, a subset of DC patients is heterozygous for mutations in the shelterin component TIN2. Here, heterozygous TIN2-DC mice showed a telomere-shortening phenotype in both telomerase-proficient and telomerase-deficient backgrounds. This study raises the possibility that some of the TIN2-DC mutations may affect telomere function in a telomerase-independent manner.
Keywords: telomere, telomerase, telomeropathy, TIN2, shelterin, dyskeratosis congenita
Abstract
The progressive bone marrow failure syndrome dyskeratosis congenita (DC) is often caused by mutations in telomerase or the factors involved in telomerase biogenesis and trafficking. However, a subset of DC patients is heterozygous for mutations in the shelterin component TIN2. To determine how the TIN2-DC mutations affect telomere function, we generated mice with the equivalent of the TIN2 K280E DC allele (TIN2DC) by gene targeting. Whereas homozygous TIN2DC/DC mice were not viable, first-generation TIN2+/DC mice were healthy and fertile. In the second and third generations, the TIN2+/DC mice developed mild pancytopenia, consistent with hematopoietic dysfunction in DC, as well as diminished fecundity. Bone marrow telomeres of TIN2+/DC mice shortened over the generations, and immortalized TIN2+/DC mouse embryonic fibroblasts (MEFs) showed telomere shortening with proliferation. Unexpectedly, telomere shortening was accelerated in TIN2+/DC mTR−/− mice and MEFs compared with TIN2+/+ mTR−/− controls, establishing that the TIN2DC telomere maintenance defect was not solely due to diminished telomerase action. The TIN2DC allele induced mild ATR kinase signaling at telomeres and a fragile telomere phenotype, suggestive of telomere replication problems. These data suggest that this TIN2-DC mutation could induce telomeric dysfunction phenotypes in telomerase-negative somatic cells and tissues that further exacerbate the telomere maintenance problems in telomerase-positive stem cell compartments.
The telomeropathies are diseases caused by defective telomeres and have a broad clinical spectrum that ranges from rare inherited syndromes with early onset and severe manifestations such as dyskeratosis congenita (DC), Hoyeraal-Hreidarrson syndrome (HHS), Revesz syndrome (RS), and Coats' plus to more prevalent diseases that often present with later onset, including idiotypic lung fibrosis, liver cirrhosis, anaplastic anemia, and myelodysplastic syndrome (for review, see Dokal and Vulliamy 2005; Mason et al. 2005; Aubert and Lansdorp 2008; Armanios 2009; Calado and Young 2009; Savage and Bertuch 2010; Anderson et al. 2012; Nelson and Bertuch 2012).
DC is a rare inherited bone marrow failure syndrome that is classically characterized by a mucocutaneous triad of oral leukoplakia, nail dystrophy, and abnormal skin pigmentation as well as a predisposition to cancer. The role of telomeres in the etiology of DC first emerged from the identification of DKC1 mutations in the X-linked form of DC (Heiss et al. 1998) and the association of the DKC1 gene product, dyskerin, with the RNA component of telomerase (Mitchell et al. 1999). Consistent with a deficiency in telomerase-mediated telomere maintenance, a common feature of DC patients is their extraordinarily short telomeres (Alter et al. 2007). Nearly half of DC cases are now known to be due to mutations in the reverse transcriptase and RNA subunits of telomerase (Vulliamy et al. 2001; Marrone et al. 2004, 2007; Armanios et al. 2005), proteins involved in telomerase biogenesis (NOP10, DKC1, and NHP2) (Heiss et al. 1998; Walne et al. 2007; Vulliamy et al. 2008; Trahan et al. 2010), and the TCAB1 factor involved in the trafficking of telomerase (Zhong et al. 2011). However, recent work has also identified mutations in shelterin components and their associated proteins in DC, HHS, RS, and Coats' plus patients (Savage et al. 2008; Walne et al. 2008, 2013; Touzot et al. 2010; Anderson et al. 2012; Ballew et al. 2013a,b; Deng et al. 2013; Le Guen et al. 2013). Notably, the second most common cause of DC is now known to arise from heterozygous mutations in the TINF2 gene, which encodes the TIN2 component of shelterin (Savage et al. 2008; Walne et al. 2008; Sarper et al. 2010). These TIN2-DC mutations are autosomal-dominant and usually arise de novo.
TIN2 is one of the six subunits of shelterin, the protein complex that protects telomeres and regulates the maintenance of telomeric DNA (Palm and de Lange 2008). TIN2 interacts with the two duplex telomeric DNA-binding proteins TRF1 and TRF2 as well as with TPP1. The binding of TIN2 to TPP1 allows the TPP1/POT1 heterodimer (TPP1/POT1a and TPP1/POT1b in mouse cells) to interact with single-stranded telomeric DNA. Due to its ability to interact with three other shelterin proteins, TIN2 plays an important role in the overall integrity of shelterin. Loss of TIN2 from mouse telomeres results in a complex phenotype that reflects the removal of the TPP1/POT1 shelterin subunits and phenotypes ascribed to partial loss of TRF2 (Takai et al. 2011). The phenotypes associated with TIN2 deletion from mouse embryo fibroblasts (MEFs) include activation of the ATM and ATR kinase signaling cascades at telomeres, an increase in the amount of ssDNA at telomere termini, and telomere fusions.
TIN2 has similarly been implicated in the stabilization of human shelterin and the protection of telomeres (Kim et al. 2004; Liu et al. 2004a; Ye et al. 2004; O'Connor et al. 2006). In addition, human TIN2 was shown to have an important role in the regulation of telomere length in telomerase-positive cells (Kim et al. 1999). Overexpression of a TIN2 truncation mutant lacking the TRF2- and TPP1-binding sites results in the rapid elongation of human telomeres and the resetting of the telomere length equilibrium. Since this TIN2 mutant does not affect telomerase expression levels and has no effect on the rate of telomere shortening in telomerase-negative human cells, its effect on telomere length homeostasis is presumed to be due to a change in telomerase recruitment or telomerase activity at telomeres. A similarly rapid telomere elongation is induced by a mutant of TPP1 that lacks the POT1-binding site and by a POT1 mutant that interacts with TPP1 but lacks the ability to bind single-stranded telomeric DNA (Loayza and de Lange 2003; Liu et al. 2004b). It was therefore proposed that telomerase is subjected to negative regulation when the TIN2–TPP1–POT1 interactions allow POT1 to engage the telomeric terminus. However, recent data have complicated this simple model, since TPP1 is also involved in the recruitment of telomerase (Abreu et al. 2010; Nandakumar et al. 2012; Zhong et al. 2012).
The majority of identified TIN2-DC mutations cluster in a highly conserved 30-amino-acid region from position 270 to 300, located eight amino acids C-terminal to the FxLxP motif that mediates binding of TIN2 to TRF1 (Chen et al. 2008; Savage and Bertuch 2010). However, despite the proximity to the TRF1-binding site, most TIN2-DC mutations do not affect the TIN2–TRF1 interaction, and the binding of the TIN2-DC mutants to telomeres appears to be normal (Yang et al. 2011; Xin and Ly 2012). The TIN2-DC mutations also do not affect the association of TIN2 with TRF2 and TPP1, which rely on the N-terminal half of TIN2 for binding. Recent data have suggested that the DC patch in TIN2 contains a binding site for HP1γ and that the loss of the TIN2–HP1γ interaction results in a cohesion defect at telomeres that contributes to diminished telomere maintenance in DC patients (Canudas et al. 2011). Another proposal is that TIN2 aids TPP1 in the recruitment of telomerase through an unknown mechanism that is disrupted by the TIN2-DC mutations, including K280E (Yang et al. 2011).
Here we report on the telomeric phenotypes associated with the mutation of lysine at position 280 of TIN2 to glutamate (K280E), the first inherited TIN2-DC mutation to be identified (Savage et al. 2008). Despite knockdown of endogenous TIN2, we failed to observe a telomere length phenotype with TIN2-K280E or other TIN2-DC mutants upon expression in human tumor cells. We therefore used gene targeting to generate a knock-in mouse strain carrying the equivalent mutation in mouse TIN2 (K267E), referred to here as TIN2-DC. Analysis of heterozygous TIN2-DC mice showed a telomere-shortening phenotype that occurred in the context of both a telomerase-proficient and a telomerase-deficient background. This result raises the possibility that some of the TIN2-DC mutations may affect telomere function in a telomerase-independent manner.
Results
No phenotypes associated with TIN2-DC mutants in HT1080 fibrosarcoma cells
We initially examined the telomere phenotypes of TIN2 K280E, TIN2 R282H, and a deletion mutant of TIN2, removing most of the DC patch by expressing these alleles in the human fibrosarcoma cell line HT1080, which has been used frequently for telomere length studies (Supplemental Fig. 1A). Consistent with previous reports (Yang et al. 2011; Xin and Ly 2012), none of the mutations affected the interaction of TIN2 with its binding partners in shelterin, TRF2, TRF1, and TPP1 (Supplemental Fig. 1B). In order to improve the detection of possibly subtle phenotypes associated with these mutations, endogenous TIN2 was depleted with an shRNA (Supplemental Fig. 1C). In cells with diminished endogenous TIN2, each of the TIN2-DC mutants localized to telomeres, as shown by immunofluorescence (IF) in conjunction with telomeric fluorescence in situ hybridization (FISH) (Supplemental Fig. 1D). Expression of these mutant alleles did not affect the growth of the cells (Supplemental Fig. 2A) and did not induce a DNA damage response at telomeres, as demonstrated by the absence of telomere dysfunction-induced foci (TIFs) (Supplemental Fig. 2B,C). Importantly, whereas the shRNA-mediated knockdown of TIN2 resulted in the expected elongation of telomeres (Ye and de Lange 2004), this phenotype was rescued to the same extent by wild-type TIN2 and the TIN2-DC mutants (Supplemental Fig. 3). Expression of the TIN2 mutants also did not result in a significant change in the telomeric overhang signals compared with expression of wild-type TIN2 (Supplemental Fig. 3).
