Snail transcription factors are key regulators of epithelial–mesenchymal transitions and tumor metastasis. Rembold et al. now report that, in addition to being a well-known transcriptional repressor, Drosophila Snail can also activate transcription. Through the integration of genetics, genomics, and bioinformatics analyses, the authors show that Snail acts directly to potentiate gene expression and identify a specific motif essential for enhancer activation. The results shed new light on complex phenotypes observed in snail mutant embryos and elucidate a new mechanism by which this transcription factor regulates development.
Keywords: transcription factor, Snail, Twist, repression, activation, spatiotemporal gene expression, Drosophila embryogenesis
Abstract
The transcription factors of the Snail family are key regulators of epithelial–mesenchymal transitions, cell morphogenesis, and tumor metastasis. Since its discovery in Drosophila ∼25 years ago, Snail has been extensively studied for its role as a transcriptional repressor. Here we demonstrate that Drosophila Snail can positively modulate transcriptional activation. By combining information on in vivo occupancy with expression profiling of hand-selected, staged snail mutant embryos, we identified 106 genes that are potentially directly regulated by Snail during mesoderm development. In addition to the expected Snail-repressed genes, almost 50% of Snail targets showed an unanticipated activation. The majority of “Snail-activated” genes have enhancer elements cobound by Twist and are expressed in the mesoderm at the stages of Snail occupancy. Snail can potentiate Twist-mediated enhancer activation in vitro and is essential for enhancer activity in vivo. Using a machine learning approach, we show that differentially enriched motifs are sufficient to predict Snail's regulatory response. In silico mutagenesis revealed a likely causative motif, which we demonstrate is essential for enhancer activation. Taken together, these data indicate that Snail can potentiate enhancer activation by collaborating with different activators, providing a new mechanism by which Snail regulates development.
The transcription factor (TF) Snail is part of a conserved Snail family of C2H2 zinc finger proteins that have been extensively studied for their role in development, cell morphogenesis, and tumor metastasis (for review, see Barrallo-Gimeno and Nieto 2005). Snail was originally identified in Drosophila, where mutant embryos are defective in mesoderm formation during gastrulation (Simpson 1983). At the onset of embryogenesis, the concerted action of Dorsal (an NFκB protein) (Roth et al. 1989; Rushlow et al. 1989), Twist (a basic helix–loop–helix [bHLH] protein) (Thisse et al. 1988), and Snail determines the presumptive mesoderm and its borders with ectodermal territories. Twist and Dorsal cooperate to activate mesodermal gene expression, while Snail promotes mesoderm development by repressing ectodermal genes within the mesodermal domain and establishes a sharp border between the mesoderm and mesectoderm (for review, see Chopra and Levine 2009). Although loss of either Twist or Snail function results in a failure of mesoderm formation (Leptin and Grunewald 1990), Snail is sufficient to promote the first steps of ventral furrow invagination (Ip et al. 1994; Seher et al. 2007). Snail therefore has an independent role in promoting mesoderm formation, but how this is achieved remains unclear.
Snail mediates transcriptional repression through the recruitment of two corepressors: the C-terminal-binding protein (dCtBP) (Nibu et al. 1998a,b) and Ebi, which recruits histone deacetylase 3 (HDAC3) (Qi et al. 2008). Mutation of any of the corepressor interaction motifs in the N terminus of Snail impairs its repressor function (Hemavathy et al. 2004; Qi et al. 2008) and, in the case of the dCtBP interaction motifs, its ability to coordinate mesoderm development (Hemavathy et al. 2004). Dissection of the repressive effects of Snail in different enhancers revealed that its function is distance-dependent. Snail was thereby classified as a short-range repressor that acts through the quenching of activators bound within 100 base pairs (bp) in the same enhancer or core promoter (Gray et al. 1994; Gray and Levine 1996).
The target sequences for Snail and Twist are very similar, and their binding has been shown to be mutually exclusive in some instances (Ip et al. 1992). This would provide one mechanism for Snail repression of Twist targets in addition to the recruitment of corepressors. Other mechanisms include inhibition of transcription by blocking the release of RNA polymerase II from the promoter (Bothma et al. 2011; McHale et al. 2011) or inhibiting enhancer–promoter looping from distal enhancers (Chopra et al. 2012).
Although Snail is generally considered to be a dedicated repressor, a number of observations hint at a potential role in transcriptional activation. Genetic studies almost 20 years ago showed that several essential mesodermal genes have reduced expression in snail mutant embryos, including Myocyte-enhancing factor 2 (Mef2) (Lilly et al. 1994), Zn finger homeodomain 1 (zfh1) (Casal and Leptin 1996; Hemavathy et al. 1997), tinman (tin) (Bodmer et al. 1990; Ip et al. 1994), and heartless (htl) (Shishido et al. 1993). This lack of expression was generally assumed to be an indirect effect caused by the derepression of an unknown repressor, a hypothesis supported by studies in mammalian cells (Jorda et al. 2005, 2007; Sun et al. 2008; Dave et al. 2011).
In addition to having an indirect role in transcriptional activation, we reasoned, based on recent chromatin immunoprecipitation (ChIP) data, that Snail may also act directly to positively regulate gene expression: Using ventralized Toll10B mutant embryos, Zeitlinger et al. (2007) showed that Snail occupies many mesodermal enhancers that are active in these embryos, which is at odds with the typical local dominant effect of a repressor cobound to an enhancer with activating TFs (Gray and Levine 1996; Zeitlinger et al. 2007). An activator role for Snail has been shown by genetic studies and reporter assays in Caenorhabditis elegans, mice, and quail—showing that Snail family members can activate B0507.1 (Reece-Hoyes et al. 2009), MMP15 (Tao et al. 2011), and Snail2 (Sakai et al. 2006)—and by in vitro studies of the p15INK4b (Hu et al. 2010) and ZNF281 genes (Hahn et al. 2013). Furthermore, Snail can increase Wnt target gene expression in human cell lines independent of direct DNA binding but via physical interaction with β-catenin (Stemmer et al. 2008). However, aside from these examples, the generality of Snail's capacity to act as an activator remains unclear; in Drosophila, where the founding member of this TF family was discovered, there is currently no evidence for a direct role in transcriptional activation. If Snail can act as an activator as well as a repressor, what regulates this functional switch in Snail's activity?
By using an integrative approach, we uncovered a new role for Drosophila Snail whereby it not only represses the activity of neuroectodermal enhancers but may also be essential for the activation of many mesodermal enhancers. Through in vivo occupancy and mutagenesis analysis of enhancer activity, we provide the first evidence that Drosophila Snail acts directly to potentiate gene expression and that a specific motif is essential for enhancer activation. Our results help explain the complex phenotypes observed in snail mutant embryos and shed new light on how this much-studied TF regulates mesoderm development.
Results
Snail binds to active mesodermal enhancers in wild-type embryos
ChIP studies in Toll10B mutant embryos identified Snail binding to enhancers of mesodermally expressed genes (Zeitlinger et al. 2007). In these embryos, Twist and Snail are ubiquitously expressed throughout the entire embryo, genetically transforming all cells to take on a mesodermal cell fate. To exclude the possibility that the observed Snail occupancy was due to the severity of developmental defects in this mutant, we first determined whether Snail binds to mesodermal enhancers in wild-type embryos.
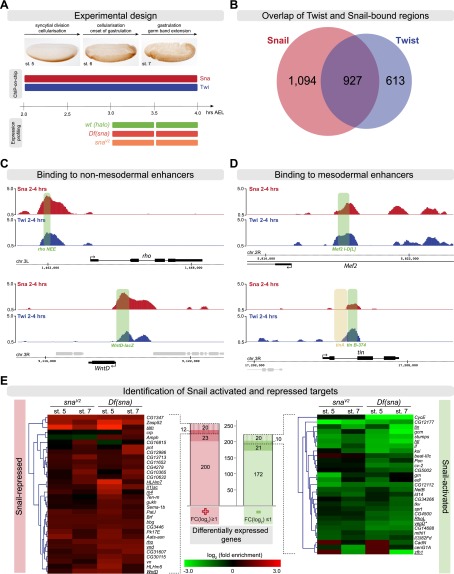
ChIP followed by hybridization to high-density tiling arrays (ChIP-on-chip) was performed on tightly staged embryos at 2–4 h of development (stages 5–7) using an antibody directed against Snail (Fig. 1A). We obtained 2021 high-confidence Snail-bound regions. Comparison with our previously generated data covering the same stages of development (Zinzen et al. 2009) showed that 46% of Snail peaks are in close proximity (300 bp) to Twist peaks (Fig. 1B). This set of 927 overlapping regions contains the majority of functionally characterized mesodermal enhancers and was therefore used to obtain a set of ChIP-defined cis-regulatory modules (ChIP-CRMs) as described previously (Supplemental Material; Zinzen et al. 2009).
Figure 1.