Generation of a TIN2-DC knock-in mouse
Since we were unable to obtain insights into the effect of TIN2-DC mutations using a human cancer cell line, we generated a knock-in allele that would allow determination of the organismal, generational, and cellular phenotypes of a TIN2-DC mutant in mice. We previously created a gene targeting construct to generate a floxed TIN2 allele (Takai et al. 2011). This construct was modified to introduce a mutation in exon 6 that leads to a lysine-to-glutamate (K-to-E) change at position 267, which is the equivalent of position 280 in human TIN2 (Supplemental Fig. 4A,B). A neo gene flanked by FRT sites downstream from exon 7 served to allow selection of targeted embryonic stem (ES) cells and was later removed by expression of Flippase in the germline. As this TIN2-DC allele might be harmful to ES cells, we prevented expression of the modified gene by insertion of a STOP cassette upstream of exon 3. The transcriptional STOP cassette was flanked by LoxP sites, allowing its removal with Cre recombinase in the mouse germline or in cells. The final product of Flippase and Cre treatment is an intact TIN2 locus with the K267E mutation in exon 6 and single LoxP and FRT sites in the introns (Supplemental Fig. 4B). Sequence analysis confirmed the introduction of the mutation in exon 6 in targeted ES cells (Supplemental Fig. 4C), and genomic blotting confirmed that Cre-mediated recombination appropriately converted the locus in ES cells (Supplemental Fig. 4D). Sequencing of the RNA from Cre-treated ES cells demonstrated that an mRNA was generated carrying the expected GAG-to-AAA change in addition to wild-type TIN2 mRNA (Supplemental Fig. 4E,F).
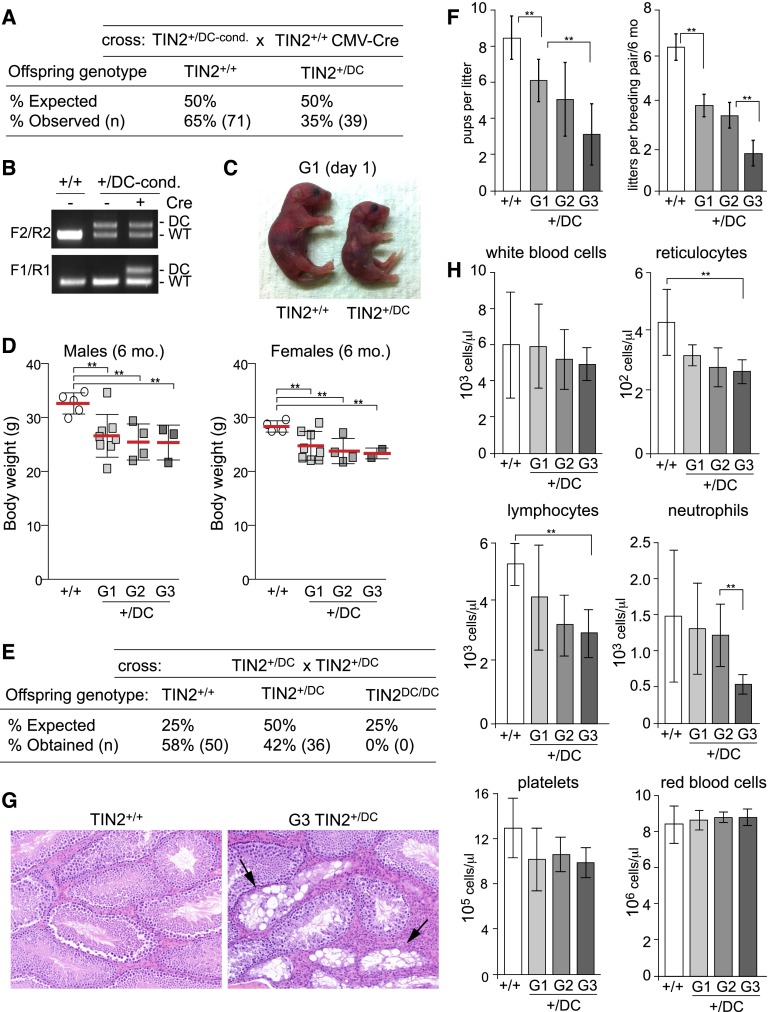
When heterozygous mice carrying the STOP cassette (TIN2+/DC-cond.) were crossed with a mouse strain expressing Cre in the germline, TIN2+/DC offspring were obtained at slightly sub-Mendelian frequency (Fig. 1A–C). The TIN2+/DC mice were smaller at birth than TIN2 wild-type littermates (Fig. 1C) and weighed slightly less than TIN2+/+ mice at weaning and at 6 mo of age (Fig. 1D; data not shown) but were otherwise healthy and fertile. However, crosses between TIN2+/DC mice failed to deliver homozygous offspring carrying only the DC allele, indicating that the TIN2DC allele is not compatible with embryonic development (Fig. 1E).
Figure 1.
TIN2-DC mice display diminished fertility and mild pancytopenia in G2/G3. (A) Genotypes of offspring from crosses of TIN2+/DC-cond. with mice expressing cytomegalovirus minimal promoter (CMV)-Cre in the germline, resulting in removal of the STOP cassette from the DC allele. (B) PCR genotyping of offspring as shown in A. (C) TIN2+/+ and TIN2+/DC pups at birth. (D) Scatter plot of the body weights of 6-mo-old male and female mice with the indicated genotypes and generations. Error bars indicate standard deviations (SDs). (**) P < 0.05 (unpaired t-tests). (E) Genotypes of offspring from intercrosses between TIN2+/DC breeding pairs. (F, left panel) Average number of pups produced per litter from breeding pairs of the indicated genotypes monitored over the course of 6 mo. (Right panel) Average number of litters produced per breeding pair of the indicated genotypes over the course of 6 mo. Error bars indicate SDs. (**) P < 0.05 (unpaired t-tests). (G) hematoxylin and eosin (H&E)-stained testis sections show testicular atrophy (arrow) in TIN2+/DC mice. (H) Average peripheral blood count values of age-matched 6-mo-old TIN2+/+ (n = 6) and TIN2+/DC (n = 6) mice with the indicated genotypes. G2 (n = 9) and G3 (n = 5) mice were generated from TIN2+/DC intercrosses. Error bars indicate SDs. (**) P < 0.05 (unpaired t-tests).
Diminished fecundity and mild pancytopenia in G2 and G3 TIN2+/DC mice
The TIN2-DC allele was associated with diminished fecundity that progressed over multiple generations, as evidenced by lower numbers of litters per breeding pair and reduced litter size (Fig. 1F). The lowered fecundity of G2 and G3 TIN2+/DC mice may in part be due to a defect in spermatogenesis, since two out of 10 TIN2-DC males examined displayed partial testicular atrophy, with a subset of seminiferous tubules showing complete atrophy (Fig. 1G; Supplemental Fig. 5).
The TIN2+/DC mice also developed a hematopoietic defect that increased in severity in later generations. Although histological examination of the bone marrow did not reveal abnormalities in most of the G2 and G3 mice analyzed, circulating peripheral blood showed evidence of mild pancytopenia affecting all hematopoietic lineages. Notably, there was a significant decrease in the numbers of reticulocytes, total lymphocytes, and neutrophils (Fig. 1H). There also was a trend toward lower numbers of platelets in G3, but this change was not statistically significant. The red blood cell count was unaffected, possibly due to extramedullary hematopoiesis. In addition, examination of G2 and G3 TIN2+/DC mice revealed myeloid hyperplasia in the bone marrow of one mouse, follicular hyperplasia in the lymph nodes of two mice, incidental nervous system abnormalities in three mice (seizures, focal gliosis in the spinal cord, and hydrocephalus), and two cases of skin dermatitis of unknown etiology (Supplemental Fig. 5). None of these phenotypes was observed in the TIN2+/+ and G1 TIN2+/DC mice examined (12 total).
Generational telomere shortening in TIN2-DC mice
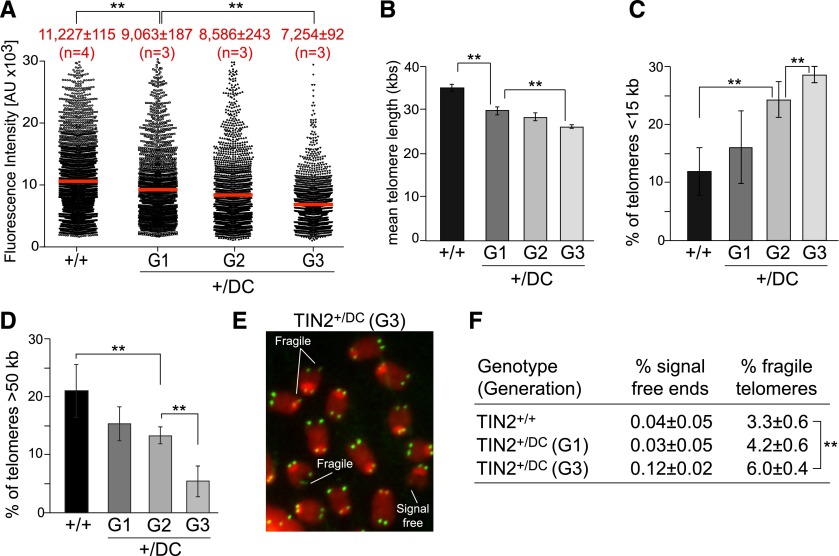
The generational anticipation of the hematological defects in heterozygous TIN2+/DC mice could be explained if the telomeres gradually shortened over the multiple generations, ultimately diminishing telomere function in the bone marrow of G3 TIN2+/DC mice. To examine possible telomere length changes in the bone marrow of G1–G3 TIN2+/DC mice, quantitative FISH (Q-FISH) was performed. The Q-FISH analysis was calibrated using an internal standard of human HeLa cells with known telomere length so that the telomeric FISH signals in different samples could be compared directly. The Q-FISH data revealed that the telomeres of TIN2+/DC mice underwent gradual and progressive shortening over three generations (Fig. 2A). The telomeres in the bone marrow of G3 TIN2+/DC mice were significantly shortened (by ∼25%) compared with wild-type TIN2 or G1 TIN2+/DC mice (Fig. 2B). The class of telomeres longer than 50 kb diminished in the later generations, and there was a concomitant increase in the fraction of telomeres shorter than 15 kb (Fig. 2C,D). There was also a slight increase in chromosome ends lacking detectable telomeric signals and a significant increase in the incidence of fragile telomeres in the bone marrow of G3 TIN2+/DC mice (Fig. 2E,F). This loss of telomeric DNA is likely the cause of the mild pancytopenia in the G2 and G3 TIN2+/DC mice, since a similar phenotype was noted in a mouse engineered to undergo rapid telomere shortening (Hockemeyer et al. 2008; He et al. 2009).
Figure 2.
Progressive telomere shortening in the bone marrow of heterozygous TIN2-DC mice. (A) Scatter plot of telomere fluorescence units (TFUs) determined by Q-FISH analysis of metaphases isolated from the bone marrow of 3-mo-old littermates with the indicated genotypes. Mean TFU values (red bars) and standard errors of the mean (SEMs) are shown. (**) P < 0.05 (unpaired t-tests). The number of mice analyzed per group (n) is denoted. (B) Graph of the mean telomere lengths in kilobases from Q-FISH analysis (as shown in A) for the indicated genotypes. Telomere length was calculated based on an internal standard of known telomere length (HeLa1.3 cells). Error bars indicate the SEM. (**) P < 0.05 (unpaired t-tests). (C) Quantification of the percentage of telomeres shorter than 15 kb from the bone marrow of mice analyzed in A. Error bars indicate SDs. (**) P < 0.05 (unpaired t-tests). (D) Quantification of the percentage of telomeres longer than 50 kb from the bone marrow of mice analyzed in A. Error bars indicate SDs. (**) P < 0.05 (unpaired t-tests). (E) Examples of telomere abnormalities observed in the bone marrow of G3 TIN2+/DC mice. (F) Quantification of telomeric abnormalities detected in metaphases isolated from the bone marrow of mice with the indicated genotypes. Averages and SDs were calculated from the analysis of individual mice (3000–4000 telomeres per genotype). (**) P < 0.05 (unpaired t-tests).
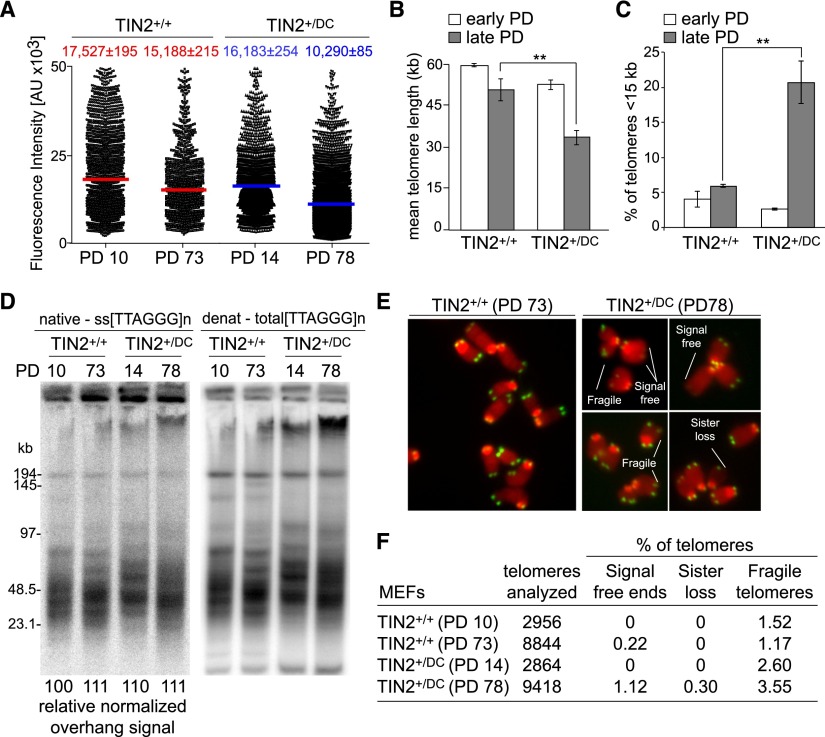
The accelerated telomere shortening in the TIN2+/DC setting was further confirmed by analysis of MEFs derived from littermate embryos of either the TIN2+/+ or TIN2+/DC-cond. genotype (Fig. 3; Supplemental Fig. 6). The MEFs were treated with Cre to remove the STOP cassette from the TIN2DC-cond allele, generating TIN2+/DC MEFs (Supplemental Fig. 6A). MEFs expressing the DC allele of TIN2 had the same growth rate as TIN2+/+ MEFs (Supplemental Fig. 6B), and immunoblotting and chromatin immunoprecipitation (ChIP) analysis did not reveal a difference in the expression or telomeric association of shelterin components between TIN2+/+ and TIN2+/DC MEFs (Supplemental Fig. 6C–G).
Figure 3.
Progressive telomere shortening in heterozygous TIN2-DC MEFs. (A) Scatter plot of TFU values from Q-FISH analysis of metaphases isolated from the indicated MEFs at the indicated PDs. Mean TFU values and SEMs are shown. Colored bars indicate mean TFU values, respectively. (B) Graph of the mean telomere lengths from Q-FISH analysis (as shown in A) in kilobases for the indicated MEFs calculated as in Figure 2B. Error bars indicate SEMs. (**) P < 0.05 (unpaired t-tests). (C) Quantification of the percentage of telomeres shorter than 15 kb from MEFs analyzed in A at early and late PDs. (**) P < 0.05 (unpaired t-tests). (D) Telomeric DNA analysis of TIN2+/+ or TIN2+/DC MEFs at the indicated PDs. (Left) Gel of native telomeric restriction fragments revealing s telomeric DNA. (Right) The same gel rehybridized after in situ denaturation of the DNA. Quantification of overhang signals were normalized to total telomeric signals and compared with TIN2+/+ MEFs at PD 10. (E) Examples of telomere abnormalities in metaphases of TIN2+/+ or TIN2+/DC MEFs at late PDs. (F) Quantification of telomeric abnormalities detected in metaphases of TIN2+/+ or TIN2+/DC MEFs at the indicated PDs.
Q-FISH analysis revealed a minor telomere-shortening phenotype in the TIN2+/+ MEFs. Although this telomere shortening might be unexpected given that the cells are telomerase-proficient, we observed slight variations in telomere dynamics in telomerase-positive mammalian cells in the past. In comparison with the TIN2+/+ MEFs, the telomeres of the TIN2+/DC MEFs shortened significantly (Fig. 3A,B). In addition, TIN2+/DC MEFs showed a significant increase in the class of telomeres shorter than 15 kb (Fig. 3C). In contrast, the telomeric overhang signals were not altered by the expression of TIN2DC (Fig. 3D). Consistent with the data on the bone marrow of TIN2+/DC mice, the TIN2+/DC MEFs showed a slight increase in the incidence of signal-free ends and fragile telomeres, especially at later population doublings (PDs) (Fig. 3E,F). Together, these data demonstrate that the TIN2-DC allele accelerates telomere shortening both in vivo and in vitro.
TIN2-DC affects telomeres in mTR-deficient mice and cells
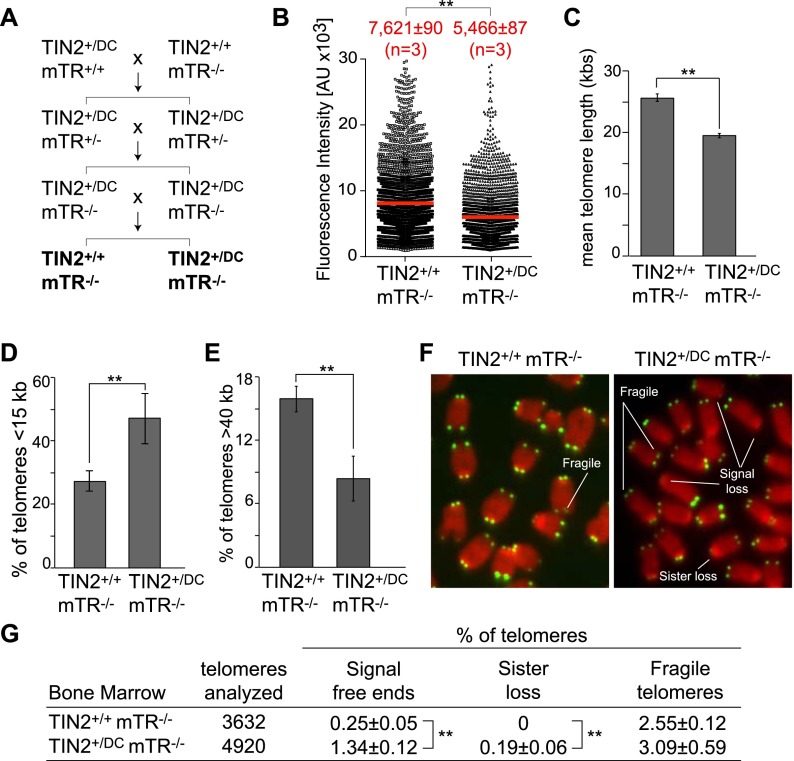
Since the majority of DC mutations affects genes in the telomerase pathway, it was anticipated that the telomere shortening in the TIN2+/DC mice is due to a defect in telomerase-mediated telomere maintenance. This mechanism would predict that the TIN2DC allele would not have an effect in the absence of telomerase. We tested this prediction by generating TIN2+/DC mTR−/− mice (Fig. 4A), which lack telomerase activity due to the deletion of the gene encoding the telomerase RNA component (Blasco et al. 1997). Interestingly, even in this telomerase-deficient setting, telomeres in the bone marrow of TIN2+/DC mTR−/− mice were significantly shorter than those of mTR−/− littermates carrying two copies of wild-type TIN2 (Fig. 4B,C). Furthermore, the class of telomeres shorter than 15 kb was significantly increased, while the subfraction of longer telomeres showed a concomitant decrease (Fig. 4D,E). Finally, the bone marrow telomeres of the TIN2+/DC mTR−/− mice showed a significant increase in chromosome ends without telomeric signals or signal loss from one of the two sisters (Fig. 4F,G).