Identification of Snail direct target genes. (A) Schematic outline of the ChIP and expression profiling experiment, highlighting the stages and genotypes used. (B) Overlap of Twist- and Snail-bound regions using 300-bp windows centered on the Snail peak summit. (C) ChIP signals (log2 mean immunoprecipitation/mock signal) showing Snail (red) and Twist (blue) occupancy on two known Snail-repressed enhancers (green bar). The gene model is shown below; thick lines indicate exons, thin lines introns, and arrows indicate direction of transcription for the gene of interest (black) and surrounding genes (gray). The chromosome arm and genome coordinates are indicated along the dashed line. (D) ChIP signals (log2 mean immunoprecipitation/mock signal) showing Snail and Twist occupancy on known active mesodermal enhancers (green bar). The tin A enhancer is not active in the mesoderm (brown bar). The gene model is indicated as in C. (E) Differentially expressed genes in two snail alleles at both time points. Central histogram: Two-hundred-fifty-five genes are up-regulated (red), and 223 genes down-regulated (green) in one or more conditions. Up-regulated genes: Twenty-three genes are associated with Snail-only CRMs, 20 with Snail-Twist cobound CRMs, and 12 with both types. Down-regulated genes: 21 are associated with Snail-only CRMs, 20 are associated with Snail–Twist cobound, and 10 are associated with both types. Heat maps show expression changes of cobound genes in snaV2 and Df(sna) at stage 5 and stage 7, respectively. The names of known Snail- or Twist-regulated genes are underlined. *(vepD) ventrally-expressed-protein-D (FBgn0053200).
To assess the quality of the data, we first examined the occupancy of Snail on its previously characterized target enhancers. For example, we found Snail and Twist binding to the 300-bp rhomboid neuroectoderm element (rho NEE) and the previously identified wnt inhibitor of Dorsal (wntD) enhancer (Fig. 1C; Sandmann et al. 2007; Zeitlinger et al. 2007), in agreement with their known roles in repressing or activating these genes' expression, respectively (Kosman et al. 1991; Ip et al. 1992; Ganguly et al. 2005). In addition, we found Snail binding to other known Snail-dependent enhancers, including vnd_348 (Markstein et al. 2004), vnd_−5.3/−4.0 (Shao et al. 2002), vn_neurogenic_ectoderm (Markstein et al. 2004), sim_mesectoderm (Markstein et al. 2004), sim_2.8sim (Kasai et al. 1992), and two regions within the sog locus.
Having confirmed that the ChIP assay reliably captures enhancers known to be repressed by Snail, we focused on actively transcribed mesodermal genes. We first examined enhancers of mesodermal genes that are genetically dependent on Snail for their expression (Fig. 1D; Supplemental Fig. S1). The expression of the myogenic regulator Mef2 is significantly reduced in snail (sna) mutant embryos (Lilly et al. 1994), and we observed cobinding of Snail and Twist on multiple putative cis-regulatory regions within the Mef2 genomic locus, one of which overlaps with the known Twist-dependent Mef2 I-D[L] enhancer element active in the early mesoderm (Fig. 1D; Cripps et al. 1998; Nguyen and Xu 1998). The early expression of tin throughout the trunk mesoderm is also reduced in sna mutant embryos, while its expression in the head mesoderm as well as later expression are unaffected (Bodmer et al. 1990). The trunk mesodermal expression of tin is mediated by a Twist-dependent 374-bp element, tin B-374 (Yin et al. 1997), which our ChIP data show to be occupied by both Twist and Snail (Fig. 1D). In addition to these two well-characterized enhancers, we also found Snail binding to known and putative cis-regulatory regions within several other Snail-dependent mesodermal genes, such as htl, ventrally-expressed-protein-D, zfh1, if, srp, and stumps (Supplemental Fig. S1; Lai et al. 1991; Shishido et al. 1993; Casal and Leptin 1996; Hemavathy et al. 1997; Morize et al. 1998).
In summary, we observed Snail occupancy on mesodermal enhancers in wild-type embryos at a time when these enhancers are active. This result suggests that either the binding of this “repressor” is not functional in particular contexts or Snail may play a direct role in their transcriptional activation and the establishment of mesodermal gene expression.
Snail and Twist directly regulate a shared pool of mesodermal genes
To examine the functional role of Snail enhancer occupancy, we first assessed the relationship between Snail enhancer binding and transcriptional changes of associated target genes upon mutation of sna. We compared the gene expression profiles of tightly stage-matched wild-type and homozygous mutant embryos for two sna alleles: a deficiency that completely removes sna function [Df(2L)TE116GW11, abbreviated as Df(sna)] and a hypomorphic sna (snaV2) allele.
To measure the earliest and most immediate effects of Snail, we collected embryos just after the onset of sna expression. Twist and sna mRNA are first detectable during nuclear cleavage cycles 12–13 at ∼2 h after egg laying (AEL) (stage 4 according to Campos-Ortega and Hartenstein 1997) (Leptin 1991), but the morphological sna mutant phenotype is not apparent until later stages. We therefore took advantage of the phenotype caused by a recessive mutation in halo (Materials and Methods; Gross et al. 2003), which we recombined onto the sna chromosome to select sna homozygous mutant embryos from their balancer siblings. halo mutant embryos are marked by a visible defect in cytoplasmic clearing during cellularization but are otherwise viable and fertile (Gross et al. 2003). We hand-selected sna homozygous mutant embryos at two time windows of development: (1) stages 5–6, at the onset of gastrulation when the mesoderm anlage is specified, and (2) stages 7–8, after gastrulation, when the mesoderm spreads out under the ectoderm.
Comparison of gene expression levels between the two sets of sna mutants to stage-matched ΔhaloAJ embryos revealed 478 differentially expressed genes in one or more conditions (“snail-pooled”) (Supplemental Material). Two-hundred-twenty-five genes (53%) were up-regulated in sna mutants at one or more conditions (log2 ≥ 1, false discovery rate [FDR] ≤ 0.05), while 223 genes (47%) were down-regulated (log2 ≤ −1, FDR ≤ 0.05) (Fig. 1E). The up-regulated set contained genes expressed in the neuroectoderm or mesectoderm that are known to be repressed by Snail in the mesoderm, including sim, vnd, wntD, rho, sog, l(1)sc, HLHm7, and m4 (our unpublished observation), demonstrating the quality and sensitivity of the data.
We defined Snail direct target genes as those that showed differential expression and had a Snail ChIP peak within 5 kb upstream of and 1 kb downstream from the annotated gene. These criteria identified 106 direct Snail target genes that are associated with 180 Snail-bound ChIP-CRMs (Supplemental Material), which were classified as “Snail-activated” or “Snail-repressed” CRMs based on the response of the target gene. Surprisingly, 51 of these genes (48%) were down-regulated in the absence of Snail, suggesting a direct role for Snail in their activation (Fig. 1E).
A subset of Snail target genes (21 activated and 23 repressed) is associated with regulatory regions that were bound only by Snail and not by Twist (“Snail-only CRMs”). However, the majority of genes appear to be regulated by both TFs, containing CRMs bound by both Twist and Snail (20 activated and 20 repressed genes) or multiple CRMs that are a mixture of Snail-only and cobound CRMs (10 activated and 12 repressed targets) (Fig. 1E). The cobound set contains all well-characterized targets of Twist and Snail, such as sim, rho, l(1)sc, wntD, and vnd (Fig. 1E, left, heat map). Snail represses these neurectodermal genes in the mesoderm, which is in agreement with its traditional repressor role.
Of particular interest here, however, are the Snail-bound enhancers associated with mesodermally active genes, whose expression appears to be activated by Snail (Fig. 1E, right, heat map). This set contains many mesodermal genes that were previously reported to be down-regulated in sna mutants; e.g., htl, tin, RhoL, ventrally-expressed-protein-D, and zfh1 (Lai et al. 1991; Shishido et al. 1993; Lilly et al. 1994; Casal and Leptin 1996; Hemavathy et al. 1997). In addition, we identified if, Cadherin-N, CG14688, and stumps as targets activated by Snail. It has generally been assumed that Snail's contribution to the expression of such genes is indirect due to Snail-mediated repression of a repressor. However, our data suggest that Snail directly contributes to the activation of these genes through Twist–Snail cobound CRMs. The majority of mesodermal genes show reduced expression as early as stage 5 in the sna-null allele, supporting the hypothesis that Snail is required to establish the initiation of their expression.
The hypomorphic snaV2 allele carries a missense mutation in a conserved region between zinc fingers 3 and 4 of Snail's DNA-binding domain. This allele has a weaker phenotype than sna-null alleles (Hemavathy et al. 1997) in that the expression of some neuroectodermal gene is only partially derepressed; the mesoderm invaginates almost normally, although it fails to differentiate properly. Various scenarios could explain this hypomorphic phenotype: (1) The nature of the lesion could exclusively affect either the activator or repressor role of Snail. However, our data exclude this scenario, as target genes of both classes are misregulated; e.g., sim and rho in the repressed group and stumps and htl in the activated group. (2) The mutation could affect the expression of all Snail target genes, but the amplitude of change is lower in snaV2 mutant embryos compared with that observed in the sna-null mutant. This scenario also does not appear to be the case because only ∼40% of Snail direct target genes that are up-regulated or down-regulated in the sna-null allele are also up-regulated or down-regulated in snaV2. The remaining 60% of Snail-dependent genes are not affected in snaV2. It is possible that the difference in expression strength of several genes is the cause of the different phenotypes, rather than complete lack of activation or repression of a single target gene.