Figure 4.
Accelerated telomere shortening in the bone marrow of TIN2+/DCmTR−/− mice. (A) Breeding scheme for the generation of TIN2+/DCmTR−/− mice. (B) Scatter plot of TFU values from Q-FISH analysis of metaphases isolated from the bone marrow of 3-mo-old littermates with the indicated genotypes. Mean TFU values (red bars) and SEMs of the indicated number of mice analyzed (n) are shown. (**) P < 0.05 (unpaired t-tests). (C) Graph of the mean telomere lengths from Q-FISH analysis (as shown in B) for the indicated genotypes. Error bars indicate the SEM. (**) P < 0.05 (unpaired t-tests). (D) Quantification of the percentage of telomeres shorter than 15 kb from the bone marrow of mice analyzed in B. Error bars indicate SDs. (**) P < 0.05 (unpaired t-tests). (E) Quantification of the percentage of telomeres longer than 40 kb from the bone marrow of mice analyzed in A. Error bars indicate SDs. (**) P < 0.05 (unpaired t-tests). (F) Examples of telomere abnormalities observed in metaphases isolated from the bone marrow of TIN2+/+ mTR−/− and TIN2+/DC mTR−/− mice. (G) Quantification of telomere abnormalities detected in metaphases isolated from the bone marrow of mice analyzed for Q-FISH. Averages were calculated from the analysis of individual mice (n = 3; 3000–4000 telomeres per genotype) and SD. (**) P < 0.05 (unpaired t-tests).
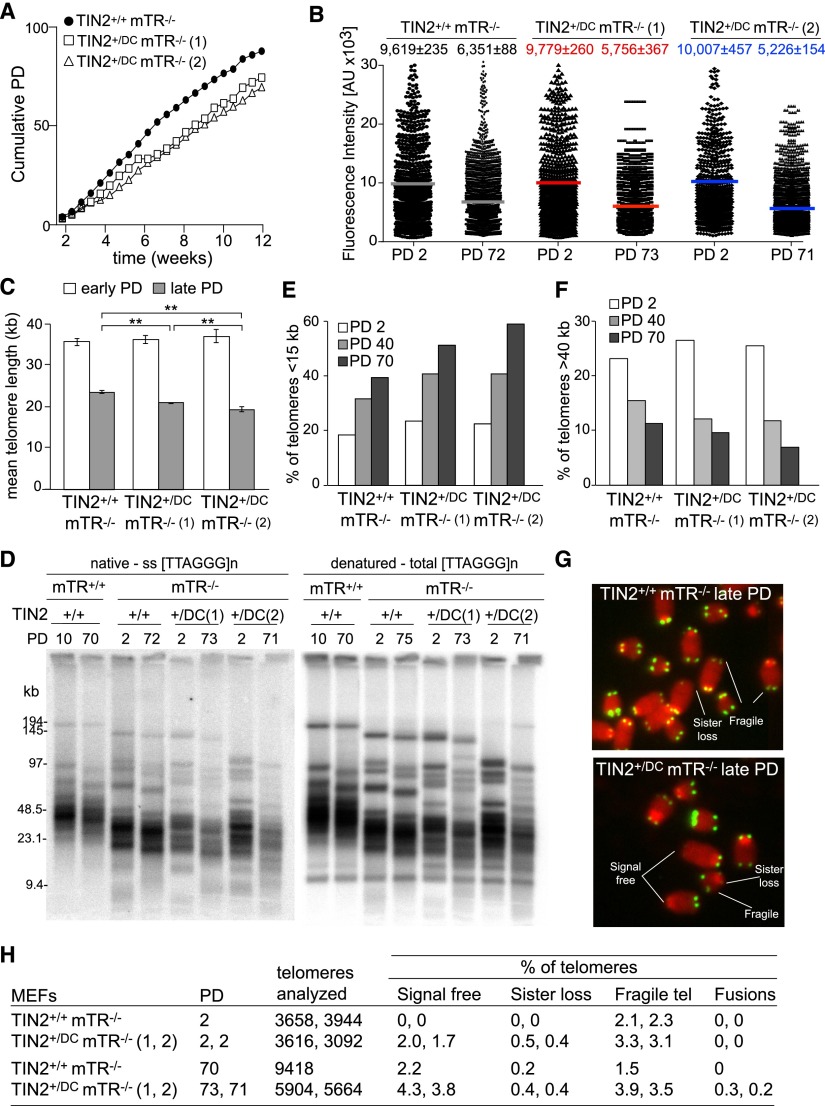
To confirm that the TIN2-DC allele accelerates telomere shortening in a telomerase-negative setting, we generated mTR−/− MEFs that were either TIN2+/DC or TIN2+/+ (Fig. 5). The MEFs were derived from littermates, immortalized with SV40 large-T antigen, and propagated for ∼70 PDs (Fig. 5A). TIN2+/DC mTR−/− MEFs proliferated at a slower rate (∼75%) than TIN2+/+ mTR−/− MEFs. Furthermore, morphological examination and staining for β-galactosidase showed evidence of senescence in a small fraction of the TIN2+/DC mTR−/− cells (Supplemental Fig. 7). Q-FISH measurements indicated that at a comparable PD, both TIN2+/DC mTR−/− MEFs had sustained greater telomere shortening than the TIN2+/+ mTR−/− MEFs, with the second TIN2+/DC mTR−/− MEF line [indicated with (2) in Fig. 5] showing the greatest reduction in mean telomere length (Fig. 5B,C). A slight difference in telomere shortening rates with greater accumulation of very short telomeres in the TIN2+/DC mTR−/− MEFs at late PDs was also apparent from examination of fractionated telomeric restriction fragments (Fig. 5D). Furthermore, the Q-FISH analysis indicated that the TIN2+/DC mTR−/− MEFs accumulated a greater number of telomeres shorter than 15 kb at late PDs and a decrease in the fraction of telomeres longer than 40 kb (Fig. 5E,F). Finally, the metaphase spreads showed an increased frequency of chromosome ends lacking telomeric signals and fragile telomeres at later PDs (Fig. 5G,H). The greater incidence of very short telomeres in the TIN2+/DC mTR−/− MEFs may explain why these cells grow slower and occasionally undergo senescence.
Figure 5.
Accelerated telomere shortening in TIN2+/DC mTR−/− MEFs. (A) Growth curve of TIN2+/+ mTR−/− and TIN2+/DC mTR−/− MEFs. (B) Scatter plot of TFU values from Q-FISH analysis of metaphases isolated from the indicated MEFs at the indicated PDs. Mean TFU values (gray, red, and blue bars) and SEMs are shown. (C) Graph of the mean telomere lengths from Q-FISH analysis (as shown in A) for the indicated MEFs. Error bars indicate the SEM. (**) P < 0.05 based on unpaired t-tests. (D) Telomere length analysis of MEFs with the indicated genotypes at the indicated PDs. (Left) Gel of native telomeric restriction fragments hybridized to a probe for the telomeric C strand. (Right) The same gel rehybridized after in situ denaturation of the DNA. (E) Quantification of the percentage of telomeres shorter than 15 kb from MEFs analyzed in B at the indicated PDs. (F) Quantification of the percentage of telomeres longer than 40 kb from MEFs analyzed in B at indicated PDs. (G) Examples of telomere abnormalities observed in metaphases isolated from TIN2+/+ mTR−/− and TIN2+/DC mTR−/− MEFs at late PDs. Telomeres were detected by FISH. (H) Quantification of telomere abnormalities detected in metaphases of TIN2+/+ mTR−/− or TIN2+/DC mTR−/− MEFs at the indicated PDs.
These data reveal that even in the mTR-deficient background, the TIN2-DC allele leads to greater telomere shortening in TIN2+/DC MEFs compared with the TIN2+/+ MEFs examined after the same number of PDs. Since the TIN2-DC allele is associated with a mild growth defect (Fig. 5A), it is possible that at a given PD, the TIN2-DC-containing cells have undergone a greater number of cell divisions. Therefore, the data do not establish whether the TIN2-DC allele leads to a greater rate of bulk telomere shortening per cell division. However, given that the TIN2+/DC mTR−/− MEFs show a greater incidence of chromosome ends lacking telomeric signals at PD 2, we consider it likely that the TIN2-DC allele leads to stochastic losses of telomeric DNA. Such telomere loss events can increase the overall shortening rate of the telomeres per cell division; lead to a skewing of the telomeric length distribution to a shorter length, as we observed (Fig. 5E,F); and result in poor growth of the cells (Fig. 5A). Thus, the data indicate that TIN2-DC allele has an effect on telomere maintenance in a telomerase-deficient setting.