In summary, these results suggest a direct relationship between Snail enhancer occupancy and the transcriptional activation of some genes. Activation appears to be not a minor part of Snail's transcriptional role but rather represents half of its regulatory input to gene expression.
Snail positively regulates enhancer activity in collaboration with Twist
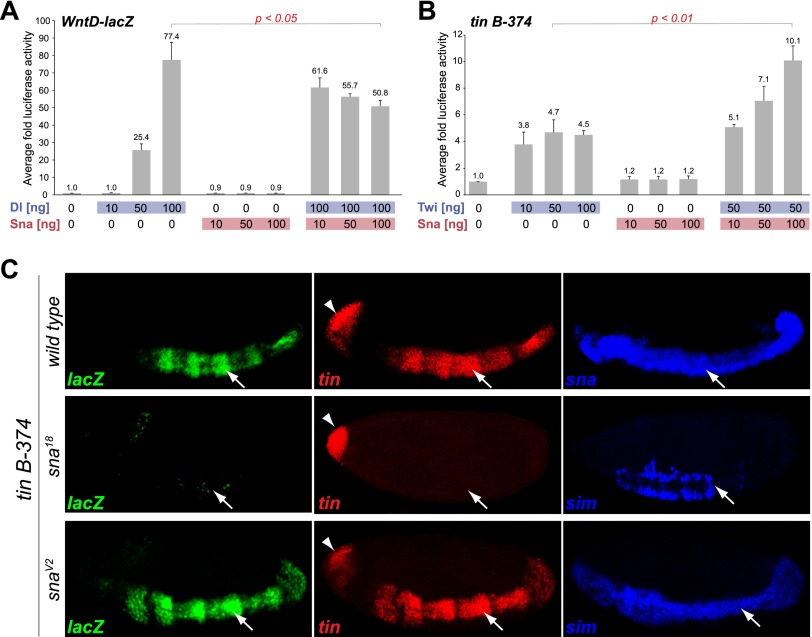
To assess the function of Snail binding to active mesodermal enhancers more directly, we performed luciferase assays in Drosophila Kc cells (Fig. 2). Kc cells, which are thought to be of hematopoietic origin (Echalier and Ohanessian 1969), do not express detectable levels of Snail, Twist, or Dorsal (data not shown), while the corepressors dCtBP and Ebi are present (Cherbas et al. 2011). First, we tested the ability of Snail to repress the Dorsal-dependent wntD-lacZ enhancer in this system (Zeitlinger et al. 2007). Transfection of Dorsal into cells carrying the wntD-lacZ-luciferase reporter alone activated the wntD enhancer up to 77-fold, while cotransfection with Snail reduced this activation by approximately a third (∼50-fold; P < 0.05 Student's t-test) (Fig. 2A) in a concentration-dependent manner. Similarly, Snail can repress the cooperative activation of the neurectodermal 300-bp rho NEE enhancer (Ip et al. 1992) by Twist and Dorsal, decreasing the response from 42-fold to 1.7-fold (Supplemental Fig. S2A). This cell culture system can therefore recapitulate in vitro the known in vivo regulation of these genes (Kosman et al. 1991; Ip et al. 1992; Ganguly et al. 2005).
Figure 2.
Snail positively regulates tin B-374 enhancer activity both in vitro and in vivo. Luciferase assays in Kc cells on the wntD-lacZ enhancer (A) and the tin B-374 enhancer (B). The X-axis indicates the amount of DNA transfected (ng), and the Y-axis is the average fold luciferase activity across replicates (n = 3), normalized to Renilla. (A) Dorsal-mediated enhancer activation is repressed upon cotransfection of Snail (Students two-tailed t-test, P < 0.05). (B) Snail augments the activating effect of Twist on the tin B-374 enhancer approximately twofold (Students two-tailed t-test, P < 0.01). (C) In vivo activity of the tin B-374 enhancer (lacZ; green) and expression of the endogenous tinman (tin) gene (red). Expression of sna (blue) marks the mesoderm (arrow), while derepression of sim (blue) in the mesoderm identifies sna mutant embryos. All embryos are orientated with anterior to the left and dorsal up. Images show a single confocal plane. Expression of tin in the anterior domain (arrowhead) is independent of sna.
Having confirmed that Snail can repress transcription in Kc cells, we selected two well-characterized mesodermal enhancers to test both in vitro and in vivo whether Snail is capable of transcriptional activation. The levels of tin transcripts are strongly reduced in sna mutant embryos (Bodmer et al. 1990; Ip et al. 1994), as confirmed by our expression profiling data [log2 = −2.77 at stage 7 in Df(sna)]. The tin B-374 enhancer is responsible for the early expression of tin in the trunk mesoderm and is directly activated by Twist in vivo (Yin et al. 1997). Consistent with these observations, this enhancer is cobound by both Twist and Snail in embryos during the time of its activity (Fig. 1D). In Kc cells, Twist increases the basal enhancer activity almost fivefold (Fig. 2B). While transfection of Snail alone does not change enhancer activity significantly, cotransfection of Snail with Twist yields an additional approximately twofold increase in reporter activity (P < 0.005 Student's t-test) (Fig. 2B), demonstrating that Snail is capable of potentiating enhancer output.
We also analyzed the response of the enhancer to Snail in vivo. In embryos, the tin B-374 enhancer recapitulates the expression of the endogenous tin gene in the trunk mesoderm but not the anterior expression domain in the head mesoderm (Fig 2C, top; Yin et al. 1997). In embryos homozygous for the amorphic sna18 allele, the activity of the enhancer is substantially reduced, in agreement with the down-regulation of the endogenous gene in the mesoderm (Fig. 2C). The expression of tin in the head mesoderm is not affected, consistent with this expression domain being regulated by a different tin A enhancer element (Yin et al. 1997), which is not bound by Snail (Fig. 1D). There are no apparent differences in the tin B-374 enhancer activity or endogenous tin expression in embryos homozygous for the hypomorphic snaV2 allele at stage 5/6, indicating that the remaining Snail activity is sufficient to initiate both the enhancer and the endogenous gene's expression at this stage.
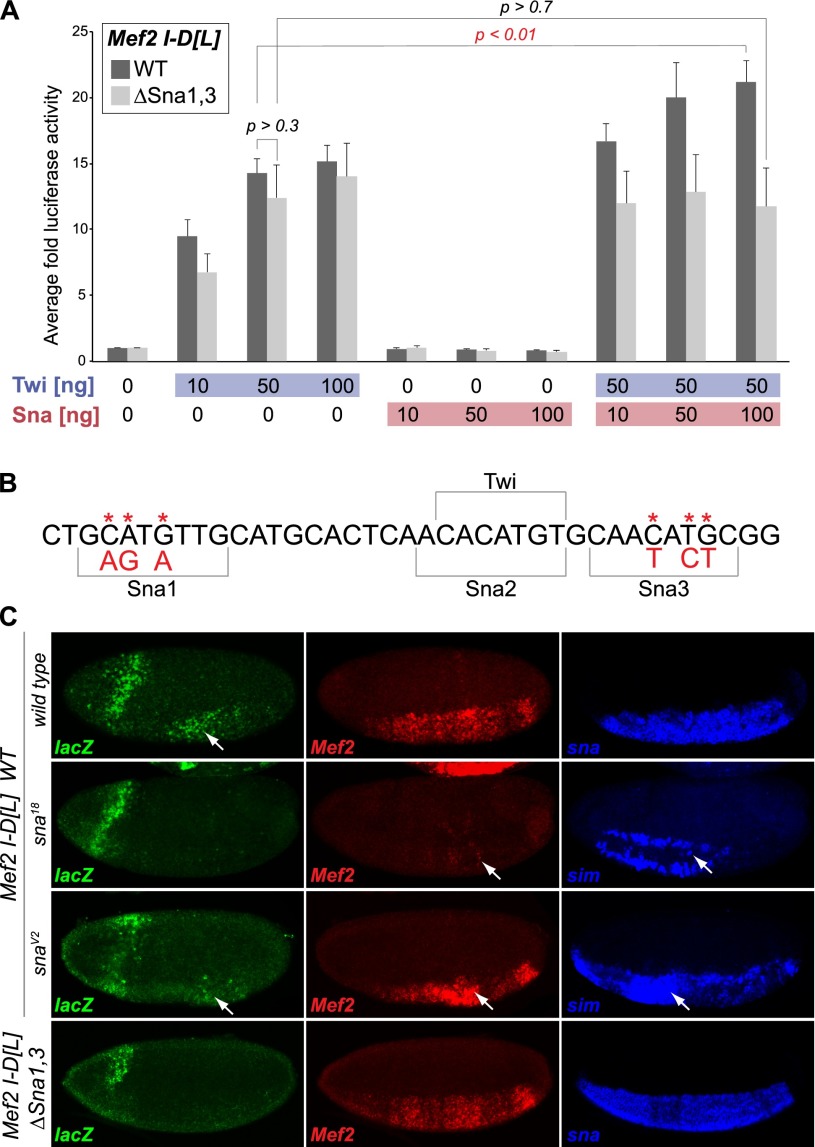
Snail also potentiates the activity of the Mef2 I-D[L] enhancer. The Mef2 I-D[L] enhancer drives expression in the early mesoderm (Nguyen and Xu 1998), where its activity is dependent on activation by Twist (Cripps et al. 1998). In Kc cells, Twist leads to an ∼14-fold increase in Mef2 I-D[L]-driven luciferase activity (Fig. 3A). While Snail alone is not sufficient to activate the enhancer, cotransfection of Snail can increase Twist enhancer activation from ∼14-fold to 21-fold (P < 0.02, Student's t-test) (Fig. 3A). Similarly, in vivo, the early mesodermal activity elicited by the enhancer is reduced in sna18 mutant embryos, as described below (Fig. 3C).