TIN2-DC induces mild ATR-dependent telomere damage signaling
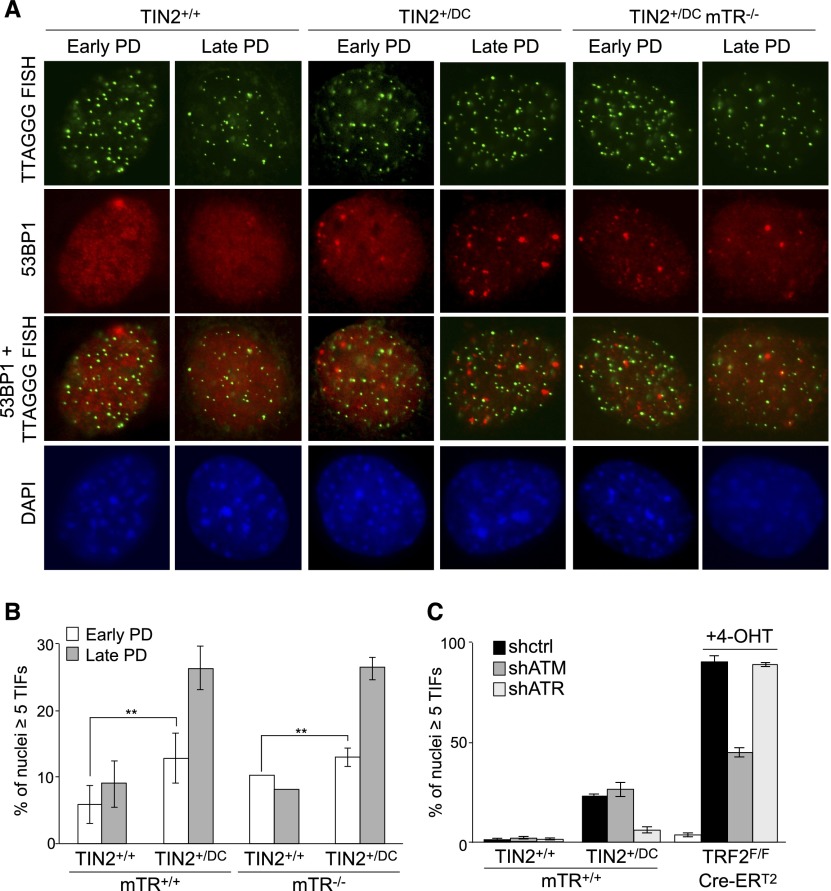
Given that the TIN2-DC allele had a telomerase-independent effect on telomeres, we sought to determine whether this TIN2 mutant induced telomere damage signaling in the MEFs described above. Expression of the TIN2-DC allele resulted in a slight but significant increase of TIF frequency at early PDs, and this effect was seen in both mTR-proficient and mTR-deficient MEFs (Fig. 6A,B). The frequency of TIF-positive cells increased at later PDs, suggesting that the shortening of the telomeric DNA aggravated the subtle telomere dysfunction phenotype.
Figure 6.
An ATR-dependent telomere damage response induced by the TIN2-DC allele. (A) IF for 53BP1 (red) and telomeric FISH (green) in TIN2+/+, TIN2+/DC, and TIN2+/DC mTR−/− MEFs at early and late PDs. DNA was stained with 4,6-diamino-2-phenylindole (DAPI; blue). (B) Quantification of TIF-positive nuclei (five or more 53BP1 TIFs per nucleus; n > 200) in TIN2+/+, TIN2+/DC, and TIN2+/DC mTR−/− MEFs. Averages and SDs are from three independent experiments. (**) P < 0.05 (paired Student's t-test). (C) Quantification of TIF-positive nuclei (scored as in B) in mTR-proficient TIN2+/+ and TIN2+/DC MEFs after transduction with shRNAs for ATM, ATR, or a negative control. TRF2F/F MEFs were included as a positive control for ATM inhibition. Cre (Cre-ERT2 in the Rosa26 locus) was induced with tamoxifen (4-OHT). Averages and SEMs are from two independent experiments.
The telomeric DNA damage response observed at early PDs in the TIN2+/DC MEFs was primarily due to ATR kinase signaling, as TIF formation was strongly decreased upon the treatment of cells with an shRNA to the ATR kinase (Fig. 6C). In contrast, an shRNA to the ATM kinase, which resulted in the expected reduction of TIFs in cells lacking TRF2, did not have an effect on the TIF response in the TIN2+/DC cells (Fig. 6C).
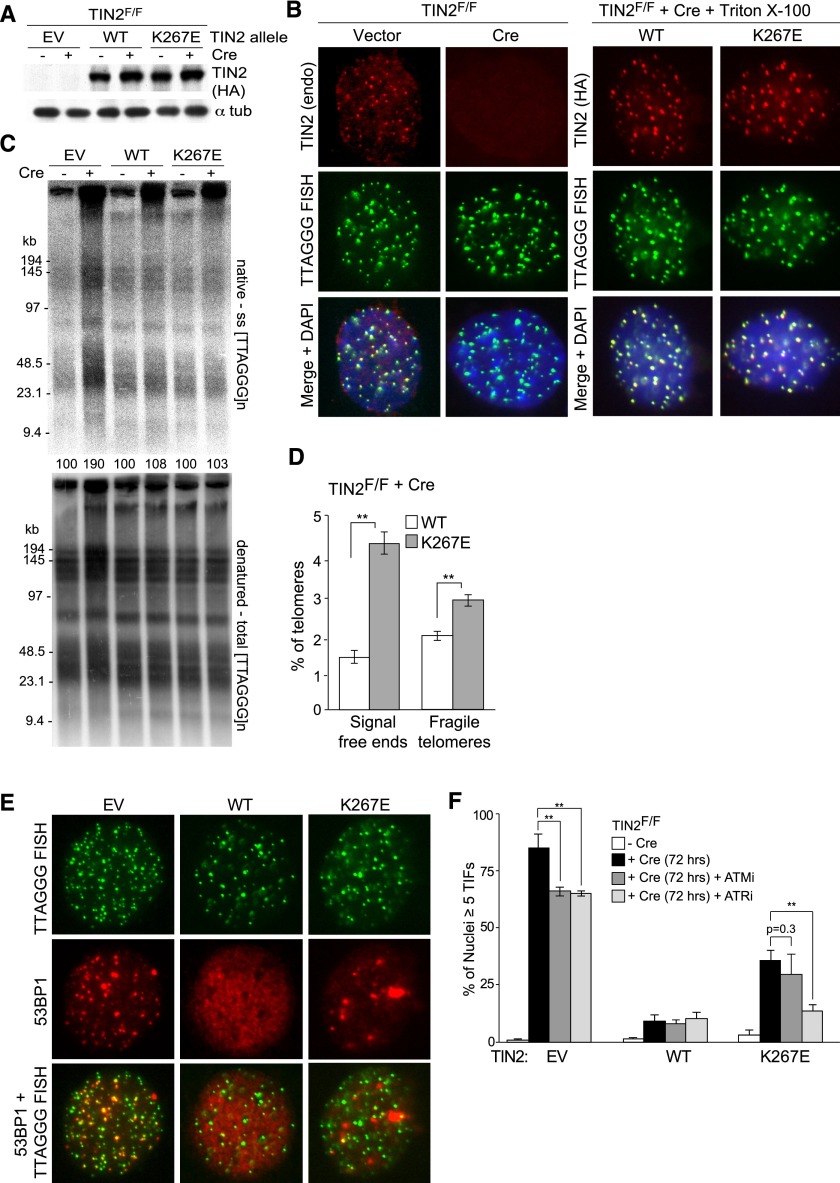
To further examine the immediate telomere dysfunction phenotypes associated with TIN2-DC expression, we introduced this allele into TIN2F/F MEFs and then deleted the endogenous TIN2 with Cre (Fig. 7A,B). Cells expressing wild-type TIN2 and cells infected with the empty vector were analyzed in parallel. As expected, the TIN2-DC allele did not have an immediate effect on the length of the telomeres in these telomerase-proficient MEFs and, consistent with the data above, was capable of repressing the increase in the overhang signal associated with TIN2 deletion (Fig. 7C). While the TIN2-DC mutant was also able to repress the telomere fusions associated with TIN2 deletion (Fig. 7C; data not shown), MEFs expressing the TIN2-DC mutant showed an increase in the incidence of signal-free ends and fragile telomeres (Fig. 7D). However, the TIN2-DC allele showed a diminished ability to repress the telomere damage signaling in TIN2-deficient cells. Interestingly, whereas both the ATM and ATR kinases are activated in the absence of TIN2, the TIN2-DC allele primarily failed in the repression of ATR-dependent telomere damage signaling (Fig. 7D,E). Thus, in addition to its effect on the maintenance and structure of the telomeric DNA, the TIN2-DC allele causes a subtle telomere damage signaling phenotype.
Figure 7.