Figure 3.
Snail-positive regulation of Mef2 I-D[L] requires the Snail motif. (A) Luciferase assay in Kc cells. The X-axis indicates the amount of DNA transfected (ng), and the Y-axis is the average fold of luciferase across replicates (n = 3), normalized to Renilla. Snail significantly potentiates Twist-mediated Mef2 I-D[L] enhancer activation (dark-gray bars) (Students two-tailed t-test, P < 0.01). Disruption of two putative Snail motifs, shown in B, abrogates Snail's effect (light-gray bars). (B) Two mutated Snail motifs, Sna1 and Sna3, are highlighted in the sequence of the Mef2 I-D[L] enhancer (asterisks and red letters indicate the base exchanges). The Sna2 motif overlaps an essential Twist motif and was therefore not mutated. (C) In vivo activity of the Mef2 I-D[L] enhancer. (Top panel) In wild-type embryos, the Mef2 I-D[L] enhancer initiates reporter gene expression (lacZ; green) in mesoderm at stage 5, similar to the endogenous Mef2 gene (red). Enhancer activity and Mef2 expression are ablated in sna18 mutant embryos but maintained in snaV2 mutant embryos. (Bottom panels) Mutation of the Snail motifs 1 and 3 (Mef2 I-D[L] ΔSna1,3) drastically reduces lacZ expression. Expression of sna (blue) marks the mesoderm, while derepression of sim (blue) in the mesoderm was used to distinguish sna mutant embryos from wild-type embryos. LacZ expression in the head fold is caused by the eve minimal promoter in the reporter vector.
Therefore, Snail can potentiate Twist-mediated activation of the tin B-374 and Mef2 I-D[L] enhancers in vitro and is necessary for their activation in vivo. Combined with Snail's in vivo binding to these enhancers at these stages of development and its requirement for the endogenous genes' expression, these data demonstrate a direct positive role for Snail in the regulation of two essential mesodermal genes.
Snail DNA binding is essential for potentiation of enhancer activity
To confirm that direct DNA binding of Snail is necessary for its activatory role, we mutated Snail sites within the Mef2 I-D[L] enhancer. There are three Snail motifs within this enhancer, with the central motif being a composite Twist/Snail site (Fig. 3B). We mutated motifs 1 and 3, while motif 2 was left intact to avoid potential interference with Twist binding (Fig. 3B; Cripps et al. 1998). The disruption of these sites did indeed abolish Snail's ability to coactivate the Mef2 I-D[L] enhancer in Kc cells. While the mutant enhancer, named Mef2 I-D[L] ΔSna1,3, can still be activated by Twist (Fig. 3A), cotransfection of Snail does not lead to a significant change in its activity (P = 0.7, Student's t-test) (Fig. 3A).
To assess the consequence of the binding site mutations in vivo, we generated transgenic reporter lines carrying wild-type or mutant versions of the enhancer linked to lacZ. Using the phiC31 integrase system (Bischof et al. 2007), each transgene was targeted to the same location within the genome to minimize positional effects. In agreement with previous studies, the wild-type Mef2 I-D[L] enhancer drives reporter gene expression in the developing mesoderm, where the expression is first detectable at stage 5 (Fig. 3C; Supplemental Fig. S3). The activity of the enhancer as well as of the endogenous Mef2 gene depends on Snail, as in sna18 mutant embryos, their expression is significantly reduced (Fig. 3C). Mutation of the Snail motifs 1 and 3 in the enhancer recapitulates this effect, rendering the enhancer inactive despite the presence of Twist-binding sites (Fig. 3C, bottom panels). The positive effect of Snail therefore depends on intact Snail-binding sites.
In addition to the Mef2 I-D[L] element, we identified a novel putative enhancer within an intron of the Mef2 gene. In transgenic reporter assays, this element is active in the mesoderm, where its activity also depends on Snail (Supplemental Fig. S4). The enhancer also drives expression in a striped pattern in the ectoderm, which is independent of Snail function, as expected. These results indicate that Snail can positively regulate Mef2 expression in vivo through at least two enhancer elements that it occupies at the stages where Mef2 initiates expression in the mesoderm.
To summarize, the positive role of Snail on the Mef2 I-D[L] enhancer is direct, as Snail must bind to the enhancer to positively regulate its activity. Our ChIP data indicate that this is likely to hold true for many more Snail-regulated enhancers.
Enhancer architecture of Snail-bound CRMs
Our results indicate that when Snail binds to the same enhancers as Twist, it can have distinct effects: On a subset of enhancers, Snail prevents Twist and other TFs from activating gene expression, while on others, it potentiates enhancer activity. How is Snail action modulated to achieve these opposing effects?
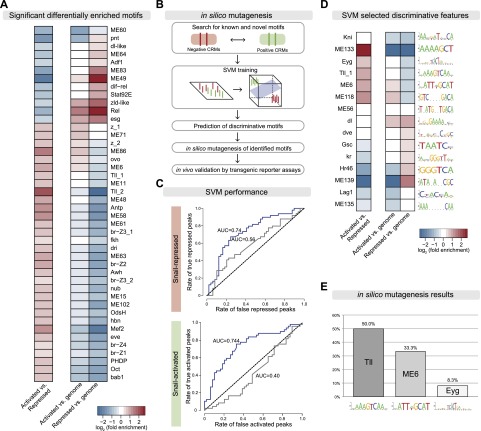
A regulatory switch in the activity of a TF can occur by many mechanisms that often depend on the cellular context, including the availability of cofactors or post-translational protein modifications (for review, see Ma 2005). In the developing mesoderm, Snail appears to be able to function in two modes within the same cell type and at the same time. We therefore reasoned that the underlying mechanism is likely to be partially encoded in the enhancer sequence itself. To assess this, we analyzed the motif content of the 102 Twist and Snail cobound CRMs (Supplemental Material) to determine whether there are motif differences between the Snail-activated (52) and Snail-repressed (50) CRMs (Fig. 1E). Snail-only CRMs (78) served as a control group.
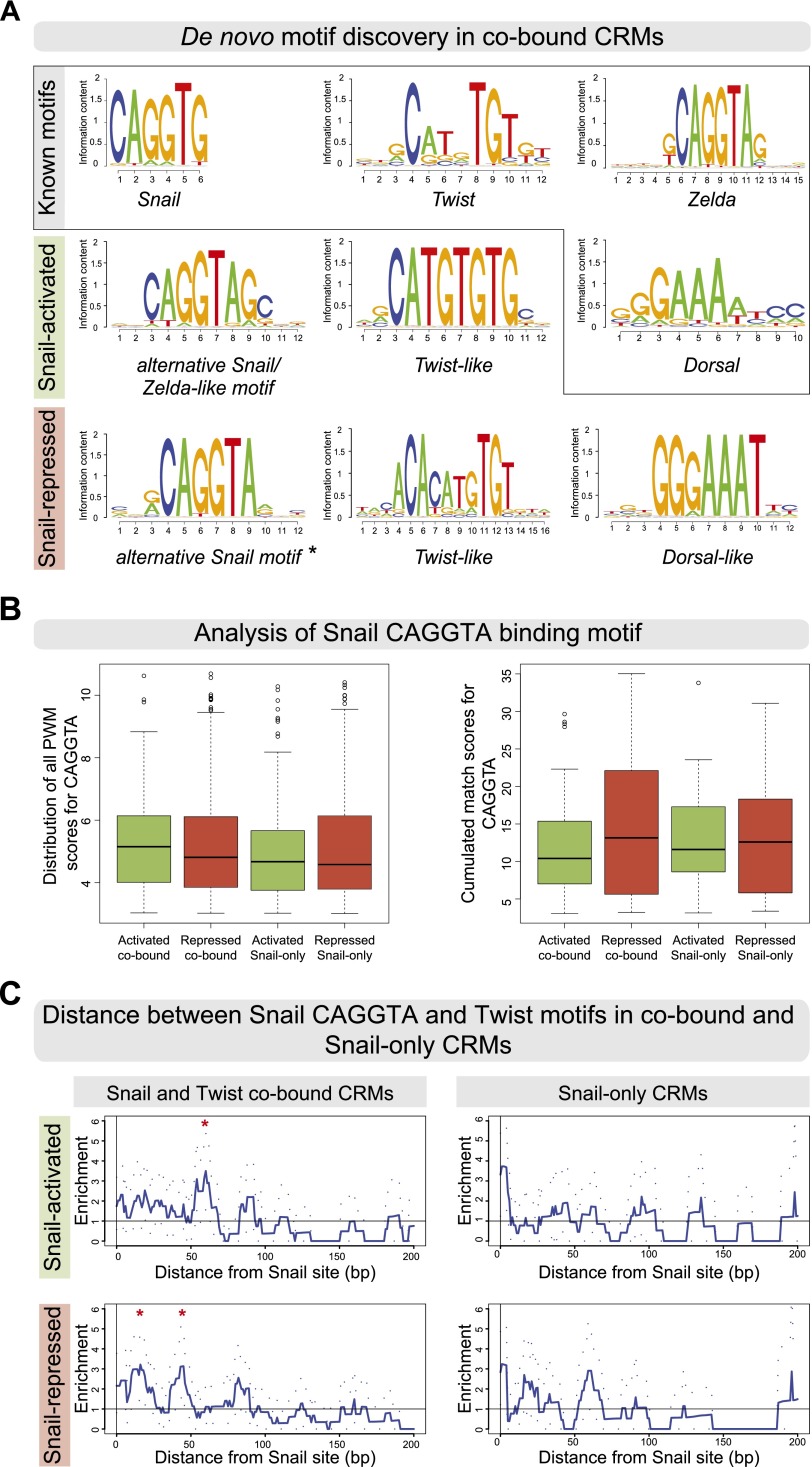
We first examined whether differences in the Snail and Twist motifs themselves could account for the different regulatory potential. De novo motif discovery using the RSAT peak motif tool (Fig. 4A; Supplemental Material; Thomas-Chollier et al. 2012) identified Twist motif signatures with the core CAc/tATG in both the activated and repressed cobound sets with a similar frequency, although the motifs show slight differences (Fig. 4A, “Twist-like”). The Twist-like motifs are not enriched in Snail-only CRMs (data not shown), consistent with the lack of Twist binding. Half-sites of the Dorsal-binding motif are specifically enriched in the repressed CRMs (Fig. 4A, “Dorsal-like”), as expected from the known role of Dorsal in activating neuroectodermal enhancers that are repressed by Snail (for review, see Chopra and Levine 2009). While the Snail CAGGTG motif (from Jaspar) (Mauhin et al. 1993) is not enriched in either cobound set, an alternative Snail motif with an adenine preference at position 6 (CAGGTA) is enriched in both the activated and repressed CRMs (Fig. 4A, “alternative Snail motif”). Snail binds to both motifs with similar affinity (Mauhin et al. 1993), and they can both substitute for each other in Snail-mediated regulation of the rho NEE enhancer (Supplemental Fig. S2A,B). In the activated CRMs, this CAGGTA motif is extended by guanidine and cytosine at positions 7 and 8, respectively (Fig. 4A); this extended motif overlaps a reported motif for Zelda, a TF required for the activation of gene expression in the early embryo (ten Bosch et al. 2006; Liang et al. 2008; Harrison et al. 2011; Nien et al. 2011).
Figure 4.
Snail-activated and -repressed enhancers contain subtle differences in their Snail and Twist motifs. (A) De novo motif discovery in cobound activated and repressed CRMs. Position weight matrices (PWMs) are shown as sequence logos, with known motifs for Snail (Jaspar MA0086.1), Twist (FlyReg), Zelda (SOLEXA_5), and Dorsal (FlyReg). An alternative Snail and a Twist-like motif are found in both sets, while only repressed CRMs are enriched for a Dorsal-like motif. (*) Alternative Snail motif used for analysis shown in B and C. (B) Distribution of PWM match scores (P-value < 1 × 10−3) for the alternative CAGGTA motif across the four classes of Snail-bound CRMs showing all PWM matches (Patser scores, left box plot) or the cumulated match scores (right box plot), summing up putative high-affinity and low-affinity sites. In both cases, no significant differences in the number of motifs were observed. (C) The base-pair distance between Twist and CAGGTA Snail motifs is greater in activated compared with repressed cobound CRMs. The Y-axis shows the mean enrichment of Snail–Twist distances over random expectations (smoothed using a 10-bp window), with 95% confidence intervals (dotted lines). Red asterisks indicate where the signal deviates from random (confidence interval remains less than one for greater than five consecutive values). (Top panel) In activated cobound CRMs, Twist motifs are preferentially enriched at a distance of 50–65 bp. (Bottom panel) In repressed cobound CRMs, Twist motifs cluster around Snail motifs at a distance of 10–20 and 40–50 bp. No enrichment of Twist motifs around CAGGTA Snail motifs is seen in Snail-only CRMs, as expected.
Aside from subtle differences in the Snail and Twist motif sequences between the two CRM sets, no clear differences in the number or quality of Snail and Twist motifs were observed between the activated and repressed CRMs (Fig. 4B; Supplemental Fig. S5). The different activities of the activated and repressed CRMs therefore do not appear to be regulated by the number or differential enrichment of high-affinity or low-affinity Snail motifs.
We next examined whether the relative positioning of the Snail and Twist sites was different between the activated and repressed CRMs. Efficient repression by Snail requires close spacing (<100 bp) between the Snail motif and the activator motif (Gray et al. 1994; Gray and Levine 1996). For Snail to potentiate enhancer activation, one might reasonably expect its motif to be located at higher distances from Twist motifs in activated CRMs compared with Snail-repressed CRMs. To assess this, we calculated the distance from each predicted Snail site to the closest Twist-binding site in each set of CRMs. The repressed CRMs show a preference in the spacing of Twist and Snail motifs, using either the CAGGTA motif (Fig. 4C) or the CAGGTG motif (Supplemental Fig. S5C), with a biphasic center-to-center distance of 10–20 bp and 40–50 bp, consistent with short-range repression. In contrast, in the activated CRMs, Twist motifs are preferentially more distant from Snail motifs, with an enrichment of a 50- to 65-bp distance for the CAGGTA motif (Fig. 4C) and an enrichment of an 80-bp distance for the CAGGTG motif (Supplemental Fig. S5C). Although a 50- to 65-bp distance may be sufficiently close for Snail to still interfere with Twist activity, an 80-bp distance should attenuate quenching (Gray et al. 1994; Gray and Levine 1996; Fakhouri et al. 2010). The longer spacing between Snail and Twist sites in activated CRMs (50–80 bp) also suggests that there is not a direct close physical interaction between Twist and Snail, which typically occurs when motifs are spaced in the range of 10 bp (for review, see Spitz and Furlong 2012).
To summarize, there are subtle differences in the Snail and Twist motifs and in their relative spacing from each other in activated versus repressed enhancers, although there are no clear differences in motif number. However, these differences in motif positioning are trends within a group of enhancers and do not represent a definitive rule that applies to every enhancer. In the Mef2 I-D[L] enhancer, for example, the Twist and Snail sites are <11 bp apart, yet this enhancer is activated by Snail. How, then, does Snail confer enhancer activation, as opposed to repression?
Cis-regulatory signatures of activating Snail binding
Given the subtle differences in Snail and Twist sites, we extended our analyses to search for differentially enriched motifs for additional TFs that may be indicative of Snail's regulatory output. Using a large collection of known and predicted TF position weight matrices (PWMs) (Stark et al. 2007), we first identified which motifs were differentially enriched in Snail–Twist-activated versus -repressed CRMs or within one set compared with the rest of the genome (Fig. 5A). This analysis revealed 32 motifs that are significantly enriched (hypergeometric P-value ≤ 0.025) in the activated compared with repressed CRMs (Fig. 5A; Supplemental Table S9), with a Tailless (Tll) motif being the most strongly enriched (Tll_2, 4.08-fold enriched; P-value = 0.016). The TF Tailless is not expressed in the mesoderm and therefore cannot be the factor involved in Snail-mediated transcriptional activation (Pignoni et al. 1990). Instead, a factor with a similar DNA-binding specificity might bind this motif, and we therefore refer to it as Tll-like motif hereafter. Twelve motifs are enriched in repressed CRMs (Fig. 5A), including the ME49 motif, for which no factor is known but which is enriched in the vicinity of genes expressed in the epidermis and foregut (Stark et al. 2007). Thus, there is a correlation between the motif content of a CRM and its likelihood to be repressed or activated by Snail.
Figure 5.
Differentially enriched motifs predict Snail's regulatory output. (A, left column) Significantly enriched motifs in activated compared with repressed cobound CRMs. The right column shows enrichment of the same motifs compared with the genome. Log2 fold enrichment values (hypergeometric P-value ≤ 0.025). (B) Work flow of machine learning approach (SVM) to discriminate between activated and repressed CRMs and the in silico mutagenesis to pinpoint the most important motifs for experimental testing. (C) Receiver–operator characteristic [ROC] curves showing SVM performance for activated and repressed CRMs. Area under the curve (AUC) is indicated. (D) The most important motifs used by the SVM to discriminate between activated and repressed CRMs (selected discriminative features). (E) In silico mutatgenesis predicts that the Tll (Tll_MA0459-1 [AAAAGTCAAM]) and ME6 (VATTWGCAT) motifs are the most important for activated cobound CRMs, affecting 50% and 33.3% of the confidently predicted activated peaks, respectively; 8.3% of CRMs depend on the Eyg motif. See Supplemental Table S9 for motif information.