An ATR-dependent telomere damage response in TIN2-deficient cells exogenously expressing the TIN2-K267E allele. (A) Immunoblots of cell extracts from TIN2F/F MEFs expressing TIN2 alleles or empty vector (EV) with or without Cre treatment (72 h). (B, left panel) Loss of endogenous TIN2 as observed by IF-FISH from TIN2F/F MEFs with or without Cre treatment (72 h). (Right panel) IF for Flag-HA-HA (FH2)-TIN2 alleles (anti-HA antibody; red) and telomeric FISH (green) following Triton X-100 extraction of soluble proteins in TIN2−/− MEFs (72 h post-Cre). (C) In-gel hybridization assay for single-stranded (ss) telomeric DNA after TIN2 deletion in cells expressing empty vector (EV), TIN2, or TIN2-K267E. (Top panel) TelC signals under the native condition. (Bottom panel) The same gel rehybridized after in situ denaturation of the DNA. Quantifications of overhang signals were normalized to total telomeric signals and compared with TIN2F/F MEF samples without Cre. (D) Quantification of telomere abnormalities detected in TIN2−/− MEFs (72 h post-Cre) expressing FH2-TIN2 alleles. Average values are from three independent experiments (3000–4000 telomeres per experiment) and SDs. (**) P < 0.05 (paired Student's t-test). (E) IF of 53BP1 (red) and telomeric FISH (green) in TIN2−/− MEFs (72 h post-Cre) expressing empty vector (EV) or FH2-TIN2 alleles. DNA was stained with DAPI (blue). (F) Quantification of TIF-positive nuclei (five or more 53BP1 foci at telomeres; n ≥ 200) in TIN2F/F MEFs transduced with an empty vector, wild-type TIN2, or TIN2-K267E and treated with ATM and ATR inhibitors (ATMi or ATRi, respectively) after Cre (72 h). Average values are from three independent experiments and SDs. (**) P < 0.05 (paired Student's t-test).
Discussion
The data presented here reveal the phenotypes associated with one of the TIN2 mutations that cause DC. Although we were unable to find any changes in telomere structure and function upon expression of this and other TIN2-DC mutants in a human cancer cell line, a knock-in of the mouse equivalent to the TIN2-DC K280E mutation conferred an obvious telomere maintenance defect. Mice heterozygous for the TIN2-DC allele showed gradual telomere shortening in telomerase-proficient mice and MEFs. After several generations, the shortening of the telomeres resulted in diminished fecundity and indices of pancytopenia that resembled a mild form of the hematological defects in DC patients. Importantly, the shortening of the telomeres was not (solely) due to a defect in telomerase-mediated telomere maintenance, since heterozygosity for TIN2-DC also resulted in accelerated telomere shortening in mice and in cells that lacked the RNA component of telomerase. Thus, this TIN2-DC mutant can exert a telomerase-independent effect on telomere maintenance that potentially contributes to the pathogenesis of DC.
DC-relevant phenotypes associated with TIN2-DC
The analysis of mice and mouse cells heterozygous for the TIN2-DC allele uncovered a number of subtle but important deficiencies associated with the mutant form of this shelterin component. The presence of one copy of TIN2-DC resulted in generational telomere shortening that led to organismal phenotypes in later generations, in keeping with the generational disease anticipation observed in DC (Vulliamy et al. 2004). Whereas the health of the G1 TIN2+/DC mice was not significantly affected, G3 mice suffered from diminished representation of lymphocytes, neutrophils, and reticulocytes in the periphery. In addition, the mice became less fertile and suffered from incidental nervous system defects. The latter phenotype is noteworthy because of one case report of a TIN2 R282H mutation associated with pancytopenia and ataxia (Tsangaris et al. 2008). We anticipate that later generations of these TIN2+/DC mice will present with even more extensive loss of fertility and bone marrow defects as well as failure of other organs, as was first noted in the mTR-deficient mice (Rudolph et al. 1999).
Notably, the data obtained with mice and MEFs of the TIN2+/DC mTR−/− genotype argue that the telomere length defect of the TIN2-DC allele is not solely due to a deficiency in telomerase-mediated telomere maintenance. The TIN2+/DC mTR−/− MEFs showed TIFs, poor proliferation, and evidence of senescence. Furthermore, even at early PDs, the TIN2+/DC mTR−/− MEFs showed a higher incidence of chromosome ends lacking telomeric signals and a skewing of the telomere length distribution toward shorter telomeres, suggestive of stochastic deletions of large blocks of telomeric DNA.
Although we were not able to determine the exact nature of the telomere maintenance defect associated with TIN2-DC, it is noteworthy that in both mTR-proficient and mTR-deficient contexts, the frequency of signal-free chromosome ends, fragile telomeres, and ATR signaling is slightly increased. The frequency of these events is too low for a detailed molecular analysis (e.g., two-dimensional [2D] gel electrophoresis to monitor t-loop cleavage) (Wang et al. 2004), hampering a full understanding of the molecular defect conferred by the TIN2-DC mutation. Since ATM signaling and nonhomologous end-joining (NHEJ) remain repressed at the telomeres, the function of TRF2 in shelterin is not likely to be compromised. However, the involvement of the ATR kinase pathway in the TIN2-DC telomere dysfunction phenotype provides an important hint. ATR kinase signaling was observed at telomeres in two settings. First, when the POT1 proteins or TPP1 are removed, the ATR kinase is activated at the telomeric termini. It is possible that the TIN2-DC mutations affect the function of the TPP1/POT1 heterodimers in a subtle way that is not detectable based on ChIP analysis, protein–protein interactions, or the telomeric overhang phenotype associated with POT1b loss. Second, ATR kinase signaling is activated when TRF1 is removed from telomeres. This signaling is proposed to be a consequence of replication problems in the telomeric DNA when TRF1 is absent. It is therefore possible that the TIN2-DC mutation compromises TRF1 function or other aspects of DNA replication at telomeres. Consistent with this mechanism, TIN2-DC expression is associated with a mild fragile telomere phenotype, an index of telomere replication problems. Interestingly, a second factor proposed to be required for efficient replication of telomeric DNA, RTEL1 (Sfeir et al. 2009; Vannier et al. 2012), was recently reported to be mutated in DC and HHS (Ballew et al. 2013b; Deng et al. 2013; Le Guen et al. 2013; Walne et al. 2013). In some of these cases, higher levels of TIFs, signal-free ends, and/or fragile telomeres were noted, consistent with the data reported here. However, it is not excluded that other processes that can compromise telomere integrity, including changes in the recombination at telomeres (Neumann et al. 2013), play a role in the phenotypes of the TIN2-DC mutation.
Modeling DC in mice
Our attempts at discerning a phenotype associated with TIN2-DC mutants using a human fibrosarcoma cell line failed. Unlike a previous report (Yang et al. 2011), we did not observe a significant change in telomere dynamics. We also did not detect a telomere DNA damage response in cells expressing this and other DC-associated TIN2 mutants even when the endogenous TIN2 was depleted. Two aspects of mutant expression experiments using a telomerase-positive tumor cell line may be to blame for the lack of detectable phenotypes. First, the high level of telomerase activity in the HT1080 cell line may have masked subtle differences in telomere length dynamics imposed by the mutant allele. Second, the overexpression of the mutant alleles, although mild (approximately fivefold), might mask the phenotypes associated with these TIN2 alleles. Thus, this study underscores the advantages of using mouse modeling for the understanding of the etiology of DC and potentially other telomeropathies. This mouse model for DC and the previously generated POT1b-deficient mouse, in which accelerated telomere shortening generates DC-related phenotypes (Hockemeyer et al. 2008), should be useful for the further understanding of this disease.
Materials and methods
Generation of TIN2-DC conditional mice and mouse strains
To generate the TIN2-K267E (TIN2DC) targeting vector, sections of the targeting vector used to generate TIN2F/F mice (Takai et al. 2011) were subcloned into peGFP-N1 for modification (Clontech). Site-directed mutagenesis (QuikChange II XL site-directed mutagenesis kit, Agilent Technologies) was used to remove the LoxP sites between exons 2 and 3 and exons 7 and 8, respectively, to introduce the K267E mutation in exon 6 (CATAAAGAG to CATGAGGAG) and place a unique SalI restriction site between exons 2 and 3. A LoxP-flanked STOP cassette (Addgene plasmid 11584) (Jackson et al. 2001) was introduced via the SalI restriction site introduced between exons 2 and 3. All introduced mutations and deletions were confirmed by DNA sequencing. K267E mutation was confirmed by amplification of exon 6 (forward, 5′-GAGAGCTCTACTTCTGGCTCAGGGAATCTG-3′; and reverse, 5′-GGAACTTCGGAATTCATTTCCAGGACAGCC-3′) and a sequencing primer (5′-CAGCATTCCACAGTCCTCTG-3′). The modified sections were subsequently subcloned into the original pBluescript II SK− vector backbone vector. Targeted ES cell clones from albino C57BL/6J were screened by genomic blotting of BgIlI-digested DNA and injected into C57BL/6J blastcysts. Two ES cell clones were used to generate chimaeras. Mice carrying the TIN2-K267E allele were generated by breeding TIN2-K267E-neo mice to C57BL/6J FLPe deleter mice (The Jackson Laboratory). Genotyping PCR was performed using standard DNA isolation techniques and Takara Taq polymerase. We used TIN2 wild-type and K267E allele genotyping PCR using primers F2 (5′-ACACTCATCACCGCCTCTTAC-3′) and R2 (5′-TGGTAATAACAGAGCTACACG-3′) at 30 cycles (30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C). To delete the LoxP-flanked STOP cassette, we used female transgenic C57BL/6 animals expressing Cre recombinase under the transcriptional control of a human cytomegalovirus minimal promoter (CMV). To confirm deletion of the STOP cassette, genotyping PCR was performed using standard DNA isolation techniques and Takara Taq polymerase. We used TIN2 wild-type and K267E allele genotyping PCR using primers F1 (5′-CGCCTGTGTATGAGCCTAAAG-3′) and F2 (5′-TCCTCTGACAGGTGCTTCACA-3′) at 30 cycles (30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C). Intercrosses with TIN2+/+ mTR−/− (maintained as mTR+/− stocks) (Blasco) were performed to obtain TIN2+/DCmTR−/− mice.