To determine whether differential motif content is sufficient to predict the regulatory output of Snail, we used an established machine learning method (a support vector machine [SVM]) for predictive discriminatory sequence analysis (Yanez-Cuna et al. 2012) based on 429 known or predicted TF PWMs (Fig. 5B; Stark et al. 2007). Using leave-one-out cross-validation, the SVM was able to discriminate between Snail-activated and Snail-repressed CRMs (71.6% of activated and 66.3% of repressed CRMs predicted correctly; area under the receiver–operator characteristic [ROC] distribution [AUC]: 0.74) (Fig. 5C) based solely on differences in their motif content using the 15 most discriminative TF motifs (Fig. 5D; Supplemental Table S9; see Yanez-Cuna et al. 2012). When we repeated the predictions after randomizing the class membership for each binding site, the predictions dropped to 56.8% (AUC: 0.56). The poor performance of this random set in addition to the cross-validation indicate that the SVM is not overfitting and that the two sets of Snail enhancers indeed contain characteristic and distinct motifs (Fig. 5C). The 15 SVM-selected discriminative motifs include motifs that are differentially enriched, as expected (Fig. 5A,D), in addition to motifs that are not strongly enriched in a given class; for example Kni, ME56, and dl. These later motifs highlight the strength of the SVM approach to take motif occurrences and their combinations in individual enhancers into account even when a motif is not enriched when enhancers are analyzed in bulk.
To determine which TF motifs were the most important for the correct prediction of each enhancer, we performed in silico mutations (Yanez-Cuna et al. 2012). The confidence of the prediction was scored by bootstrapping the data selected in model training for each individual binding site in wild-type and mutant CRMs after all instances of a given motif were computationally ablated (Supplemental Material). Each CRM was thereby classified 100 times using 100 different training sets, excluding the respective test CRM, yielding a score between 0 and 100 for the number of correct predictions. This analysis ranked the Tll-like motif (Tll_1) as the most important motif for the prediction of activated enhancers, followed by the ME6 motif (Fig. 5E). While it is not known which TF binds to the ME6 motif, the motif is depleted from loci of ubiquitously expressed genes (Stark et al. 2007).
This suggests that the Tll-like motif contributes to a positive regulatory output for Snail-bound regions, a hypothesis that we tested experimentally below.
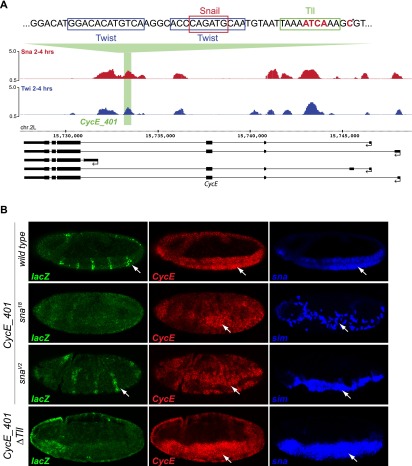
The Tll-like motif determines the mode of Snail enhancers' activity
Two CRMs predicted in silico to depend on the Tll-like motif are located within an intron of the CyclinE (CycE) gene (Fig. 6A) and upstream of CG14688 (Supplemental Fig. S7). In transgenic enhancer assays, the wild-type CycE enhancer, termed CycE_401, is first active in a striped pattern in the mesoderm and ectoderm at the onset of gastrulation (stage 6) (data not shown), and the mesodermal expression becomes gradually stronger during germ band extension (Fig. 6B, stage 7); this expression partially recapitulates the mesodermal and ectodermal expression of the endogenous CycE gene (Fig. 6B). In addition, the enhancer mediates lacZ expression in delaminating neuroblasts, in which CycE is also expressed, but this activity is not dependent on Snail (Supplemental Fig. S6).
Figure 6.
The Tll motif is essential for the Snail-activated CycE_401 enhancer. (A) An intronic region of the Cyclin E gene is cobound by Twist and Snail (blue and red ChIP signal [log2 mean immunoprecipitation/mock], respectively). Tll-like, Twist, and Snail motifs are indicated above the ChIP signal; bold red letters mark mutated nucleotides, including a base mutated based on an earlier version of predictions (asterisk). The gene model is shown below; thick lines indicate exons, thin lines indicate introns, and arrows indicate direction of transcription. (B) In vivo activity of the CycE_401 enhancer (genome coordinates are in the Supplemental Material). In situ hybridization of the reporter lacZ gene (green), endogenous CyclinE gene (red), and sna or sim (blue) in wild-type and sna mutant embryos, as indicated. (Top panel) The CycE_401 enhancer drives lacZ expression in a striped pattern in mesoderm (white arrows), partially recapitulating Cyclin E expression. Enhancer activity is dramatically reduced in sna18 mutant embryos (amorph, second row) compared with snaV2 (hypomorph). (Bottom panel) Mutation of the Tll-like motif reduces enhancer activity in mesoderm and ectoderm. All embryos are stage 7, with anterior shown to the left, and are single confocal planes. LacZ expression in the head fold is caused by the eve minimal promoter used in the reporter vector.
Mesodermal expression of CycE_401-lacZ depends on sna activity, as placing the enhancer in a sna18 mutant leads to reduced lacZ expression (Fig. 6B). The expression of the endogenous CycE gene is also reduced in the mesoderm in sna18 mutant embryos. The residual Snail function in the snaV2 mutation appears to be sufficient for transcriptional activation of the enhancer (Fig. 6B), although the CycE gene's expression is slightly reduced. In summary, the enhancer CycE_401 directs Snail-dependent expression in the mesoderm and additional Snail-independent patterns in the ectoderm and neuroblasts.
We next assessed the requirement of the Tll-like motif for transcriptional activation of the CycE_401 by mutating the motif (CycE_401_ΔTll). Loss of the Tll-like motif is sufficient to severely reduce enhancer activity in the mesoderm despite the presence of intact Twist and Snail motifs (Fig. 6B, bottom panel). Similarly, although the wild-type CG14688_400 enhancer activates expression in the presumptive mesoderm at stages 5 and 6, its activity is reduced upon mutation of the Tll-like motif (Supplemental Fig. S7). These results indicate that the Tll-like motif is essential for the activity of both Snail-activated enhancers.
In summary, removal of either of the two components by the genetic perturbation of Snail function in sna mutant embryos, shown for the CycE enhancer, or by mutagenesis of the Tll-like motif in the enhancer reduces the activation of reporter gene expression. Efficient activation can be achieved only when both factors are present.
Discussion
The Drosophila TF Snail has been considered to be a dedicated transcriptional repressor. By integrating genomic, bioinformatic, and genetic approaches, we uncovered an unknown role for Snail as a positive modulator of enhancer activity. Considering its well-characterized role as a repressor, the extent to which Snail positively regulates mesodermal gene expression was surprising, accounting for almost 50% of its regulatory activity when acting with Twist.
Motif prediction identifies a new motif essential for Snail activation
The motif content of enhancers that are either activated or repressed by Snail differs significantly from each other. Our machine learning approach and in silico mutagenesis showed that (1) these motifs are sufficient to predict the transcriptional output of Snail and (2) the Tll-like motif is a strong candidate for “switching” the activity of Snail. In the context of the CycE_401 and CG14688_400 enhancers, full activation in the mesoderm by Snail and Twist requires an intact Tll-like motif. We do not know the identity of the TF that regulates Snail-activated enhancers through this motif. The TF Tailless itself is expressed in the anterior and posterior pole but not in the mesoderm and therefore cannot account for this activity (Pignoni et al. 1990).
Searching for TFs that recognize motifs that either overlap or are highly similar to the Tll-like motif and that are expressed in the early Drosophila embryo identified three candidate TFs. First, the pair-rule gene fushi tarazu (ftz). However, the expression pattern of ftz does not overlap CycE_401-lacZ expression, ruling out an involvement of Ftz in this enhancer's regulation. Second, dTCF (pangolin), which is expressed maternally in the early embryo (Brunner et al. 1997; van de Wetering et al. 1997), activates gene expression in response to wingless (wg) signaling (van de Wetering et al. 1997), and wg-expressing stripes are located adjacent to and partially overlapping the CycE_401-lacZ expression domain (Supplemental Fig. S8B). To assess a potential role of dTCF, we blocked its function with a dominant-negative form of dTCF (UAS-DN-TCF) (van de Wetering et al. 1997), which we expressed either ubiquitously or specifically in the mesoderm (using twist-Gal4); neither had a detectable effect on enhancer activity (Supplemental Fig. S8C), ruling out a role for dTCF. Third, the motif for Tll and Eve are similar, and stripes of eve expression are located anterior to and partially overlapping the CycE_401-lacZ expression domain. Reducing eve expression by injecting dsRNA specific for eve indicated an involvement of Eve in restricting the width of the lacZ-expressing stripes but not in the activation of the stripes per se (data not shown). The identity of the TF occupying the Tll-like motif in mesoderm therefore remains a mystery at this point.
What is the mechanism responsible for switching the regulatory mode of Snail?
Although the TF contributing to Snail activation remains unknown, it is interesting to speculate how it might influence the transcriptional activity of Snail.
In general, the requirement for partner factors seems to be a common theme in Snail-mediated transcriptional regulation. Drosophila Snail acts synergistically with Twist and the factor binding to the Tll-like motif in the mesoderm during early embryonic development. Similar observations have been made in epithelial–mesenchymal transition-related processes, such as tumor formation or neural crest migration. In human HepG2 cells, Snail associates and acts in concert with EGR-1 and SP-1 to participate in p15INK4b activation induced by tetradecanoyl phorbol acetate (TPA) (Hu et al. 2010). A similar synergistic effect was reported for Snail2 and Sox9 in the autoactivation of Snail2 in quail (Sakai et al. 2006). While Snail2 and Sox9 individually activated a Snail2 reporter only moderately, the activity was potentiated when both TFs were cotransfected in neural plate explants. In both cases, the binding sites are closely spaced in the enhancer, and the proteins interact physically. The short distance between the Tll-like and Snail motifs in the CycE_401 enhancer would allow a direct physical interaction as well. Such protein interactions can change the conformation and hence activity of proteins (for review, see Dyson and Wright 2005). It is interesting in this context that the deletion of the N-terminal SNAG repressor domain converted hSlug/SNA2 from a repressor to a potent activator in luciferase assays in HEK293T cells (Hemavathy et al. 2000). A small fragment in the N terminus was sufficient to activate reporter activity when fused to the Gal4 DNA-binding domain. Whether this activator domain is functional in vivo and is normally masked by the repressor domain is unknown. A similar conformational change of the Drosophila Snail protein, induced by the factor binding to the Tll-like motif, might expose a domain that interacts with coactivators. Alternatively, the presence of the factor might allow the formation of a higher-order complex that includes Twist.
Although the studies in HepG2 cells and in quail show that full activation by Snail requires a partner factor, vertebrate Snail proteins can also activate on their own in vitro (Huang et al. 2009; Tao et al. 2011; Wels et al. 2011; Hahn et al. 2013). Potential synergistic effects have not been analyzed in these studies. Snail is also sufficient for a weak activation of the p15INK4b and Snail2 promoter (Sakai et al. 2006; Hu et al. 2010). Drosophila Snail instead acts in a synergistic manner for which the presence of all three factors—Twist, Snail, and the factor binding the Tll-like motif—are required for enhancer activation in vivo. A key requisite for understanding the mechanisms by which Snail activates gene expression will be to identify the TF that occupies sequence elements such as the Tll-like motif.
A new view of how Snail regulates diverse developmental processes
The function of Snail in distinguishing mesodermal from ectodermal fates has been traditionally seen as a repressor of the ectodermal differentiation program. This study demonstrates that Drosophila Snail can also activate part of the program specific for the mesoderm. The role of Snail in gastrulation is thus dual and involves a balance of repression and activation.
One of the functions of Snail is to enable the formation of the ventral furrow together with Twist. Whereas the target genes of Twist that mediate furrow formation are known, it is completely unclear which genes act downstream from Snail. Only one such gene has been identified so far (Chanet and Schweisguth 2012). The gene bearded, which is repressed in the mesoderm by Snail, is partly responsible for allowing adherens junctions in the mesoderm to be relocalized, but this is not sufficient for furrow formation. Therefore, there must be other genes that fulfill essential functions in gastrulation downstream from Snail. The snaV2 mutant might give some hints of what genes these might be, since it is still able to make a furrow, although many Snail target genes are misregulated. Stepwise reduction of only the repressive activity of Snail by mutation of one or two corepressor-binding sites results in a stepwise increase in the strength of the gastrulation phenotype (Hemavathy et al. 2004). Thus, the repressive activity of Snail is certainly required. However, the 60% of Snail-dependent genes that are not or are only weakly affected in the snaV2 mutant (i.e., those most likely to be responsible for mediating furrow formation) do not fall into a uniform category; they contain both up-regulated and down-regulated genes. This might be an indication that misregulation of a larger set of both repressed and activated genes leads to the failure in furrow formation. This is also consistent with the fact that simply reducing the level of Snail by half leads to a delay in gastrulation (Seher et al. 2007).
In summary, our study revealed a direct activator role for Drosophila Snail, a function that is seemingly conserved from flies to humans (Sakai et al. 2006; Stemmer et al. 2008; Reece-Hoyes et al. 2009; Hu et al. 2010; Tao et al. 2011; Wels et al. 2011; Hahn et al. 2013) and places the Snail family of proteins in the category of dually acting TFs.
Materials and methods
Drosophila strains
The Drosophila melanogaster snail amorphic allele Df(2L)TE116GW11 [Df(sna)] (Ashburner et al. 1990) and the hypomorphic allele snailV2 (Hemavathy et al. 1997) were recombined with ΔhaloAJ and balanced over SM1. ΔhaloAJ is a deletion of ∼19 kb, encompassing the entire halo gene, made using FLP/FRT-mediated recombination between Exilixis insertions PBac{WH}f04301 and PBac{WH}f07557. Transgenic flies for enhancer assays were established with the landing site line attP40 (w P{nos-phiC31\int.NLS};P{CaryP}attP40) (Markstein et al. 2008) for CycE_401, Mef2_401, and CG1488_400-lacZ or attP51C (y w M{eGFP.vas-int.Dm}ZH-2A; M{RFP.attP}ZH-51C) (Bischof et al. 2007) for Mef2 I-D[L] and tin B-374. Transgenes for enhancer assays were recombined with the loss-of-function EMS-induced allele sna18 (snaIIG05) (Grau et al. 1984) and the hypomorphic snaV2 allele. The matα4-GAL4:VP16 (V32) stock was provided by Daniel St. Johnston, the twi-Gal4 stock was from Michael Akam (Greig and Akam 1993), and the UAS-TCF-DN stock was generated by the Hans Clevers laboratory (van de Wetering et al. 1997).
ChIP
Embryo collections and ChIPs were performed as described previously (Sandmann et al. 2006; Zinzen et al. 2009). Two independently staged wild-type Oregon-R embryo collections were obtained at 2–4 h AEL and fixed with formaldehyde. Chromatin was precipitated with guinea pig anti-Snail antiserum (Zeitlinger et al. 2007) as well as preimmune sera for mock ChIPs. All ChIPs were assayed for enrichment quality by quantitative real-time PCR. DNA amplification, labeling, and hybridizations to high-density Affymetrix Drosophila 2.0 tiling arrays were performed as described previously (Sandmann et al. 2006; Zinzen et al. 2009). All data analysis is described in the Supplemental Material.
Expression profiling analysis of snail mutant embryos
Embryos were collected from ΔhaloAJ Df(snail)/SM1, ΔhaloAJ snailV2/SM1, and ΔhaloAJ /ΔhaloAJ control flies for 1 h on apple juice agar plates at 25°C. After dechorionation, embryos were washed, transferred to a fresh apple juice agar plate, and overlaid with PBS (pH 7.4). Homozygous mutant embryos were hand-sorted under a stereomicroscope for the visible Δhalo phenotype. At the desired stage, 150–200 mutant or wild-type embryos were transferred to Eppendorf tubes and flash-frozen in liquid nitrogen. RNA was extracted using the RNeasy minikit (Qiagen) as described in Sandmann et al. ( 2007). Two micrograms of RNA of each sample was amplified using the Affymetrix GeneChip One-Cycle cDNA synthesis kit according to the manufacturer. The amplified cDNA was fragmented and hybridized using standard protocols to the Affymetrix GeneChip Drosophila Genome array version 2, covering 18,500 transcripts. For each time point, three independent populations of sna mutant and stage-matched ΔhaloAJ embryos were collected, aged, and assessed by microarray analysis, leading to 18 hybridizations. All data analysis is described in the Supplemental Material.
Data availability
All ChIP data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-TABM-651 (Twist) and E-MTAB-1589 (Snail). The high-confidence TF-binding information, including CRM coordinates and occupancy by different TFs, is provided in Supplemental Tables S4–S7 and also on the Furlong laboratory Web page at http://furlonglab.embl.de.
All gene expression hybridization data are available at ArrayExpress under accession number E-MTAB-1598.
Acknowledgments
We thank Lisa Vogelsang for technical assistance, and all members of the Furlong and Leptin laboratories for discussions. This work was technically supported by the EMBL Advanced Light Microscopy Facility and Genomics Core Facility. This work was supported by a Deutsche Forschungsgemeinschaft (DFG) grant (FU 750/1) and Human Frontier Science Program grant to E.E.F., and a DFG grant (LE546) to M.L. M.R. was supported by an EMBO long-term fellowship.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.230953.113.