Cell lines
HT1080, HeLa1.3, 293T, and SV40-LT immortalized TIN2F/F cells were previously published (Celli et al. 2006; Takai et al. 2010, 2011) and maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with L-glutamine, penicillin–streptomycin, nonessential amino acids, and 10% Hyclone BCS serum. Primary TIN2+/+, TIN2+/DC-cond., TIN2+/+mTR−/−, and TIN2+/DCmTR−/− MEFs were isolated from embryonic day 13.5 (E13.5) embryos and immortalized by retroviral infection with pBabeSV40-LT (a gift from Greg Hannon). Immortalized MEFs were grown in DMEM supplemented with L-glutamine, penicillin–streptomycin, nonessential amino acids, and 15% fetal bovine serum (Invitrogen). Sodium pyruvate and 2-mercaptoethanol were added to medium for primary MEFs. To delete the STOP cassette, TIN2+/DC-cond. MEFs were infected with pWZL-hygromycin retroviruses expressing Cre recombinase and selected with hygromycin for 2–4 d. RT–PCR analysis was conducted on total RNA isolated from MEFs: forward primer (spanning exons 5 and 6) (5′-TGGACATTTCCAGAGAGCTCTACT-3′) and reverse primer (spanning exons 6 and 7) (5′-TTCACACTGTTCTCCTGCTCTGCA-3′). For ChIP analysis, the hygromycin-selected MEFs were subsequently infected with pLPC-puromycin retroviruses expressing Flag-HA-HA (FH2)-tagged TPP1 or MYC-tagged POT1a and selected with puromycin for 2–4 d. TIN2F/F MEFs were infected with pWZL-hygromycin retroviruses expressing FH2-tagged mouse TIN2 alleles or an empty vector and selected with hygromycin for 2–4 d. Human HT1080 fibrosarcoma cells were first infected with pWZL-hygromycin retroviruses expressing shRNA-insensitive FH2-TIN2 alleles or an empty vector. TIN2i constructs possess silent mutations to generate shRNA resistance (AGACAATATGGTGTGGACAT to AGGCAGTACGGCGTGGACAT). Following selection with hygromycin, HT1080 cells were infected with a previously characterized shRNA (puromycin-resistant lentiviral vector) against human TIN2 and selected with puromycin (Takai et al. 2011). For immunoprecipitation studies, 293T cells were transiently cotransfected with MYC-tagged TRF1, untagged TRF1 and TRF2 plasmids (pLPC-based), and FH2-tagged human TIN2 alleles (pLPC-based) using the calcium phosphate coprecipitation method. All introduced mutations and deletions in TIN2 were made using a QuikChange II site-directed mutagenesis kit (Agilent Technologies) and confirmed by DNA sequencing. The shRNAs used to inhibit the ATM and ATR kinases were described previously (Denchi and de Lange 2007; Wu et al. 2010). The ATM inhibitor (ATMi) was KU55933 (TOCRIS Biosciences), used at 2.5 μM. The ATR inhibitor (ATRi) was ETP46464 (Toledo et al. 2011), used at 1.0 μM. ATMi and ATRi were added to the TC medium 24 h after the Cre infection and 48 h before the analysis of the TIF phenotype at 72 h post-Cre.
Immunoblotting
Cell extracts were made from MEFs using 450 mM NaCl lysis buffer (50 mM Tris-HCl at pH 7.4, 1% Triton X-100, 0.1% SDS, 450 mM NaCl, 1 mM EDTA, 1 mM DTT) and protease inhibitors. Lysates were centrifuged at 13,200 rpm for 10 min. Protein concentrations from supernatants were determined using the Bradford protein assay (Bio-Rad). Samples were suspended in 2× sample buffer (75 mM Tris-Cl at pH 6.8, 10% glycerol, 2% SDS, 0.05% bromophenol blue, 2.5% β-mercaptoethanol) prior to loading on 8% or 10% SDS-PAGE gels and then transferred to nitrocellulose membranes in transfer buffer (25 mM Tris, 0.192 M glycine, 20% methanol). After blocking with 5% nonfat dry milk in PBST (PBS/0.1% Tween 20) for 20 min at room temperature, membranes were incubated in PBST for 1 h with the following primary antibodies: rabbit polyclonal anti-mTRF1 antibody (#1449), rabbit polyclonal anti-mTRF2 antibody (#1254), rabbit polyclonal anti-mRap1 antibody (#1252), rabbit polyclonal anti-mPOT1a antibody (#1220), rabbit polyclonal anti-mPOT1b antibody (#1222), rabbit polyclonal anti-hTRF1 antibody (#370), rabbit polyclonal anti-hTRF2 antibody (#647), mouse monoclonal anti-α-tubulin (Sigma-Aldrich), mouse monoclonal anti-MYC antibody (9E10, Calbiochem), and mouse monoclonal anti-HA antibody (12CA5; Roche). Membranes were developed with enhanced chemiluminescence (ECL) (Amersham).
Coimmunoprecipitation
Cell extracts were made from 293T cells using 150 mM NaCl lysis buffer (50 mM Tris-HCl at pH 7.4, 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) and protease inhibitors. Lysates were centrifuged at 13,200 rpm for 10 min. One percent of the supernatant was taken to serve as input. Following preclearing with protein G beads (GE Healthcare) for 30 min at 4°C, supernatants were incubated with anti-HA Affinity Matrix (Roche) overnight at 4°C on a rotator wheel. Following three 10-min washes with lysis buffer, beads were pelleted by centrifugation and resuspended in 2× sample buffer (75 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS, 0.05% bromophenol blue, 2.5% β-mercaptoethanol) prior to loading on 8% or 10% SDS-PAGE gels and then transferred to nitrocellulose membranes in transfer buffer (25 mM Tris, 0.192 M glycine, 20% methanol). Membranes were immunoblotted as described above.
Indirect IF
Cells were grown on coverslips and fixed with 2% paraformaldehyde (PFA) in PBS for 10 min. Telomeric DNA FISH was combined with IF as described previously (Denchi and de Lange 2007) using the following antibodies: polyclonal rabbit anti-mTIN2 (#1447), polyclonal rabbit anti-human 53BP1 antibody NB 100-304 (Novus), and anti-HA antibody (12CA5, Roche). Secondary antibodies were Alexa Fluor 555 goat anti-rabbit IgG (Molecular Probe) or RRX-conjugated donkey anti-mouse IgG (Jackson Laboratories). A FITC-TelC (FITC-OO-CCCTAACCCTAACCCTAA; Applied Biosystems) probe was used to detect telomeric DNA using the protocol developed by Sedivy and colleagues (Herbig et al. 2004). DNA was stained with 4,6-diamino-2-phenylindole (DAPI). Where noted, nucleoplasmic proteins were removed using Triton X-100 buffer (0.5% Triton X-100, 20 mM Hepes-KOH at pH7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) for 2 min on ice prior to fixation with 2% PFA in PBS. Digital images were captured with a Zeiss Axioplan II microscope with a Hamamatsu C4742-95 camera using Improvision OpenLab software.
ChIP
ChIP was performed as described (Loayza and de Lange 2003) with minor modifications. Cells were fixed in medium with 1% PFA for 60 min at room temperature. Glycine was added to 0.2 M to stop the cross-linking. Cells were pelleted by centrifugation and washed once with cold PBS followed by a final wash in PBS/1 mM PMSF. The cells were resuspended in cell lysis buffer (5 mM PIPES at pH 8.0, 85 mM KCl, 0.5% NP-40, 1 mM PMSF, Complete protease inhibitor cocktail [Roche]) and incubated for 15 min on ice. After sonication, the lysates were centrifuged at 13,200 rpm for 10 min. The supernatants were incubated with crude rabbit antibody serum raised against mTRF1 (#1448), mTRF2 (#1254), or mTIN2 (#1447) or with commercial purified antibodies against MYC (9B11, Cell Signaling) or HA (Abcam). Samples were incubated overnight at 4°C and for 45 min with ChIP-grade protein G magnetic beads (Invitrogen Dynal). The immunoprecipitated DNA was collected and washed using a 16-tube magnetic separation rack (Cell Signaling) and precipitated with ethanol after reversal of the cross-links. The DNA samples were dissolved in TE, blotted using a slot blotter, and hybridized with an 800-base-pair (bp) probe labeled with Klenow and a primer for the C-rich telomeric repeat strand. The signal intensity, measured by ImageQuant software, was normalized to the signals of the input DNA on the same blot.
Telomere analysis
Telomeric overhang signals and telomeric restriction fragment patterns were analyzed by in-gel analysis as previously described (Ye and de Lange 2004; Celli and de Lange 2005). Briefly, a [CCCTAA]4 oligonucleotide was hybridized to native MboI-digested genomic DNA fractionated on CHEF gels to determine the overhang signal. After capture of the signal, the DNA was denatured in situ, neutralized, and then rehybridized with the same probe to determine the total telomeric DNA signals. The overhang signal in each lane was normalized to the duplex telomeric signal so that comparisons of these ratios revealed changes in the overhang status. The procedures for telomeric FISH on metaphase spreads were as described previously (Sfeir et al. 2009). Briefly, cells at the indicated time points and treatments were incubated for 2 h with 0.2 μg/mL colcemid. The cells were harvested, swollen in 75 mM KCl, fixed in methanol/acetic acid (3:1), and dropped onto glass slides. After aging overnight, the slides were washed in 1× PBS for 5 min followed by consecutive incubation with 75%, 95%, and 100% ethanol. The slides were allowed to air dry before applying hybridizing solution (70% formamide, 1 mg/mL blocking reagent [Roche], 10 mM Tris-HCl at pH 7.2) containing FITC-OO-[CCCTAA]3 PNA probe (Applied Biosystems). The spreads were denatured for 3 min at 80°C on a heat block and hybridized for 2 h at room temperature. The slides were washed twice for 15 min with 70% formamide/10 mM Tris-HCl (pH 7.0) followed by three 5-min washes in 0.1 M Tris-HCl (pH 7.0)/0.15 M NaCl/0.08% Tween-20. The chromosomal DNA was counterstained with DAPI, added to the second wash. Slides were mounted in anti-fade reagent (ProLong Gold, Invitrogen), and digital images were captured with a Zeiss Axioplan II microscope with a Hamamatsu C4742-95 camera using Improvision OpenLab software.