References
- Ashburner M, Thompson P, Roote J, Lasko PF, Grau Y, el Messal M, Roth S, Simpson P 1990. The genetics of a small autosomal region of Drosophila melanogaster containing the structural gene for alcohol dehydrogenase. VII. Characterization of the region around the snail and cactus loci. Genetics 126: 679–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA 2005. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 132: 3151–3161 [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R, Jan LY, Jan YN 1990. A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation of Drosophila. Development 110: 661–669 [DOI] [PubMed] [Google Scholar]
- Bothma JP, Magliocco J, Levine M 2011. The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr Biol 21: 1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K 1997. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385: 829–833 [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V 1997. The embryonic development of Drosophila melanogaster, 2nd edition. Springer Verlag, Berlin. [Google Scholar]
- Casal J, Leptin M 1996. Identification of novel genes in Drosophila reveals the complex regulation of early gene activity in the mesoderm. Proc Natl Acad Sci 93: 10327–10332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet S, Schweisguth F 2012. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nat Cell Biol 14: 467–476 [DOI] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. 2011. The transcriptional diversity of 25 Drosophila cell lines. Genome Res 21: 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Levine M 2009. Combinatorial patterning mechanisms in the Drosophila embryo. Brief Funct Genomics Proteomic 8: 243–249 [DOI] [PubMed] [Google Scholar]
- Chopra VS, Kong N, Levine M 2012. Transcriptional repression via antilooping in the Drosophila embryo. Proc Natl Acad Sci 109: 9460–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN 1998. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev 12: 422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, de Herreros AG 2011. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem 286: 12024–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Echalier G, Ohanessian A 1969. Isolation, in tissue culture, of Drosophila melangaster cell lines. C R Acad Sci Hebd Seances Acad Sci D 268: 1771–1773 [PubMed] [Google Scholar]
- Fakhouri WD, Ay A, Sayal R, Dresch J, Dayringer E, Arnosti DN 2010. Deciphering a transcriptional regulatory code: Modeling short-range repression in the Drosophila embryo. Mol Syst Biol 6: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Jiang J, Ip YT 2005. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development 132: 3419–3429 [DOI] [PubMed] [Google Scholar]
- Grau Y, Carteret C, Simpson P 1984. Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics 108: 347–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Levine M 1996. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev 10: 700–710 [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M 1994. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev 8: 1829–1838 [DOI] [PubMed] [Google Scholar]
- Greig S, Akam M 1993. Homeotic genes autonomously specify one aspect of pattern in the Drosophila mesoderm. Nature 362: 630–632 [DOI] [PubMed] [Google Scholar]
- Gross SP, Guo Y, Martinez JE, Welte MA 2003. A determinant for directionality of organelle transport in Drosophila embryos. Curr Biol 13: 1660–1668 [DOI] [PubMed] [Google Scholar]
- Hahn S, Jackstadt R, Siemens H, Hunten S, Hermeking H 2013. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial–mesenchymal transition. EMBO J 32: 3079–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 7: e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K, Meng X, Ip YT 1997. Differential regulation of gastrulation and neuroectodermal gene expression by Snail in the Drosophila embryo. Development 124: 3683–3691 [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Guru SC, Harris J, Chen JD, Ip YT 2000. Human Slug is a repressor that localizes to sites of active transcription. Mol Cell Biol 20: 5087–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K, Hu X, Ashraf SI, Small SJ, Ip YT 2004. The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu. Dev Biol 269: 411–420 [DOI] [PubMed] [Google Scholar]
- Hu CT, Chang TY, Cheng CC, Liu CS, Wu JR, Li MC, Wu WS 2010. Snail associates with EGR-1 and SP-1 to upregulate transcriptional activation of p15INK4b. FEBS J 277: 1202–1218 [DOI] [PubMed] [Google Scholar]
- Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, Wu KJ, Yang MH 2009. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia 11: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M 1992. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev 6: 1728–1739 [DOI] [PubMed] [Google Scholar]
- Ip YT, Maggert K, Levine M 1994. Uncoupling gastrulation and mesoderm differentiation in the Drosophila embryo. EMBO J 13: 5826–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda M, Olmeda D, Vinyals A, Valero E, Cubillo E, Llorens A, Cano A, Fabra A 2005. Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J Cell Sci 118: 3371–3385 [DOI] [PubMed] [Google Scholar]
- Jorda M, Vinyals A, Marazuela A, Cubillo E, Olmeda D, Valero E, Cano A, Fabra A 2007. Id-1 is induced in MDCK epithelial cells by activated Erk/MAPK pathway in response to expression of the Snail and E47 transcription factors. Exp Cell Res 313: 2389–2403 [DOI] [PubMed] [Google Scholar]
- Kasai Y, Nambu JR, Lieberman PM, Crews ST 1992. Dorsal–ventral patterning in Drosophila: DNA binding of snail protein to the single-minded gene. Proc Natl Acad Sci 89: 3414–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Ip YT, Levine M, Arora K 1991. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 254: 118–122 [DOI] [PubMed] [Google Scholar]
- Lai ZC, Fortini ME, Rubin GM 1991. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev 34: 123–134 [DOI] [PubMed] [Google Scholar]
- Leptin M 1991. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 5: 1568–1576 [DOI] [PubMed] [Google Scholar]
- Leptin M, Grunewald B 1990. Cell shape changes during gastrulation in Drosophila. Development 110: 73–84 [DOI] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN 1994. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci 91: 5662–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J 2005. Crossing the line between activation and repression. Trends Genet 21: 54–59 [DOI] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M 2004. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131: 2387–2394 [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauhin V, Lutz Y, Dennefeld C, Alberga A 1993. Definition of the DNA-binding site repertoire for the Drosophila transcription factor SNAIL. Nucleic Acids Res 21: 3951–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale P, Mizutani CM, Kosman D, Mackay DL, Belu M, Hermann A, McGinnis W, Bier E, Hwa T 2011. Gene length may contribute to graded transcriptional responses in the Drosophila embryo. Dev Biol 360: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morize P, Christiansen AE, Costa M, Parks S, Wieschaus E 1998. Hyperactivation of the folded gastrulation pathway induces specific cell shape changes. Development 125: 589–597 [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Xu X 1998. Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev Biol 204: 550–566 [DOI] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M 1998a. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J 17: 7009–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M 1998b. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280: 101–104 [DOI] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C 2011. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7: e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA 1990. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62: 151–163 [DOI] [PubMed] [Google Scholar]
- Qi D, Bergman M, Aihara H, Nibu Y, Mannervik M 2008. Drosophila Ebi mediates Snail-dependent transcriptional repression through HDAC3-induced histone deacetylation. EMBO J 27: 898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Barrasa MI, Hatzold J, Smit RB, Arda HE, Pope PA, Gaudet J, Conradt B, Walhout AJ 2009. The C. elegans Snail homolog CES-1 can activate gene expression in vivo and share targets with bHLH transcription factors. Nucleic Acids Res 37: 3689–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Stein D, Nusslein-Volhard C 1989. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59: 1189–1202 [DOI] [PubMed] [Google Scholar]
- Rushlow CA, Han K, Manley JL, Levine M 1989. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell 59: 1165–1177 [DOI] [PubMed] [Google Scholar]
- Sakai D, Suzuki T, Osumi N, Wakamatsu Y 2006. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development 133: 1323–1333 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jakobsen JS, Furlong EE 2006. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1: 2839–2855 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE 2007. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev 21: 436–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seher TC, Narasimha M, Vogelsang E, Leptin M 2007. Analysis and reconstitution of the genetic cascade controlling early mesoderm morphogenesis in the Drosophila embryo. Mech Dev 124: 167–179 [DOI] [PubMed] [Google Scholar]
- Shao X, Koizumi K, Nosworthy N, Tan DP, Odenwald W, Nirenberg M 2002. Regulatory DNA required for vnd/NK-2 homeobox gene expression pattern in neuroblasts. Proc Natl Acad Sci 99: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E, Higashijima S, Emori Y, Saigo K 1993. Two FGF-receptor homologues of Drosophila: One is expressed in mesodermal primordium in early embryos. Development 117: 751–761 [DOI] [PubMed] [Google Scholar]
- Simpson P 1983. Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics 105: 615–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE 2012. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet 13: 613–626 [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer V, de Craene B, Berx G, Behrens J 2008. Snail promotes Wnt target gene expression and interacts with β-catenin. Oncogene 27: 5075–5080 [DOI] [PubMed] [Google Scholar]
- Sun L, Diamond ME, Ottaviano AJ, Joseph MJ, Ananthanarayan V, Munshi HG 2008. Transforming growth factor-β 1 promotes matrix metalloproteinase-9-mediated oral cancer invasion through snail expression. Mol Cancer Res 6: 10–20 [DOI] [PubMed] [Google Scholar]
- Tao G, Levay AK, Gridley T, Lincoln J 2011. Mmp15 is a direct target of Snai1 during endothelial to mesenchymal transformation and endocardial cushion development. Dev Biol 359: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW 2006. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133: 1967–1977 [DOI] [PubMed] [Google Scholar]
- Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F 1988. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J 7: 2175–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J 2012. RSAT peak-motifs: Motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res 40: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799 [DOI] [PubMed] [Google Scholar]
- Wels C, Joshi S, Koefinger P, Bergler H, Schaider H 2011. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial–mesenchymal transition-like phenotype in melanoma. J Invest Dermatol 131: 1877–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A 2012. Uncovering cis-regulatory sequence requirements for context specific transcription factor binding. Genome Res 22: 2018–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M 1997. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124: 4971–4982 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M 2007. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21: 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All ChIP data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-TABM-651 (Twist) and E-MTAB-1589 (Snail). The high-confidence TF-binding information, including CRM coordinates and occupancy by different TFs, is provided in Supplemental Tables S4–S7 and also on the Furlong laboratory Web page at http://furlonglab.embl.de.
All gene expression hybridization data are available at ArrayExpress under accession number E-MTAB-1598.