Q-FISH analysis
Q-FISH analysis was performed on metaphase spreads that were collected and processed as described above from MEFs or the bone marrow of mice (femurs), and TFL-Telo image analysis software was used as previously published (Poon and Lansdorp 2001). As recommended, at least 15 metaphase spreads were used for analysis of each sample. Carboxylate-modified FluoSpheres (0.2 μM; Molecular Probes) were used for system calibration. Metaphase spreads from HeLa1.3 cells were mixed with experimental samples and used as internal controls. Telomere fluorescence hybridization intensity (telomere fluorescence unit [TFU]) values for HeLa1.3 cells (collected from each individual sample) were averaged and normalized to 5000 TFUs. TFU values for MEFs and bone marrow samples were subjected to normalization based on HeLa1.3 ratios. The telomere lengths of HeLa1.3 cells were identical to HeLa1.2.11 cells (parental cell line) (Takai et al. 2010). To convert TFU values to kilobases, HeLa1.3 cells were assigned the mean value of 17 kb, as previously published for HeLa1.2.11 (Canela et al. 2007). Prism 6 (GraphPad) and Excel (Microsoft) were used to process and graph data.
Pathology and peripheral blood analysis
After euthanasia by CO2, smears of femoral bone marrow were prepared and stained with Wright-Giemsa, and all organs were examined grossly and fixed in 10% neutral-buffered formalin. Fixed tissues were processed in alcohol and xylene, embedded in paraffin, sectioned at 4-μm thickness, stained with hematoxylin and eosin, and examined. The following tissues were examined: heart, lungs, thymus, mediastinal lymph node, kidneys, liver, pancreas, spleen, gallbladder, stomach, duodenum, jejunum, ileum, cecum, colon, salivary glands, mesenteric lymph node, mandibular lymph node, testes, epididymides, seminal vesicles, prostate, uterus, cervix, ovaries, oviducts, skin, urinary bladder, adrenal, thyroid, parathyroid, esophagus, trachea, femur, tibia, stifle joint, skeletal muscles, vertebrae, sternum, spinal cord, bone marrow, brain, eye, ears, nasal and oral cavities, and teeth. For hematology analysis, blood was collected in tubes containing EDTA. Automated analysis was performed by an IDEXX Procyte DX hematology analyzer, and the following parameters were determined: red blood cell count, hemoglobin concentration, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, red blood cell distribution width standard deviation and coefficient of variance, reticulocyte count and percentage, platelet count, platelet distribution width, mean platelet volume, and relative and absolute counts of white blood cells, neutrophils, lymphocytes, monocytes, eosinophils, and basophils. A blood smear was prepared, stained with Wright-Giemsa stain, and manually examined for morphology of red blood cells, white blood cells, and platelets.
Acknowledgments
We are extremely grateful to Devon White for his exceptional mouse husbandry. We thank members of the de Lange laboratory and Agata Smogorzewska for discussion and technical advice. We thank Oscar Fernandez-Capetillo for providing the ATR inhibitor. We thank the Tri-Institutional Laboratory for Comparative Pathology for their analysis of the mice. This research was supported by grants from the National Institutes of Health (5R37GM49046 and 5RO1AG16642) and an American Cancer Society post-doctoral fellowship to D.F. T.d.L. is an American Cancer Society Research Professor.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.233395.113.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, Peters JA, Giri N, Lansdorp PM 2007. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood 110: 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O'Sullivan J, et al. 2012. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 44: 338–342 [DOI] [PubMed] [Google Scholar]
- Armanios M 2009. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet 10: 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, et al. 2005. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci 102: 15960–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G, Lansdorp PM 2008. Telomeres and aging. Physiol Rev 88: 557–579 [DOI] [PubMed] [Google Scholar]
- Ballew BJ, Joseph V, De S, Sarek G, Vannier JB, Stracker T, Schrader KA, Small TN, O'Reilly R, Manschreck C, et al. 2013a. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of hoyeraal hreidarsson syndrome. PLoS Genet 9: e1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballew BJ, Yeager M, Jacobs K, Giri N, Boland J, Burdett L, Alter BP, Savage SA 2013b. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in dyskeratosis congenita. Hum Genet 132: 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 [DOI] [PubMed] [Google Scholar]
- Calado RT, Young NS 2009. Telomere diseases. N Engl J Med 361: 2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Vera E, Klatt P, Blasco MA 2007. High-throughput telomere length quantification by FISH and its application to human population studies. Proc Natl Acad Sci 104: 5300–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, Smith S 2011. A role for heterochromatin protein 1γ at human telomeres. Genes Dev 25: 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Celli GB, Lazzerini Denchi E, de Lange T 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8: 885–890 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M 2008. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Deng Z, Glousker G, Molczan A, Fox AJ, Lamm N, Dheekollu J, Weizman OE, Schertzer M, Wang Z, Vladimirova O, et al. 2013. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc Natl Acad Sci 110: E3408–E3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I, Vulliamy T 2005. Telomerase deficiency and human disease. In Telomeres. (ed. de Lange T, et al.), pp. 139–162. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- He H, Wang Y, Guo X, Ramchandani S, Ma J, Shen MF, Garcia DA, Deng Y, Multani AS, You MJ, et al. 2009. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol 29: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 14: 501–513 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T 2008. Engineered telomere degradation models dyskeratosis congenita. Genes Dev 22: 1773–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15: 3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J 1999. TIN2, a new regulator of telomere length in human cells. Nat Genet 23: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J 2004. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem 279: 43799–43804 [DOI] [PubMed] [Google Scholar]
- Le Guen T, Jullien L, Touzot F, Schertzer M, Gaillard L, Perderiset M, Carpentier W, Nitschke P, Picard C, Couillault G, et al. 2013. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum Mol Genet 22: 3239–3249 [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z 2004a. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z 2004b. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 424: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Marrone A, Stevens D, Vulliamy T, Dokal I, Mason PJ 2004. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood 104: 3936–3942 [DOI] [PubMed] [Google Scholar]
- Marrone A, Walne A, Tamary H, Masunari Y, Kirwan M, Beswick R, Vulliamy T, Dokal I 2007. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood 110: 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PJ, Wilson DB, Bessler M 2005. Dyskeratosis congenita—a disease of dysfunctional telomere maintenance. Curr Mol Med 5: 159–170 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ND, Bertuch AA 2012. Dyskeratosis congenita as a disorder of telomere maintenance. Mutat Res 730: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann AA, Watson CM, Noble JR, Pickett HA, Tam PP, Reddel RR 2013. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev 27: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor MS, Safari A, Xin H, Liu D, Songyang Z 2006. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci 103: 11874–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Poon SS, Lansdorp PM 2001. Quantitative fluorescence in situ hybridization (Q-FISH). Curr Protoc Cell Biol 12: 18.4.1–18.4.21 [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA 1999. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96: 701–712 [DOI] [PubMed] [Google Scholar]
- Sarper N, Zengin E, Kilic SC 2010. A child with severe form of dyskeratosis congenita and TINF2 mutation of shelterin complex. Pediatr Blood Cancer 55: 1185–1186 [DOI] [PubMed] [Google Scholar]
- Savage SA, Bertuch AA 2010. The genetics and clinical manifestations of telomere biology disorders. Genet Med 12: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP 2008. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet 82: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T 2010. In vivo stoichiometry of shelterin components. J Biol Chem 285: 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T 2011. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell 44: 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O 2011. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzot F, Callebaut I, Soulier J, Gaillard L, Azerrad C, Durandy A, Fischer A, de Villartay JP, Revy P 2010. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc Natl Acad Sci 107: 10097–10102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahan C, Martel C, Dragon F 2010. Effects of dyskeratosis congenita mutations in dyskerin, NHP2 and NOP10 on assembly of H/ACA pre-RNPs. Hum Mol Genet 19: 825–836 [DOI] [PubMed] [Google Scholar]
- Tsangaris E, Adams SL, Yoon G, Chitayat D, Lansdorp P, Dokal I, Dror Y 2008. Ataxia and pancytopenia caused by a mutation in TINF2. Hum Genet 124: 507–513 [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I 2004. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet 36: 447–449 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, Dokal I 2008. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci 105: 8073–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, Masunari Y, Al-Qurashi FH, Aljurf M, Dokal I 2007. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum Mol Genet 16: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I 2008. TINF2 mutations result in very short telomeres: Analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 112: 3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I 2013. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet 92: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin ZT, Ly H 2012. Characterization of interactions between naturally mutated forms of the TIN2 protein and its known protein partners of the shelterin complex. Clin Genet 81: 301–302 [DOI] [PubMed] [Google Scholar]
- Yang D, He Q, Kim H, Ma W, Songyang Z 2011. TIN2 protein dyskeratosis congenita missense mutants are defective in association with telomerase. J Biol Chem 286: 23022–23030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, de Lange T 2004. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet 36: 618–623 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, Van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271 [DOI] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE 2011. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 25: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]